Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction
Abstract
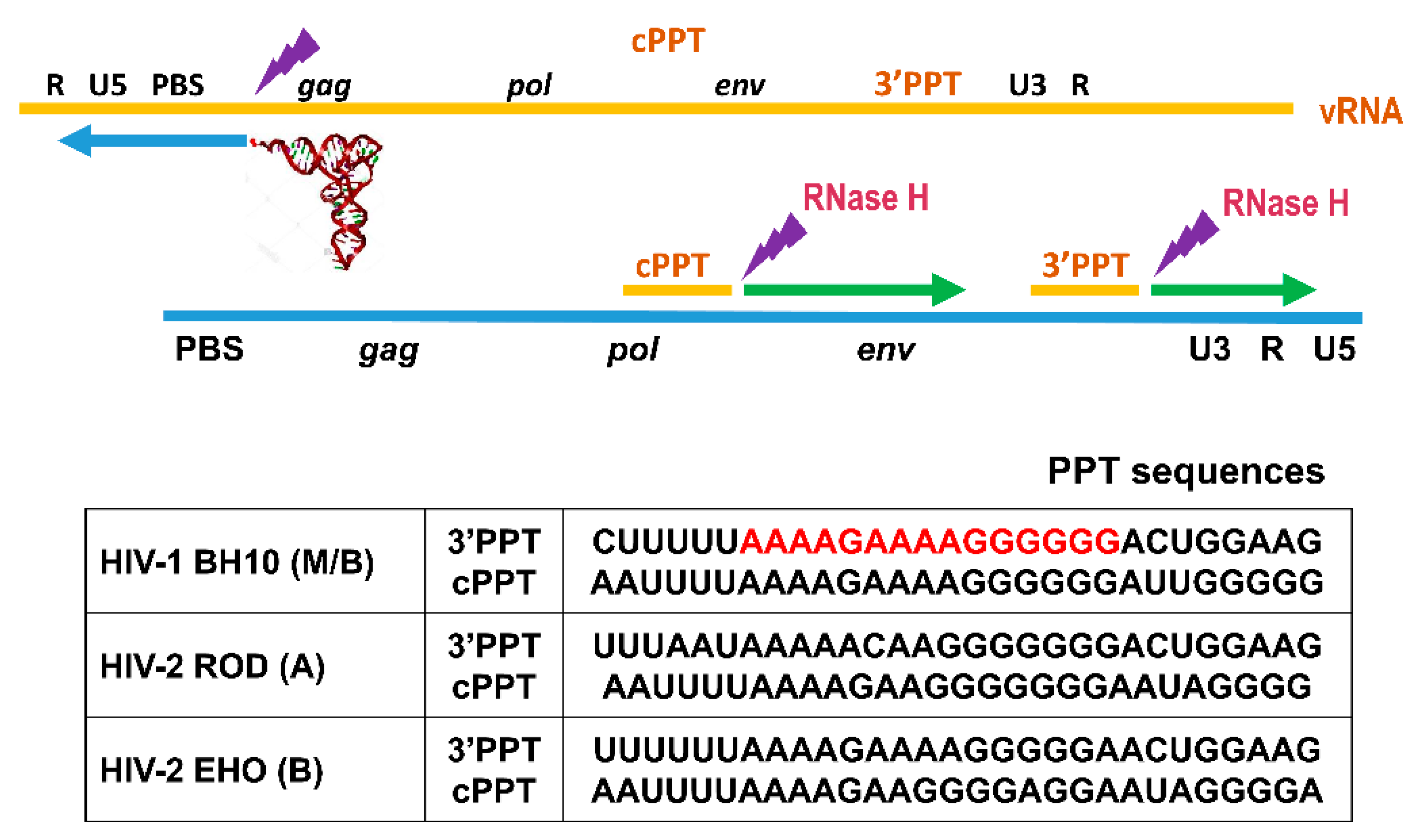
:1. Introduction
2. Materials and Methods
2.1. Expression and Purification of Recombinant RTs
2.2. Mutagenesis
2.3. Nucleotides, Template-Primers and Antiretroviral Drugs
2.4. RNase H Activity Assays
3. Results
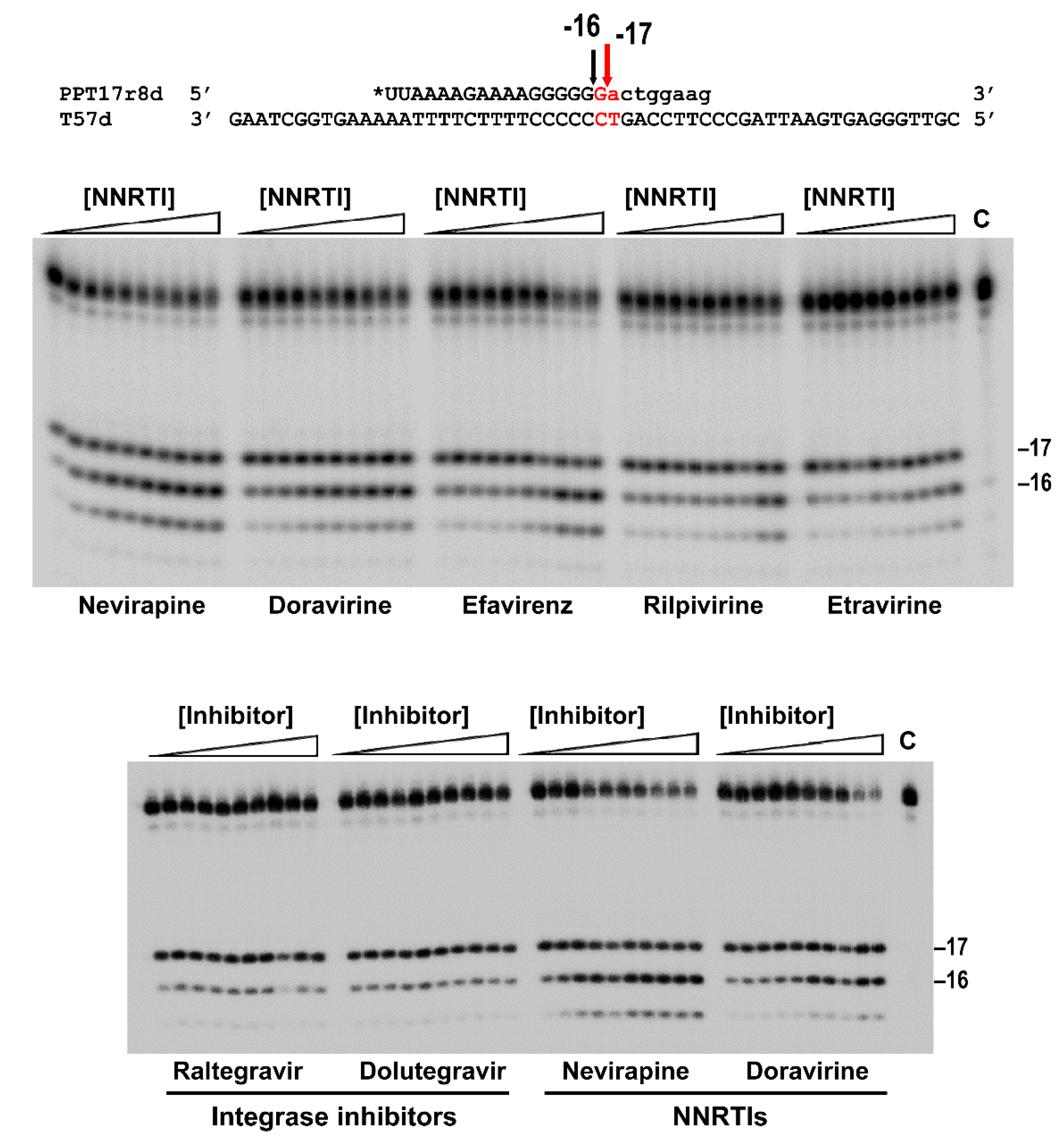
3.1. Effects of NNRTIs and Integrase Inhibitors on PPT Removal by HIV-1BH10 RT
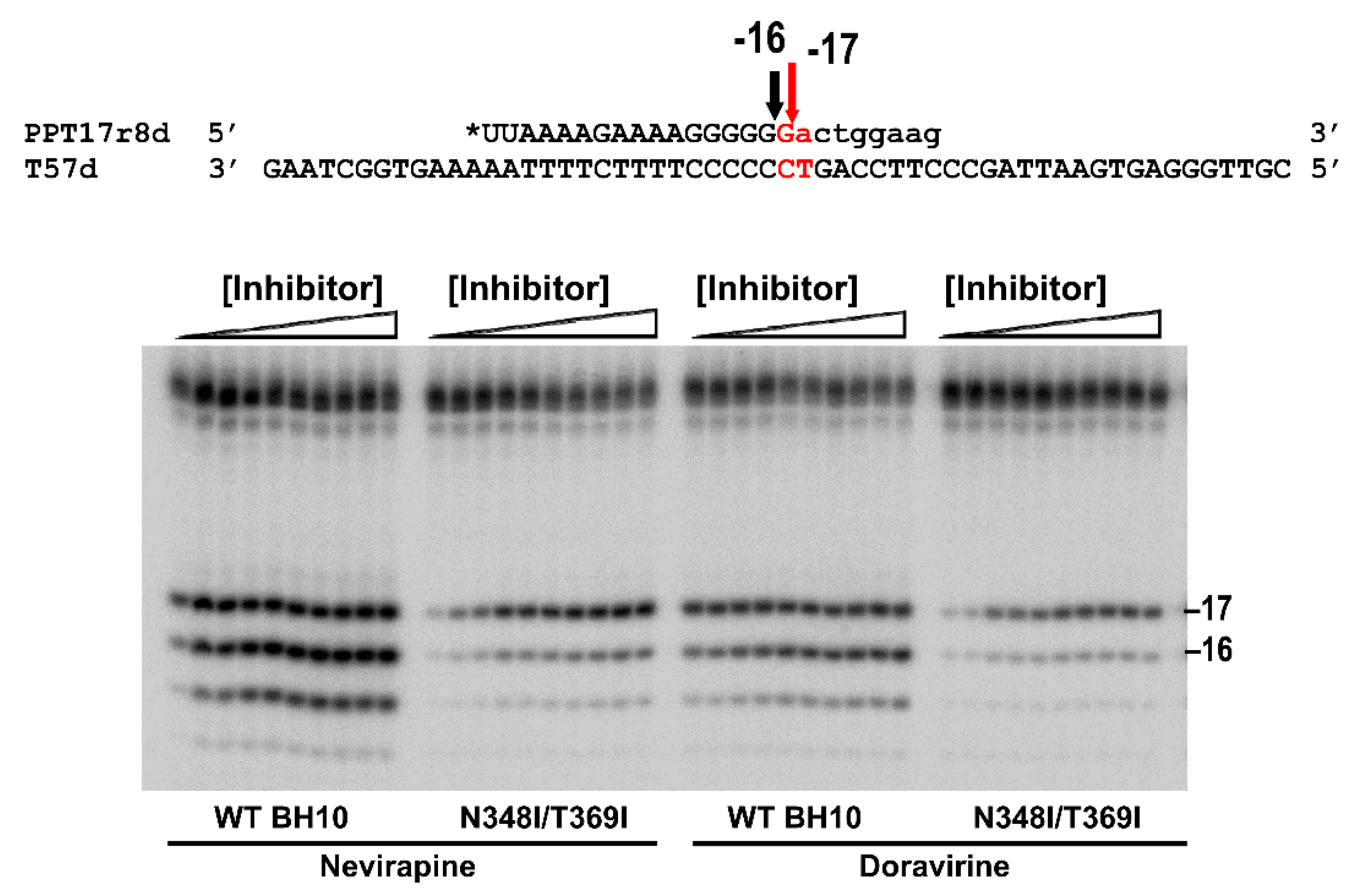
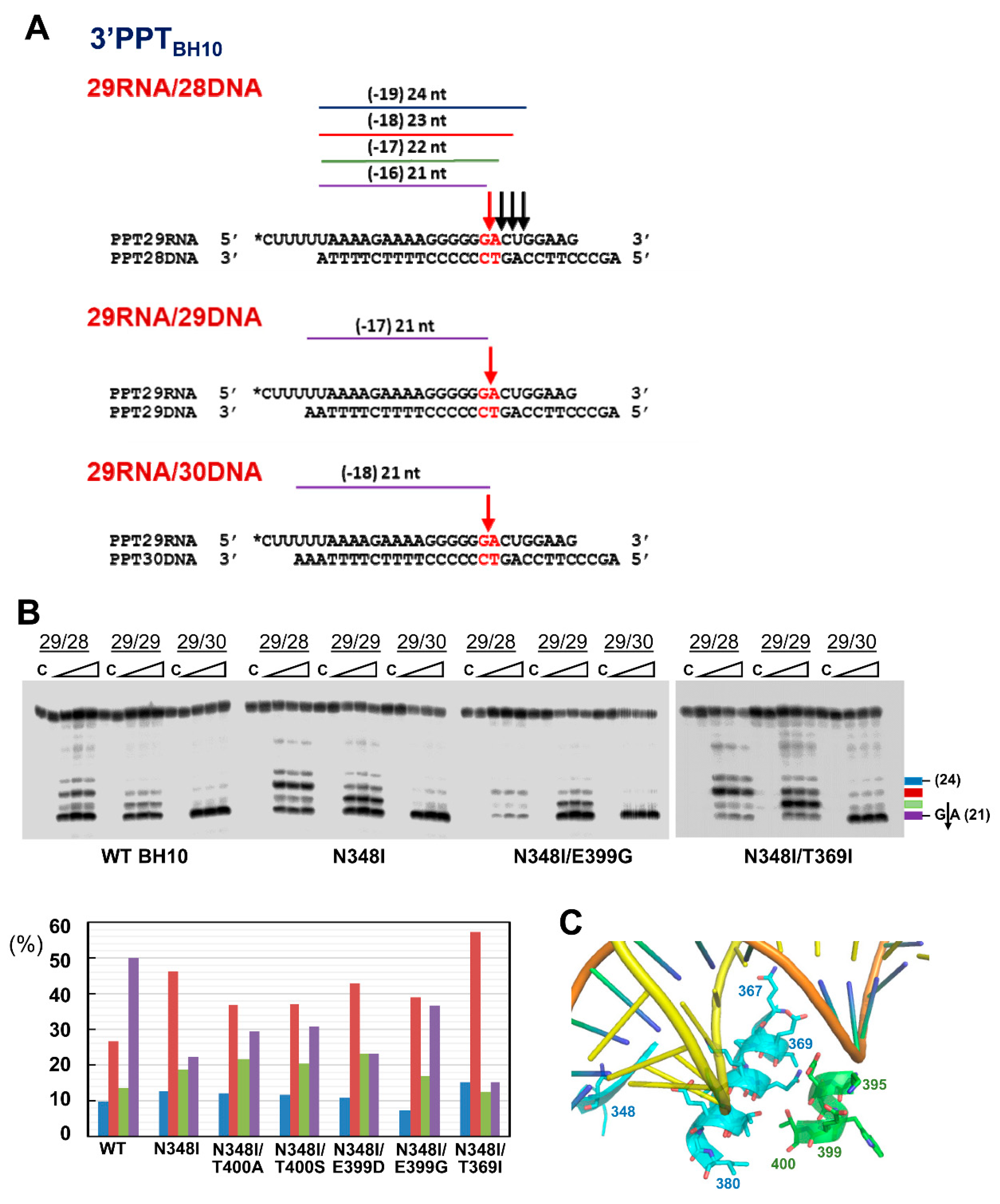
3.2. Dominant Effects of N348I/T369I on the RNase H Cleavage Window at the PPT/U3 Junction
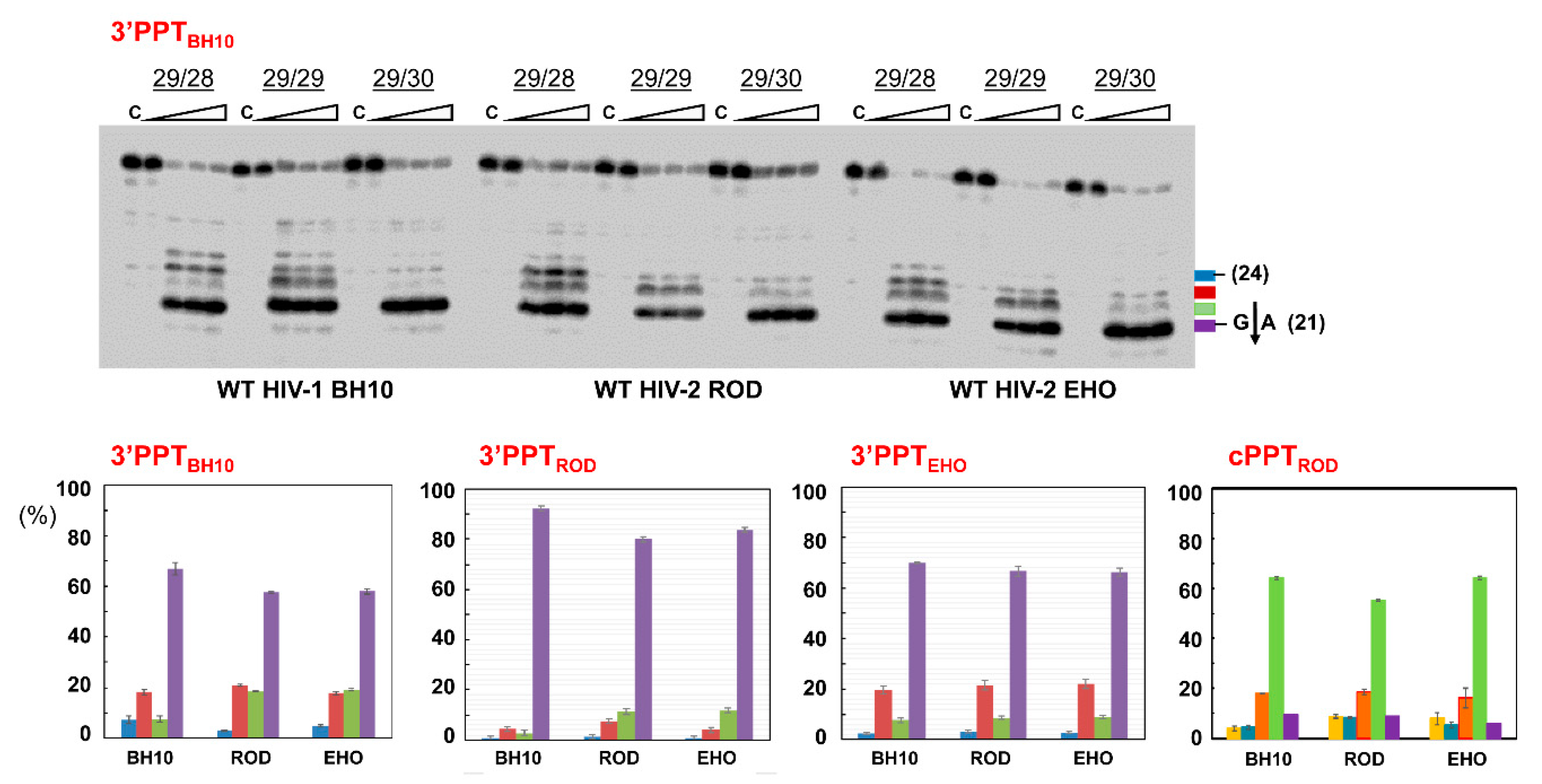
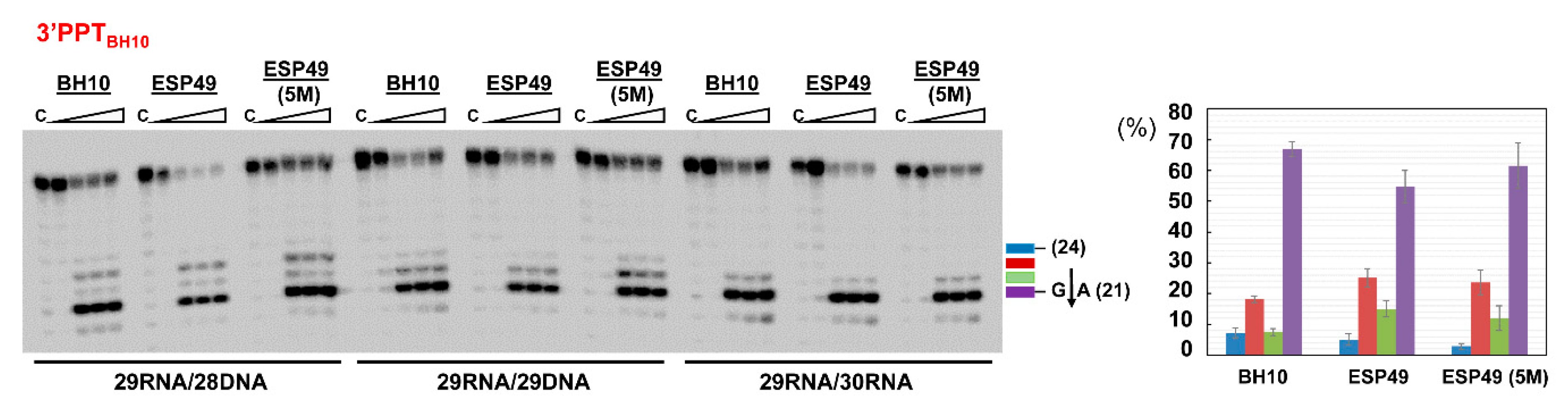
3.3. RNase H Cleavage Patterns of Prototypic HIV RTs and Representative PPTs
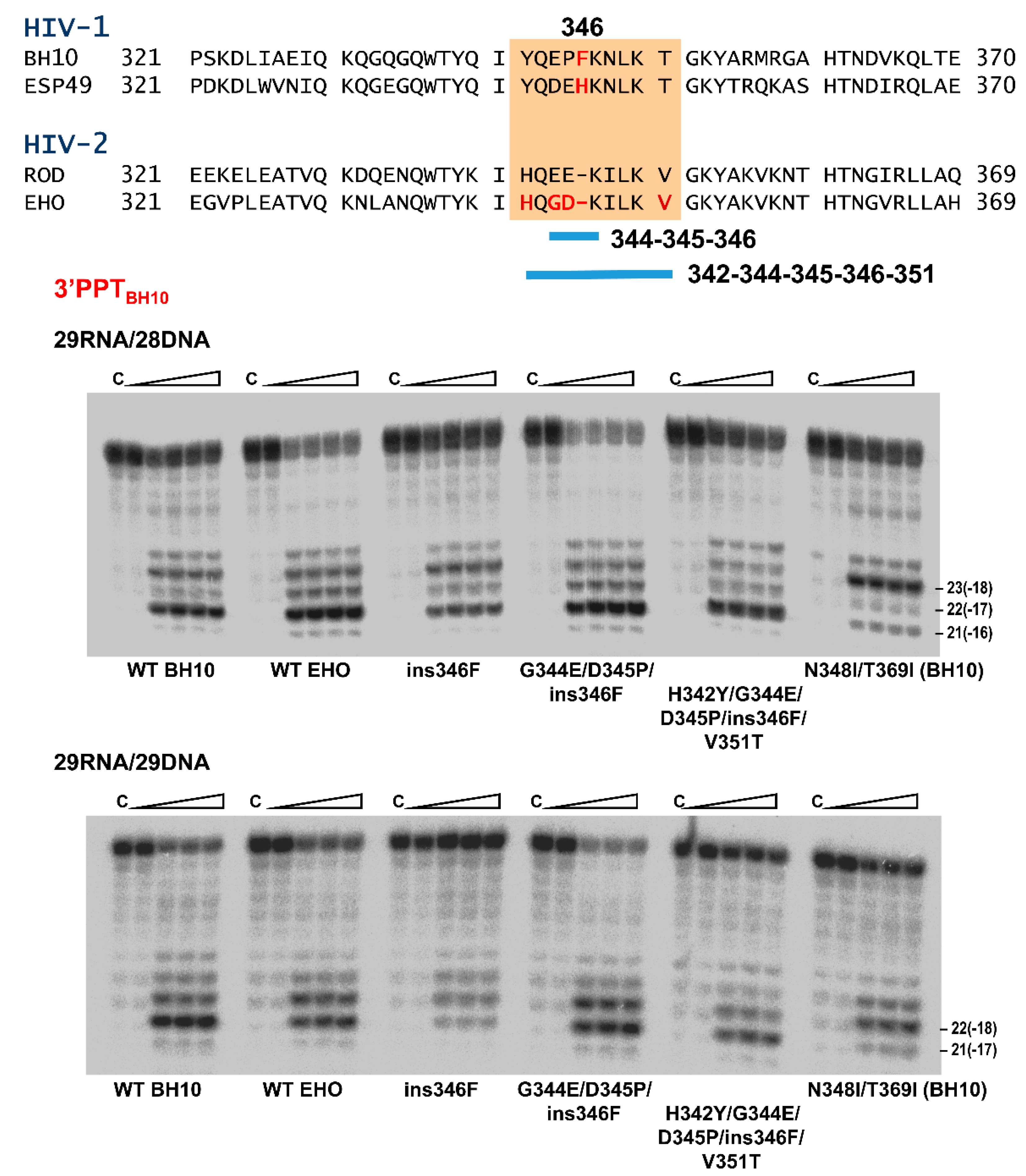
3.4. Characterization of Connection Subdomain Mutants of HIV-2EHO RT and Their Effect on PPT Processing
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hu, W.S.; Hughes, S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012, 2, a006882. [Google Scholar] [CrossRef] [Green Version]
- Menéndez-Arias, L.; Sebastián-Martín, A.; Álvarez, M. Viral reverse transcriptases. Virus Res. 2017, 234, 153–176. [Google Scholar] [CrossRef]
- Zennou, V.; Petit, C.; Guetard, D.; Nerhbass, U.; Montagnier, L.; Charneau, P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 2000, 101, 173–185. [Google Scholar] [CrossRef] [Green Version]
- Arhel, N.J.; Souquere-Besse, S.; Charneau, P. Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: Intracellular visualization at ultrastructural resolution levels. Retrovirology 2006, 3, 38. [Google Scholar] [CrossRef] [Green Version]
- Figiel, M.; Krepl, M.; Park, S.; Poznański, J.; Skowronek, K.; Gołąb, A.; Ha, T.; Šponer, J.; Nowotny, M. Mechanism of polypurine tract primer generation by HIV-1 reverse transcriptase. J. Biol. Chem. 2018, 293, 191–202. [Google Scholar] [CrossRef] [Green Version]
- Rausch, J.W.; Le Grice, S.F.J. ‘Binding, bending and bonding’: Polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int. J. Biochem. Cell Biol. 2004, 36, 1752–1766. [Google Scholar] [CrossRef]
