Emergence of Novel Reassortant H1N1 Avian Influenza Viruses in Korean Wild Ducks in 2018 and 2019
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Virus Isolation
2.3. Viral Genome Extraction and RT-PCR to Confirm the Virus and Identify the Host
2.4. Next-Generation Sequencing (NGS) Using Illumina HiSeq X
2.5. Viral Genomic and Phylogenetic Tree Analyses
2.6. Determination of 50% Tissue Culture Infectious Dose (TCID50) and 50% Egg Infectious Dose (EID50)
2.7. Viral Growth Kinetics in MDCK Cells
2.8. Animal Study
2.9. Statistical Analysis
3. Results
3.1. Genome Characterization of Two H1N1 Isolates
3.2. A/Greater White-Fronted Goose/South Korea/KNU18-64/2018(H1N1) and A/Wild Bird/South Korea/WKU19-4/2019(H1N1) were Generated as a Result of Reassortment Events
3.3. Molecular Characterization of the H1N1 Isolate
3.4. Replication of H1N1 in Mammalian Cells
3.5. Pathogenicity in Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alexander, D.J. An overview of the epidemiology of avian influenza. Vaccine 2007, 25, 5637–5644. [Google Scholar] [CrossRef] [PubMed]
- CDC. Influenza Type A Viruses. Available online: https://www.cdc.gov/flu/avianflu/influenza-a-virus-subtypes.htm (accessed on 19 April 2017).
- Zhou, J.F.; Yang, L.; Lan, Y.; Li, Z.; Zhao, X.; Wang, M.; Guo, Y.J.; Li, D.X.; Shu, Y.L. Review on the etiological property of 1918/1919 Spainsh flu virus (H1N1). Bing Du Xue Bao 2009, 25, 8–11. [Google Scholar] [PubMed]
- CDC. Past Pandemics. Available online: https://www.cdc.gov/flu/pandemic-resources/basics/past-pandemics.html (accessed on 10 August 2018).
- CDC. Influenza Historic Timeline. Available online: https://www.cdc.gov/flu/pandemic-resources/pandemic-timeline-1930-and-beyond.htm (accessed on 30 January 2019).
- Babiuk, S.; Albrecht, R.; Berhane, Y.; Marszal, P.; Richt, J.A.; Garcia-Sastre, A.; Pasick, J.; Weingartl, H. 1918 and 2009 H1N1 influenza viruses are not pathogenic in birds. J. Gen. Virol. 2010, 91, 339–342. [Google Scholar] [CrossRef] [PubMed]
- Scholtissek, C.; Burger, H.; Bachmann, P.A.; Hannoun, C. Genetic relatedness of hemagglutinins of the H1 subtype of influenza A viruses isolated from swine and birds. Virology 1983, 129, 521–523. [Google Scholar] [CrossRef]
- Kilbourne, E.D. Influenza pandemics of the 20th century. Emerg. Infect. Dis. 2006, 12, 9. [Google Scholar] [CrossRef] [PubMed]
- Schnitzler, S.U.; Schnitzler, P. An update on swine-origin influenza virus A/H1N1: A review. Virus Genes 2009, 39, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y. The 2009 H1N1 pandemic influenza in Korea. Tuberc. Respir. Dis. 2016, 79, 70–73. [Google Scholar] [CrossRef] [Green Version]
- Garten, R.J.; Davis, C.T.; Russell, C.A.; Shu, B.; Lindstrom, S.; Balish, A.; Sessions, W.M.; Xu, X.; Skepner, E.; Deyde, V.; et al. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009, 325, 197–201. [Google Scholar] [CrossRef] [Green Version]
- Ku, K.B.; Park, E.H.; Yum, J.; Kim, J.A.; Oh, S.K.; Seo, S.H. Highly pathogenic avian influenza A(H5N8) virus from waterfowl, South Korea, 2014. Emerg. Infect. Dis. 2014, 20, 1587–1588. [Google Scholar] [CrossRef]
- Lee, Y.J.; Kang, H.M.; Lee, E.K.; Song, B.M.; Jeong, J.; Kwon, Y.K.; Kim, H.R.; Lee, K.J.; Hong, M.S.; Jang, I.; et al. Novel reassortant influenza A(H5N8) viruses, South Korea, 2014. Emerg. Infect. Dis. 2014, 20, 1087–1089. [Google Scholar] [CrossRef]
