Viral Metagenomic Analysis of Aedes albopictus Mosquitos from Southern Switzerland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Sample Processing and Sequencing
2.3. Quality Control, Pre-Processing, and Assembly of Metagenomics Reads
3. Results
3.1. Mosquito-Associated Viruses
3.2. Other Insect-Associated Viruses
3.3. Other Viruses
3.4. Wolbachia Sp. Detection
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Medlock, J.M.; Hansford, K.M.; Schaffner, F.; Versteirt, V.; Hendrickx, G.; Zeller, H.; Van Bortel, W. A review of the invasive mosquitoes in Europe: Ecology, public health risks, and control options. Vector Borne Zoonotic Dis. 2012, 12, 435–447. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paupy, C.; Delatte, H.; Bagny, L.; Corbel, V.; Fontenille, D. Aedes albopictus, an arbovirus vector: From the darkness to the light. Microbes Infect. 2009, 11, 1177–1185. [Google Scholar] [CrossRef] [PubMed]
- Wymann, M.N.; Flacio, E.; Radczuweit, S.; Patocchi, N.; Luthy, P. Asian tiger mosquito (Aedes albopictus)—A threat for Switzerland? Eurosurveillance 2008, 13, 3–4. [Google Scholar] [CrossRef] [PubMed]
- Flacio, E.; Engeler, L.; Tonolla, M.; Lüthy, P.; Patocchi, N. Strategies of a thirteen year surveillance programme on Aedes albopictus (Stegomyia albopicta) in southern Switzerland. Parasites Vectors 2015, 8, 208. [Google Scholar] [CrossRef] [Green Version]
- Flacio, E.; Engeler, L.; Tonolla, M.; Müller, P. Spread and establishment of Aedes albopictus in southern Switzerland between 2003 and 2014: An analysis of oviposition data and weather conditions. Parasites Vectors 2016, 9, 304. [Google Scholar] [CrossRef] [Green Version]
- Suter, T.T.; Flacio, E.; Feijoó Fariña, B.; Engeler, L.; Tonolla, M.; Regis, L.N.; de Melo Santos, M.A.; Müller, P. Surveillance and Control of Aedes albopictus in the Swiss-Italian Border Region: Differences in Egg Densities between Intervention and Non-intervention Areas. PLoS Negl. Trop. Dis. 2016, 10, e0004315. [Google Scholar] [CrossRef] [Green Version]
- Ravasi, D.; Guidi, V.; Flacio, E.; Lüthy, P.; Perron, K.; Lüdin, S.; Tonolla, M. Investigation of temperature conditions in Swiss urban and suburban microhabitats for the overwintering suitability of diapausing Aedes albopictus eggs. Parasites Vectors 2018, 11, 212. [Google Scholar] [CrossRef]
- Sambri, V.; Cavrini, F.; Rossini, G.; Pierro, A.; Landini, M.P. The 2007 epidemic outbreak of Chikungunya virus infection in the Romagna region of Italy: A new perspective for the possible diffusion of tropical diseases in temperate areas? New Microbiol. 2008, 31, 303–304. [Google Scholar]