- Julias, J.G.; McWilliams, M.J.; Sarafianos, S.G.; Arnold, E.; Hughes, S.H. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 9515–9520. [Google Scholar] [CrossRef] [Green Version]
- Rausch, J.W.; Lener, D.; Miller, J.T.; Julias, J.G.; Hughes, S.H.; Le Grice, S.F.J. Altering the RNase H primer grip of human immunodeficiency virus reverse transcriptase modifies cleavage specificity. Biochemistry 2002, 41, 4856–4865. [Google Scholar] [CrossRef]
- McWilliams, M.J.; Julias, J.G.; Sarafianos, S.G.; Alvord, W.G.; Arnold, E.; Hughes, S.H. Combining mutations in HIV-1 reverse transcriptase with mutations in the HIV-1 polypurine tract affects RNase H cleavages involved in PPT utilization. Virology 2006, 348, 378–388. [Google Scholar] [CrossRef] [Green Version]
- Sarafianos, S.G.; Marchand, B.; Das, K.; Himmel, D.M.; Parniak, M.A.; Hughes, S.H.; Arnold, E. Structure and function of HIV-1 reverse transcriptase: Molecular mechanisms of polymerization and inhibition. J. Mol. Biol. 2009, 385, 693–713. [Google Scholar] [CrossRef] [Green Version]
- Tian, L.; Kim, M.S.; Li, H.; Wang, J.; Yang, W. Structure of HIV-1 reverse transcriptase cleaving RNA in an RNA/DNA hybrid. Proc. Natl. Acad. Sci. USA 2018, 115, 507–512. [Google Scholar] [CrossRef] [Green Version]
- Xavier Ruiz, F.; Arnold, E. Evolving understanding of HIV-1 reverse transcriptase structure, function, inhibition, and resistance. Curr. Opin. Struct. Biol. 2020, 61, 113–123. [Google Scholar] [CrossRef]
- Li, A.; Li, J.; Johnson, K.A. HIV-1 reverse transcriptase polymerase and RNase H (ribonuclease H) active sites work simultaneously and independently. J. Biol. Chem. 2016, 291, 26566–26585. [Google Scholar] [CrossRef] [Green Version]
- Figiel, M.; Krepl, M.; Poznanski, J.; Golab, A.; Šponer, J.; Nowotny, M. Coordination between the polymerase and RNase H activity of HIV-1 reverse transcriptase. Nucleic Acids Res. 2017, 45, 3341–3352. [Google Scholar] [CrossRef]