- Lee, E.K.; Song, B.M.; Lee, Y.N.; Heo, G.B.; Bae, Y.C.; Joh, S.J.; Park, S.C.; Choi, K.S.; Lee, H.J.; Jang, I.; et al. Multiple novel H5N6 highly pathogenic avian influenza viruses, South Korea, 2016. Infect. Genet. Evol. 2017, 51, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.-k.; Sung, H.-w.; Joh, S.-j.; Lee, Y.-j.; Kim, M.-c.; Choi, J.-g.; Lee, E.-k.; Wee, S.-H.; Kim, J.-h. An outbreak of highly pathogenic avian influenza subtype H5N1 in broiler breeders, Korea. J. Vet. Med Sci. 2005, 67, 1193–1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, D.-H.; Park, J.-K.; Youn, H.-N.; Lee, Y.-N.; Lim, T.-H.; Kim, M.-S.; Lee, J.-B.; Park, S.-Y.; Choi, I.-S.; Song, C.-S. Surveillance and isolation of HPAI H5N1 from wild Mandarin Ducks (Aix galericulata). J. Wildl. Dis. 2011, 47, 994–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-J.; Choi, Y.-K.; Kim, Y.-J.; Song, M.-S.; Jeong, O.-M.; Lee, E.-K.; Jeon, W.-J.; Jeong, W.; Joh, S.-J.; Choi, K.-s. Highly pathogenic avian influenza virus (H5N1) in domestic poultry and relationship with migratory birds, South Korea. Emerg. Infect. Dis. 2008, 14, 487. [Google Scholar] [CrossRef]
- Jeong, O.-M.; Kim, M.-C.; Kim, M.-J.; Kang, H.-M.; Kim, H.-R.; Kim, Y.-J.; Joh, S.-J.; Kwon, J.-H.; Lee, Y.-J. Experimental infection of chickens, ducks and quails with the highly pathogenic H5N1 avian influenza virus. J. Vet. Sci. 2009, 10, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.; Kang, H.-M.; Lee, E.-K.; Song, B.-M.; Kwon, Y.-K.; Kim, H.-R.; Choi, K.-S.; Kim, J.-Y.; Lee, H.-J.; Moon, O.-K. Highly pathogenic avian influenza virus (H5N8) in domestic poultry and its relationship with migratory birds in South Korea during 2014. Vet. Microbiol. 2014, 173, 249–257. [Google Scholar] [CrossRef]
- Lee, D.-H.; Kwon, J.-H.; Noh, J.-Y.; Park, J.-K.; Yuk, S.-S.; Erdene-Ochir, T.-O.; Lee, J.-B.; Park, S.-Y.; Choi, I.-S.; Lee, S.-W. Pathogenicity of the Korean H5N8 highly pathogenic avian influenza virus in commercial domestic poultry species. Avian Pathol. 2016, 45, 208–211. [Google Scholar] [CrossRef] [Green Version]
- VanDalen, K.K.; Franklin, A.B.; Mooers, N.L.; Sullivan, H.J.; Shriner, S.A. Shedding light on avian influenza H4N6 infection in mallards: Modes of transmission and implications for surveillance. PLoS ONE 2010, 5, e12851. [Google Scholar] [CrossRef] [Green Version]
- Tuong, H.T.; Nguyen, N.M.; Sung, H.W.; Park, H.; Yeo, S.J. Genetic Characterization of Avian Influenza A (H11N9) Virus Isolated from Mandarin Ducks in South Korea in 2018. Viruses 2020, 12, 203. [Google Scholar] [CrossRef] [Green Version]
- Killian, M.L. Hemagglutination assay for influenza virus. Methods Mol. Biol. 2014, 1161, 3–9. [Google Scholar]
- World Health Organization. CDC protocol of realtime RTPCR for swine influenza A (H1N1); World Health Organization (WHO): Geneva, Switzerland, 2009. [Google Scholar]
- Brown, J.D.; Berghaus, R.D.; Costa, T.P.; Poulson, R.; Carter, D.L.; Lebarbenchon, C.; Stallknecht, D.E. Intestinal excretion of a wild bird-origin H3N8 low pathogenic avian influenza virus in mallards (Anas platyrhynchos). J. Wildl. Dis. 2012, 48, 991–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, P.D.; Stoeckle, M.Y.; Zemlak, T.S.; Francis, C.M. Identification of Birds through DNA Barcodes. PLoS Biol. 2004, 2, e312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambardar, S.; Gupta, R.; Trakroo, D.; Lal, R.; Vakhlu, J. High throughput sequencing: An overview of sequencing chemistry. Indian J. Microbiol. 2016, 56, 394–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.M.; Sung, H.W.; Yun, K.-J.; Park, H.; Yeo, S.-J. Genetic Characterization of a Novel North American-Origin Avian Influenza A (H6N5) Virus Isolated from Bean Goose of South Korea in 2018. Viruses 2020, 12, 774. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- World Health Organization. Manual for the Laboratory Diagnosis and Virological Surveillance of Influenza; World Health Organization (WHO): Geneva, Switzerland, 2011. [Google Scholar]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Jiménez-Bluhm, P.; Karlsson, E.A.; Ciuoderis, K.A.; Cortez, V.; Marvin, S.A.; Hamilton-West, C.; Schultz-Cherry, S.; Osorio, J.E. Avian H11 influenza virus isolated from domestic poultry in a Colombian live animal market. Emerg. Microbes Infect. 2016, 5, 1–9. [Google Scholar] [CrossRef]