- Delisle, E.; Rousseau, C.; Broche, B.; Leparc-Goffart, I.; L’Ambert, G.; Cochet, A.; Prat, C.; Foulongne, V.; Ferre, J.B.; Catelinois, O.; et al. Chikungunya outbreak in Montpellier, France, September to October 2014. Eurosurveillance 2015, 20, 21108. [Google Scholar] [CrossRef] [Green Version]
- La Ruche, G.; Souarès, Y.; Armengaud, A.; Peloux-Petiot, F.; Delaunay, P.; Desprès, P.; Lenglet, A.; Jourdain, F.; Leparc-Goffart, I.; Charlet, F.; et al. First two autochthonous dengue virus infections in metropolitan France, September 2010. Eurosurveillance 2010, 15, 19676. [Google Scholar]
- Hall, R.A.; Bielefeldt-Ohmann, H.; McLean, B.J.; O’Brien, C.A.; Colmant, A.M.; Piyasena, T.B.; Harrison, J.J.; Newton, N.D.; Barnard, R.T.; Prow, N.A.; et al. Commensal Viruses of Mosquitoes: Host Restriction, Transmission, and Interaction with Arboviral Pathogens. Evol. Bioinform. Online 2016, 12 (Suppl. 2), 35–44. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Tesh, R.B. Insect-specific viruses and their potential impact on arbovirus transmission. Curr. Opin. Virol. 2015, 15, 69–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall-Mendelin, S.; McLean, B.J.; Bielefeldt-Ohmann, H.; Hobson-Peters, J.; Hall, R.A.; van den Hurk, A.F. The insect-specific Palm Creek virus modulates West Nile virus infection in and transmission by Australian mosquitoes. Parasites Vectors 2016, 9, 414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goenaga, S.; Kenney, J.L.; Duggal, N.K.; Delorey, M.; Ebel, G.D.; Zhang, B.; Levis, S.C.; Enria, D.A.; Brault, A.C. Potential for Co-Infection of a Mosquito-Specific Flavivirus, Nhumirim Virus, to Block West Nile Virus Transmission in Mosquitoes. Viruses 2015, 7, 5801–5812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolling, B.G.; Olea-Popelka, F.J.; Eisen, L.; Moore, C.G.; Blair, C.D. Transmission dynamics of an insect-specific flavivirus in a naturally infected Culex pipiens laboratory colony and effects of co-infection on vector competence for West Nile virus. Virology 2012, 427, 90–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hobson-Peters, J.; Yam, A.W.; Lu, J.W.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray Valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Montgomery, B.L.; Popovici, J.; Iturbe-Ormaetxe, I.; Johnson, P.H.; Muzzi, F.; Greenfield, M.; Durkan, M.; Leong, Y.S.; Dong, Y.; et al. Successful establishment of Wolbachia in Aedes populations to suppress dengue transmission. Nature 2011, 476, 454–457. [Google Scholar] [CrossRef]