- Götte, M.; Kameoka, M.; McLellan, N.; Cellai, L.; Wainberg, M.A. Analysis of efficiency and fidelity of HIV-1 (+)-strand DNA synthesis reveals a novel rate-limiting step during retroviral reverse transcription. J. Biol. Chem. 2001, 276, 6711–6719. [Google Scholar] [CrossRef] [Green Version]
- Grobler, J.A.; Dornadula, G.; Rice, M.R.; Simcoe, A.L.; Hazuda, D.J.; Miller, M.D. HIV-1 reverse transcriptase plus-strand initiation exhibits preferential sensitivity to non-nucleoside reverse transcriptase inhibitors in vitro. J. Biol. Chem. 2007, 282, 8005–8010. [Google Scholar] [CrossRef]
- Betancor, G.; Álvarez, M.; Marcelli, B.; Andrés, C.; Martínez, M.A.; Menéndez-Arias, L. Effects of HIV-1 reverse transcriptase connection subdomain mutations on polypurine tract removal and initiation of (+)-strand DNA synthesis. Nucleic Acids Res. 2015, 43, 2259–2270. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Fransen, S.; Paxinos, E.E.; Stawiski, E.; Huang, W.; Petropoulos, C.J. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: Assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob. Agents Chemother. 2010, 54, 1973–1980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Vingerhoets, J.; Fransen, S.; Tambuyzer, L.; Azijn, H.; Frantzell, A.; Paredes, R.; Coakley, E.; Nijs, S.; Clotet, B.; et al. Connection domain mutations in HIV-1 reverse transcriptase do not impact etravirine susceptibility and virologic responses to etravirine-containing regimens. Antimicrob. Agents Chemother. 2011, 55, 2872–2879. [Google Scholar] [CrossRef] [Green Version]
- Menéndez-Arias, L.; Betancor, G.; Matamoros, T. HIV-1 reverse transcriptase connection subdomain mutations involved in resistance to approved non-nucleoside inhibitors. Antiviral Res. 2011, 92, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Menéndez-Arias, L. Molecular basis of human immunodeficiency virus type 1 drug resistance: Overview and recent developments. Antiviral Res. 2013, 98, 93–120. [Google Scholar] [CrossRef] [PubMed]
- Yap, S.H.; Sheen, C.W.; Fahey, J.; Zanin, M.; Tyssen, D.; Lima, V.D.; Wynhoven, B.; Kuiper, M.; Sluis-Cremer, N.; Harrigan, P.R.; et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med. 2007, 4, e335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paredes, R.; Puertas, M.C.; Bannister, W.; Kisic, M.; Cozzi-Lepri, A.; Pou, C.; Bellido, R.; Betancor, G.; Bogner, J.; Gargalianos, P.; et al. A376S in the connection subdomain of HIV-1 reverse transcriptase confers increased risk of virological failure to nevirapine therapy. J. Infect. Dis. 2011, 204, 741–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palaniappan, C.; Fay, P.J.; Bambara, R.A. Nevirapine alters the cleavage specificity of ribonuclease H of human immunodeficiency virus 1 reverse transcriptase. J. Biol. Chem. 1995, 270, 4861–4869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hang, J.Q.; Li, Y.; Yang, Y.; Cammack, N.; Mirzadegan, T.; Klumpp, K. Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs). Biochem. Biophys. Res. Commun. 2007, 352, 341–350. [Google Scholar] [CrossRef]
- Radzio, J.; Sluis-Cremer, N. Efavirenz accelerates HIV-1 reverse transcriptase ribonuclease H cleavage, leading to diminished zidovudine excision. Mol. Pharmacol. 2008, 73, 601–606. [Google Scholar] [CrossRef] [PubMed]
- Nikolenko, G.N.; Delviks-Frankenberry, K.A.; Pathak, V.K. A novel molecular mechanism of dual resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J. Virol. 2010, 84, 5238–5249. [Google Scholar] [CrossRef] [Green Version]
- Delviks-Frankenberry, K.A.; Nikolenko, G.N.; Pathak, V.K. The “connection” between HIV drug resistance and RNase H. Viruses 2010, 2, 1476–1503. [Google Scholar] [CrossRef] [Green Version]
- Lapkouski, M.; Tian, L.; Miller, J.T.; Le Grice, S.F.J.; Yang, W. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat. Struct. Mol. Biol. 2013, 20, 230–236. [Google Scholar] [CrossRef] [Green Version]
- Biondi, M.J.; Beilhartz, G.L.; McCormick, S.; Götte, M. N348I in HIV-1 reverse transcriptase can counteract the nevirapine-mediated bias toward RNase H cleavage during plus-strand initiation. J. Biol. Chem. 2010, 285, 26966–26975. [Google Scholar] [CrossRef] [Green Version]