- Flacio, E.; Bricalli-Rossi-Pedruzzi, A.; Bernasconi-Casati, E.; Patocchi, N. Culicidae fauna from Canton Ticino and report of three new species for Switzerland. Mitt. Schweiz. Entomol. Gesell. 2014, 87, 163–182. [Google Scholar]
- Becker, N.; Petric, D.; Zgomba, M.; Boase, C.; Madon, M.B.; Dahl, C.; Kaiser, A. Mosquitoes and Their Control, 2nd ed.; Springer: Berlin/Heidelberg, Germany, 2010; p. 577. [Google Scholar]
- Romi, R.; Pontuale, G.; Sabatinelli, G. Le zanzare italiane: Generalità e identificazione degli stadi preimaginali (Diptera: Culicidae). Fragm. Entomol. 1997, 29, 1–141. [Google Scholar]
- Schaffner, F.; Angel, G.; Geoffroy, B.; Hervy, J.-P.; Rhaiem, A.; Brunhes, J. The Mosquitoes of Europe. An Identification and Training Programme; IRD Editions: Montpellier, France, 2001. [Google Scholar]
- Severini, F.; Toma, L.; Di Luca, M.; Romi, R. Italian Mosquitoes: General Information and Identification of Adults (Diptera, Culicidae). Fragm. Entomol. 2009, 41, 213–372. [Google Scholar] [CrossRef] [Green Version]
- Stojanovich, C.J.; Scott, H.G. Llustrated Key to the Adult Male Mosquitoes of America (North of Mexico); P. Stojanovich and H. G. Scott: Madison, WI, USA, 1997. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Liu, C.M.; Luo, R.; Sadakane, K.; Lam, T.W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huson, D.H.; Albrecht, B.; Bağcı, C.; Bessarab, I.; Górska, A.; Jolic, D.; Williams, R.B.H. MEGAN-LR: New algorithms allow accurate binning and easy interactive exploration of metagenomic long reads and contigs. Biol. Direct 2018, 13, 6. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Haddow, A.D.; Guzman, H.; Popov, V.L.; Wood, T.G.; Widen, S.G.; Haddow, A.D.; Tesh, R.B.; Weaver, S.C. First isolation of Aedes flavivirus in the Western Hemisphere and evidence of vertical transmission in the mosquito Aedes (Stegomyia) albopictus (Diptera: Culicidae). Virology 2013, 440, 134–139. [Google Scholar] [CrossRef] [Green Version]
- Crochu, S.; Cook, S.; Attoui, H.; Charrel, R.N.; De Chesse, R.; Belhouchet, M.; Lemasson, J.J.; de Micco, P.; de Lamballerie, X. Sequences of flavivirus-related RNA viruses persist in DNA form integrated in the genome of Aedes spp. mosquitoes. J. Gen. Virol. 2004, 85 Pt 7, 1971–1980. [Google Scholar] [CrossRef]
- Birnberg, L.; Temmam, S.; Aranda, C.; Correa-Fiz, F.; Talavera, S.; Bigot, T.; Eloit, M.; Busquets, N. Viromics on Honey-Baited FTA Cards as a New Tool for the Detection of Circulating Viruses in Mosquitoes. Viruses 2020, 12, 274. [Google Scholar] [CrossRef] [Green Version]
- Vasilakis, N.; Castro-Llanos, F.; Widen, S.G.; Aguilar, P.V.; Guzman, H.; Guevara, C.; Fernandez, R.; Auguste, A.J.; Wood, T.G.; Popov, V.; et al. Arboretum and Puerto Almendras viruses: Two novel rhabdoviruses isolated from mosquitoes in Peru. J. Gen. Virol. 2014, 95 Pt 4, 787–792. [Google Scholar] [CrossRef] [PubMed]
- Pauvolid-Corrêa, A.; Solberg, O.; Couto-Lima, D.; Nogueira, R.M.; Langevin, S.; Komar, N. Novel Viruses Isolated from Mosquitoes in Pantanal, Brazil. Genome Announc. 2016, 4, e01195-16. [Google Scholar]
- Dombrovsky, A.; Luria, N. The Nerium oleander aphid Aphis nerii is tolerant to a local isolate of Aphid lethal paralysis virus (ALPV). Virus Genes 2013, 46, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Dayaram, A.; Galatowitsch, M.L.; Argüello-Astorga, G.R.; van Bysterveldt, K.; Kraberger, S.; Stainton, D.; Harding, J.S.; Roumagnac, P.; Martin, D.P.; Lefeuvre, P.; et al. Diverse circular replication-associated protein encoding viruses circulating in invertebrates within a lake ecosystem. Infect. Genet. Evol. 2016, 39, 304–316. [Google Scholar] [CrossRef]
- Shi, M.; Lin, X.D.; Tian, J.H.; Chen, L.J.; Chen, X.; Li, C.X.; Qin, X.C.; Li, J.; Cao, J.P.; Eden, J.S.; et al. Redefining the invertebrate RNA virosphere. Nature 2016, 540, 539–543. [Google Scholar] [CrossRef]
- Li, C.X.; Shi, M.; Tian, J.H.; Lin, X.D.; Kang, Y.J.; Chen, L.J.; Qin, X.C.; Xu, J.; Holmes, E.C.; Zhang, Y.Z. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 2015, 4, e05378. [Google Scholar] [CrossRef]
- Olson, J.G.; Ksiazek, T.G.; Suhandiman; Triwibowo. Zika virus, a cause of fever in Central Java, Indonesia. Trans. R. Soc. Trop. Med. Hyg. 1981, 75, 389–393. [Google Scholar] [CrossRef]
- Delatte, H.; Paupy, C.; Dehecq, J.S.; Thiria, J.; Failloux, A.B.; Fontenille, D. Aedes albopictus, vector of chikungunya and dengue viruses in Reunion Island: Biology and control. Parasite 2008, 15, 3–13. [Google Scholar] [CrossRef] [Green Version]
- Delatte, H.; Gimonneau, G.; Triboire, A.; Fontenille, D. Influence of temperature on immature development, survival, longevity, fecundity, and gonotrophic cycles of Aedes albopictus, vector of chikungunya and dengue in the Indian Ocean. J. Med. Entomol. 2009, 46, 33–41. [Google Scholar] [CrossRef]
- Chang, C.; Ortiz, K.; Ansari, A.; Gershwin, M.E. The Zika outbreak of the 21st century. J. Autoimmun. 2016, 68, 1–13. [Google Scholar] [CrossRef]
- Yang, C.F.; Hou, J.N.; Chen, T.H.; Chen, W.J. Discriminable roles of Aedes aegypti and Aedes albopictus in establishment of dengue outbreaks in Taiwan. Acta Trop. 2014, 130, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Rezza, G. Dengue and chikungunya: Long-distance spread and outbreaks in naïve areas. Pathog. Glob. Health 2014, 108, 349–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taylor, L.H.; Latham, S.M.; Woolhouse, M.E. Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2001, 356, 983–989. [Google Scholar] [CrossRef] [PubMed]