- Poveda, E.; de Mendoza, C.; Pattery, T.; González, M.M.; Villacian, J.; Soriano, V. Phenotypic impact of resistance mutations on etravirine susceptibility in HIV patients with prior failure to nonnucleoside analogues. AIDS 2008, 22, 2395–2398. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.F.; Lengruber, R.B.; Soares, E.A.; Jere, A.; Sprinz, E.; Martinez, A.M.; Silveira, J.; Sion, F.S.; Pathak, V.K.; Soares, M.A. Conservation patterns of HIV-1 RT connection and RNase H domains: Identification of new mutations in NRTI-treated patients. PLoS ONE 2008, 3, e1781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lengruber, R.B.; Delviks-Frankenberry, K.A.; Nikolenko, G.N.; Baumann, J.; Santos, A.F.; Pathak, V.K.; Soares, M.A. Phenotypic characterization of drug resistance-associated mutations in HIV-1 RT connection and RNase H domains and their correlation with thymidine analogue mutations. J. Antimicrob. Chemother. 2011, 66, 702–708. [Google Scholar] [CrossRef] [Green Version]
- Wright, D.W.; Deuzing, I.P.; Flandre, P.; van den Eede, P.; Govaert, M.; Setiawan, L.; Coveney, P.V.; Marcelin, A.G.; Calvez, V.; Boucher, C.A.; et al. A polymorphism at position 400 in the connection subdomain of HIV-1 reverse transcriptase affects sensitivity to NNRTIs and RNaseH activity. PLoS ONE 2013, 8, e74078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez, M.; Matamoros, T.; Menéndez-Arias, L. Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 2009, 392, 872–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matamoros, T.; Barrioluengo, V.; Abia, D.; Menéndez-Arias, L. Major groove binding track residues of the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase enhance cDNA synthesis at high temperatures. Biochemistry 2013, 52, 9318–9328. [Google Scholar] [CrossRef]
- Boretto, J.; Longhi, S.; Navarro, J.M.; Selmi, B.; Sire, J.; Canard, B. An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 2001, 292, 139–147. [Google Scholar] [CrossRef]
- Matamoros, T.; Deval, J.; Guerreiro, C.; Mulard, L.; Canard, B.; Menéndez-Arias, L. Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by alpha-phosphate-modified thymidine analogues. J. Mol. Biol. 2005, 349, 451–463. [Google Scholar] [CrossRef]