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef] [PubMed]
- Sanborn, M.A.; Klein, T.A.; Kim, H.C.; Fung, C.K.; Figueroa, K.L.; Yang, Y.; Asafo-Adjei, E.A.; Jarman, R.G.; Hang, J. Metagenomic Analysis Reveals Three Novel and Prevalent Mosquito Viruses from a Single Pool of Aedes vexans nipponii Collected in the Republic of Korea. Viruses 2019, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Sõmera, M.; Sarmiento, C.; Truve, E. Overview on Sobemoviruses and a Proposal for the Creation of the Family Sobemoviridae. Viruses 2015, 7, 3076–3115. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, K.; Isawa, H.; Tsuda, Y.; Sawabe, K.; Kobayashi, M. Isolation and characterization of a new insect flavivirus from Aedes albopictus and Aedes flavopictus mosquitoes in Japan. Virology 2009, 391, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Cook, S.; Moureau, G.; Kitchen, A.; Gould, E.A.; de Lamballerie, X.; Holmes, E.C.; Harbach, R.E. Molecular evolution of the insect-specific flaviviruses. J. Gen. Virol. 2012, 93 Pt 2, 223–234. [Google Scholar] [CrossRef] [Green Version]
- Calzolari, M.; Zé-Zé, L.; Vázquez, A.; Sánchez Seco, M.P.; Amaro, F.; Dottori, M. Insect-specific flaviviruses, a worldwide widespread group of viruses only detected in insects. Infect. Genet. Evol. 2016, 40, 381–388. [Google Scholar] [CrossRef]
- Roiz, D.; Vázquez, A.; Rosso, F.; Arnoldi, D.; Girardi, M.; Cuevas, L.; Perez-Pastrana, E.; Sánchez-Seco, M.P.; Tenorio, A.; Rizzoli, A. Detection of a new insect flavivirus and isolation of Aedes flavivirus in Northern Italy. Parasites Vectors 2012, 5, 223. [Google Scholar] [CrossRef] [Green Version]
- Grisenti, M.; Vázquez, A.; Herrero, L.; Cuevas, L.; Perez-Pastrana, E.; Arnoldi, D.; Rosà, R.; Capelli, G.; Tenorio, A.; Sánchez-Seco, M.P.; et al. Wide detection of Aedes flavivirus in north-eastern Italy—A European hotspot of emerging mosquito-borne diseases. J. Gen. Virol. 2015, 96 Pt 2, 420–430. [Google Scholar] [CrossRef] [Green Version]
- Schultz, M.J.; Frydman, H.M.; Connor, J.H. Dual Insect specific virus infection limits Arbovirus replication in Aedes mosquito cells. Virology 2018, 518, 406–413. [Google Scholar] [CrossRef]
- Baidaliuk, A.; Miot, E.F.; Lequime, S.; Moltini-Conclois, I.; Delaigue, F.; Dabo, S.; Dickson, L.B.; Aubry, F.; Merkling, S.H.; Cao-Lormeau, V.M.; et al. Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo. J. Virol. 2019, 93, e00705-19. [Google Scholar] [CrossRef] [Green Version]