- Álvarez, M.; Sebastián-Martín, A.; García-Marquina, G.; Menéndez-Arias, L. Fidelity of classwide-resistant HIV-2 reverse transcriptase and differential contribution of K65R to the accuracy of HIV-1 and HIV-2 reverse transcriptases. Sci. Rep. 2017, 7, 44834. [Google Scholar] [CrossRef] [Green Version]
- Álvarez, M.; Nevot, M.; Mendieta, J.; Martínez, M.A.; Menéndez-Arias, L. Amino acid residues in HIV-2 reverse transcriptase that restrict the development of nucleoside analogue resistance through the excision pathway. J. Biol. Chem. 2018, 293, 2247–2259. [Google Scholar] [CrossRef] [Green Version]
- Kati, W.M.; Johnson, K.A.; Jerva, L.F.; Anderson, K.S. Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 1992, 267, 25988–25997. [Google Scholar] [CrossRef]
- Betancor, G.; Puertas, M.C.; Nevot, M.; Garriga, C.; Martínez, M.A.; Martinez-Picado, J.; Menéndez-Arias, L. Mechanisms involved in the selection of HIV-1 reverse transcriptase thumb subdomain polymorphisms associated with nucleoside analogue therapy failure. Antimicrob. Agents Chemother. 2010, 54, 4799–4811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarafianos, S.G.; Das, K.; Tantillo, C.; Clark, A.D., Jr.; Ding, J.; Whitcomb, J.M.; Boyer, P.L.; Hughes, S.H.; Arnold, E. Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J. 2001, 20, 1449–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, K.; Martinez, S.E.; Bauman, J.D.; Arnold, E. HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat. Struct. Mol. Biol. 2012, 19, 253–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehteshami, M.; Beilhartz, G.L.; Scarth, B.J.; Tchesnokov, E.P.; McCormick, S.; Wynhoven, B.; Harrigan, P.R.; Götte, M. Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J. Biol. Chem. 2008, 283, 22222–22232. [Google Scholar] [CrossRef] [Green Version]
- Schuckmann, M.M.; Marchand, B.; Hachiya, A.; Kodama, E.N.; Kirby, K.A.; Singh, K.; Sarafianos, S.G. The N348I mutation at the connection subdomain of HIV-1 reverse transcriptase decreases binding to nevirapine. J. Biol. Chem. 2010, 285, 38700–38709. [Google Scholar] [CrossRef] [Green Version]
- Deeks, E.D. Doravirine: First global approval. Drugs 2018, 78, 1643–1650. [Google Scholar] [CrossRef]
- Martin, E.A.; Lai, M.T.; Ngo, W.; Feng, M.; Graham, D.; Hazuda, D.J.; Kumar, S.; Hwang, C.; Sklar, P.; Asante-Appiah, E. Review of doravirine resistance patterns identified in participants during clinical development. J. Acquir. Immune Defic. Syndr. 2020, 85, 635–642. [Google Scholar] [CrossRef]
- Feng, M.; Sachs, N.A.; Xu, M.; Grobler, J.; Blair, W.; Hazuda, D.J.; Miller, M.D.; Lai, M.T. Doravirine suppresses common nonnucleoside reverse transcriptase inhibitor-associated mutants at clinically relevant concentrations. Antimicrob. Agents Chemother. 2016, 60, 2241–2247. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.; Lai, M.T.; Hazuda, D. Rational design of doravirine: From bench to patients. ACS Infect. Dis. 2020, 6, 64–73. [Google Scholar] [CrossRef]
- Malet, I.; Subra, F.; Charpentier, C.; Collin, G.; Descamps, D.; Calvez, V.; Marcelin, A.G.; Delelis, O. Mutations located outside the integrase gene can confer resistance to HIV-1 integrase strand transfer inhibitors. mBio 2017, 8, e00922-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wijting, I.E.A.; Lungu, C.; Rijnders, B.J.A.; van der Ende, M.E.; Pham, H.T.; Mesplede, T.; Pas, S.D.; Voermans, J.J.C.; Schuurman, R.; van de Vijver, D.A.M.C.; et al. HIV-1 resistance dynamics in patients with virologic failure to dolutegravir maintenance monotherapy. J. Infect. Dis. 2018, 218, 688–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.T.; Berkhout, B. How polypurine tract changes in the HIV-1 RNA genome can cause resistance against the integrase inhibitor dolutegravir. mBio 2018, 9, e00006-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malet, I.; Subra, F.; Richetta, C.; Charpentier, C.; Collin, G.; Descamps, D.; Calvez, V.; Marcelin, A.G.; Delelis, O. Reply to Das and Berkhout; “How polypurine tract changes in the HIV-1 RNA genome can cause resistance against the integrase inhibitor dolutegravir”. mBio 2018, 9, e00623-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijayan, R.S.K.; Arnold, E.; Das, K. Molecular dynamics study of HIV-1 RT-DNA-nevirapine