- Cammisa-Parks, H.; Cisar, L.A.; Kane, A.; Stollar, V. The complete nucleotide sequence of cell fusing agent (CFA): Homology between the nonstructural proteins encoded by CFA and the nonstructural proteins encoded by arthropod-borne flaviviruses. Virology 1992, 189, 511–524. [Google Scholar] [CrossRef]
- Kuno, G.; Chang, G.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Chouin-Carneiro, T.; Ant, T.H.; Herd, C.; Louis, F.; Failloux, A.B.; Sinkins, S.P. Wolbachia strain wAlbA blocks Zika virus transmission in Aedes aegypti. Med. Vet. Entomol. 2020, 34, 116–119. [Google Scholar] [CrossRef] [Green Version]
- Ahantarig, A.; Trinachartvanit, W.; Kittayapong, P. Relative Wolbachia density of field-collected Aedes albopictus mosquitoes in Thailand. J. Vector Ecol. 2008, 33, 173–177. [Google Scholar] [CrossRef]
- Dutton, T.J.; Sinkins, S.P. Strain-specific quantification of Wolbachia density in Aedes albopictus and effects of larval rearing conditions. Insect Mol. Biol. 2004, 13, 317–322. [Google Scholar] [CrossRef]
- Kittayapong, P.; Baisley, K.J.; Sharpe, R.G.; Baimai, V.; O’Neill, S.L. Maternal transmission efficiency of Wolbachia superinfections in Aedes albopictus populations in Thailand. Am. J. Trop. Med. Hyg. 2002, 66, 103–107. [Google Scholar] [CrossRef] [Green Version]
- Sadeghi, M.; Altan, E.; Deng, X.; Barker, C.M.; Fang, Y.; Coffey, L.L.; Delwart, E. Virome of >12 thousand Culex mosquitoes from throughout California. Virology 2018, 523, 74–88. [Google Scholar] [CrossRef]
- Saiyasombat, R.; Bolling, B.G.; Brault, A.C.; Bartholomay, L.C.; Blitvich, B.J. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae). J. Med. Entomol. 2011, 48, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
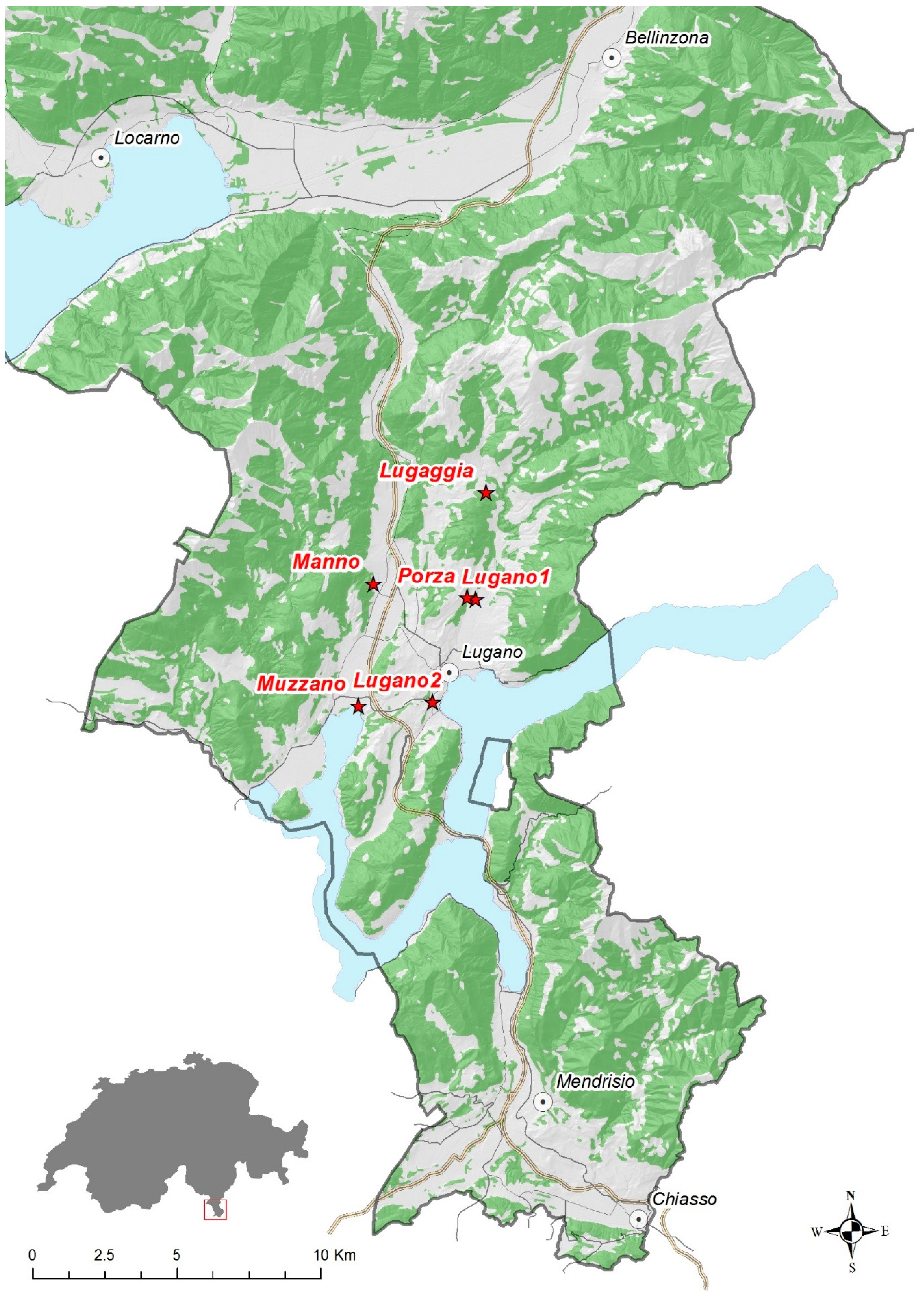



| Pool Name | Sample Location | Position | Number of Mosquitos | Gender | Sampling Time Point |
|---|---|---|---|---|---|
| MO1 | Lugaggia | 46.062 N 8.969 E | 5 | female | August |
| MO2 | Manno | 46.034 N 8.918 E | 16 | female | August |
| MO3 | Lugano1 | 46.028 N 8.964 E | 42 | female | August |
| MO4 | Porza | 46.029 N 8.960 E | 21 | female | August |
| MO5 | Lugano1 | 46.028 N 8.964 E | 48 | female | September |
| MO6 | Lugano1 | 34 | female | September | |
| MO7 | Lugano1 | 35 | female | September | |
| MO8 | Lugano1 | 40 | female | September | |
| MO9 | Muzzano | 45.996 N 8.911 E | 40 | female | September |
| MO10 | Muzzano | 40 | female | September | |
| MO11 | Muzzano | 40 | female | September | |
| MO12 | Muzzano | 50 | female | September | |
| MO13 | Lugano2 | 45.997 N 8.944 E | 59 | female | September |
| MO14 | Lugano2 | 28 | female | September | |