complexes explains NNRTI inhibition and resistance by connection mutations. Proteins 2014, 82, 815–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, H.T.; Colby-Germinario, S.P.; Oliveira, M.; Han, Y.; Quan, Y.; Zanichelli, V.; Wainberg, M.A. The connection domain mutation N348I in HIV-1 reverse transcriptase enhances resistance to etravirine and rilpivirine but restricts the emergence of the E138K resistance mutation by diminishing viral replication capacity. J. Virol. 2014, 88, 1536–1547. [Google Scholar] [CrossRef] [Green Version]
- Ren, J.; Bird, L.E.; Chamberlain, P.P.; Stewart-Jones, G.B.; Stuart, D.I.; Stammers, D.K. Structure of HIV-2 reverse transcriptase at 2.35-A resolution and the mechanism of resistance to non-nucleoside inhibitors. Proc. Natl. Acad. Sci. USA 2002, 99, 14410–14415. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.J.; Wang, R.R.; Chen, H.; Luo, R.H.; Yang, L.M.; Liu, J.P.; Sun, H.D.; Zhang, H.B.; Xiao, W.L.; Zheng, Y.T. SJP-L-5 inhibits HIV-1 polypurine tract primed plus-strand DNA elongation; indicating viral DNA synthesis initiation at multiple sites under drug pressure. Sci. Rep. 2018, 8, 2574. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Álvarez, M.; Sapena-Ventura, E.; Luczkowiak, J.; Martín-Alonso, S.; Menéndez-Arias, L. Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction. Viruses 2021, 13, 131. https://doi.org/10.3390/v13010131
Álvarez M, Sapena-Ventura E, Luczkowiak J, Martín-Alonso S, Menéndez-Arias L. Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction. Viruses. 2021; 13(1):131. https://doi.org/10.3390/v13010131
Chicago/Turabian StyleÁlvarez, Mar, Enrique Sapena-Ventura, Joanna Luczkowiak, Samara Martín-Alonso, and Luis Menéndez-Arias. 2021. "Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction" Viruses 13, no. 1: 131. https://doi.org/10.3390/v13010131
APA StyleÁlvarez, M., Sapena-Ventura, E., Luczkowiak, J., Martín-Alonso, S., & Menéndez-Arias, L. (2021). Analysis and Molecular Determinants of HIV RNase H Cleavage Specificity at the PPT/U3 Junction. Viruses, 13(1), 131. https://doi.org/10.3390/v13010131