| MO15 | Mix | 40 | male | mix |
| Virus | MO1 | MO2 | MO3 | MO4 | MO5 | MO6 | MO7 | MO8 | MO9 | MO10 | MO11 | MO12 | MO13 | MO14 | MO15 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aedes albopictus cell fusing agent virus | 0.003% | 0.0002% | - | - | - | 0.0002% | 0.0003% | 0.0001% | 0.0006% | 0.0001% | 0.0002% | 0.0002% | - | - | - |
| Aedes flavivirus | - | 0.5% | 0.06% | 0.005% | 0.03% | 0.06% | 0.01% | 0.02% | 0.007% | 0.04% | 0.02% | 0.04% | 0.04% | 0.002% | 0.153% |
| Aphid lethal paralysis virus | - | - | - | 0.001% | - | - | - | - | - | - | - | - | - | - | - |
| Arboretum virus | - | - | - | - | - | - | - | 0.002% | - | 0.001% | - | - | - | - | - |
| Culex Iflavi-like virus 4 | - | - | - | - | - | - | - | - | - | - | - | - | - | - | 4.6% |
| Fig fleck-associated virus | - | - | - | - | - | - | - | - | - | 0.004% | 0.008% | - | - | - | - |
| Guato virus | - | 0.002% | - | 0.0004% | - | 0.0003% | 0.001% | - | - | 0.001% | 0.002% | 0.0003% | - | - | - |
| Hubei mosquito virus 2 | 1.3% | 0.058% | - | 0.42% | 0.77% | 0.19% | 0.3% | 0.2% | - | 0.037% | - | 0.2% | 1.2% | - | - |
| Kaiowa virus | - | 0.0002% | - | - | - | 0.0002% | - | - | - | - | - | - | - | - | - |
| Lake Sarah-associated circular virus-48 | - | - | - | - | - | - | - | - | - | - | - | 0.002% | - | - | - |
| Plant associated genomovirus 3 | - | - | - | - | - | - | - | - | - | - | - | - | 0.003% | - | - |
| Wenzhou sobemo-like virus 4 | 21.4% | 1.3% | 33.1% | 19.9% | 29.0% | 13.8% | 17.9% | 15.7% | 20.5% | 17.3% | 12.6% | 15.5% | 25.2% | 14.4% | 31.4% |
| Whidbey virus | - | 0.0002% | - | - | - | - | - | - | - | - | - | - | - | - | - |
| Total % of viruses | 22.7% | 1.9% | 33.2% | 20.3% | 29.8% | 14.0% | 18.2% | 16.0% | 20.5% | 17.4% | 12.6% | 15.7% | 26.5% | 14.4% | 36.1% |
| Reads generated in million | 4.2 | 9.4 | 6.7 | 7.5 | 7.1 | 9.9 | 12.2 | 9.5 | 8.1 | 20.6 | 6.7 | 8.9 | 6.8 | 3.5 | 3.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kubacki, J.; Flacio, E.; Qi, W.; Guidi, V.; Tonolla, M.; Fraefel, C. Viral Metagenomic Analysis of Aedes albopictus Mosquitos from Southern Switzerland. Viruses 2020, 12, 929. https://doi.org/10.3390/v12090929
Kubacki J, Flacio E, Qi W, Guidi V, Tonolla M, Fraefel C. Viral Metagenomic Analysis of Aedes albopictus Mosquitos from Southern Switzerland. Viruses. 2020; 12(9):929. https://doi.org/10.3390/v12090929
Chicago/Turabian StyleKubacki, Jakub, Eleonora Flacio, Weihong Qi, Valeria Guidi, Mauro Tonolla, and Cornel Fraefel. 2020. "Viral Metagenomic Analysis of Aedes albopictus Mosquitos from Southern Switzerland" Viruses 12, no. 9: 929. https://doi.org/10.3390/v12090929