- Su, Y.; Yang, H.-Y.; Zhang, B.-J.; Jia, H.-L.; Tien, P. Analysis of a point mutation in H5N1 avian influenza virus hemagglutinin in relation to virus entry into live mammalian cells. Arch. Virol. 2008, 153, 2253–2261. [Google Scholar] [CrossRef]
- Henningson, J.N.; Rajao, D.S.; Kitikoon, P.; Lorusso, A.; Culhane, M.R.; Lewis, N.S.; Anderson, T.K.; Vincent, A.L. Comparative virulence of wild-type H1N1pdm09 influenza A isolates in swine. Vet. Microbiol. 2015, 176, 40–49. [Google Scholar] [CrossRef]
- Carbone, V.; Schneider, E.K.; Rockman, S.; Baker, M.; Huang, J.X.; Ong, C.; Cooper, M.A.; Yuriev, E.; Li, J.; Velkov, T. Molecular characterisation of the haemagglutinin glycan-binding specificity of egg-adapted vaccine strains of the pandemic 2009 H1N1 swine influenza A virus. Molecules 2015, 20, 10415–10434. [Google Scholar] [CrossRef]
- Jampangern, W.; Chunsutthiwat, S.; Pittayawonganon, C.-T.G.; Hiramatsu, H.; Pooruk, P.; Thitithanyanont, A.; Ungchusak, C.; Louisirirotchanakul, S.; Lerdsamran, H. An Avian Influenza H5N1 Virus That Binds to a Human-Type Receptor. J. Virol. 2007, 81, 9950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elderfield, R.A.; Watson, S.J.; Godlee, A.; Adamson, W.E.; Thompson, C.; Dunning, J.; Fernandez-Alonso, M.; Blumenkrantz, D.; Hussell, T.; Zambon, M. Accumulation of human-adapting mutations during circulation of A (H1N1) pdm09 influenza in humans in the UK. J. Virol. 2014, 88, 13269–13283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matrosovich, M.; Gambaryan, A.; Teneberg, S.; Piskarev, V.; Yamnikova, S.; Lvov, D.; Robertson, J.; Karlsson, K.-A. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 1997, 233, 224–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matrosovich, M.; Zhou, N.; Kawaoka, Y.; Webster, R. The surface glycoproteins of H5 influenza viruses isolated from humans, chickens, and wild aquatic birds have distinguishable properties. J. Virol. 1999, 73, 1146–1155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Yu, Z.; Liu, L.; Wang, T.; Sun, W.; Wang, C.; Xia, Z.; Gao, Y.; Zhou, B.; Qian, J. Adaptive amino acid substitutions enhance the virulence of a novel human H7N9 influenza virus in mice. Vet. Microbiol. 2016, 187, 8–14. [Google Scholar] [CrossRef]
- Prokopyeva, E.; Sobolev, I.; Prokopyev, M.; Shestopalov, A. Adaptation of influenza A (H1N1) pdm09 virus in experimental mouse models. Infect. Genet. Evol. 2016, 39, 265–271. [Google Scholar] [CrossRef]
- Chen, H.; Bright, R.A.; Subbarao, K.; Smith, C.; Cox, N.J.; Katz, J.M.; Matsuoka, Y. Polygenic virulence factors involved in pathogenesis of 1997 Hong Kong H5N1 influenza viruses in mice. Virus Res. 2007, 128, 159–163. [Google Scholar] [CrossRef]
- Correia, V.; Santos, L.A.; Gíria, M.; Almeida-Santos, M.M.; Rebelo-de-Andrade, H. Influenza A (H1N1) pdm09 resistance and cross-decreased susceptibility to oseltamivir and zanamivir antiviral drugs. J. Med. Virol. 2015, 87, 45–56. [Google Scholar] [CrossRef]
- Koo, B.S.; Kim, H.K.; Song, D.; Na, W.; Song, M.S.; Kwon, J.J.; Wong, S.S.; Noh, J.Y.; Ahn, M.J.; Kim, D.J.; et al. Virological and pathological characterization of an avian H1N1 influenza A virus. Arch. Virol. 2018, 163, 1153–1162. [Google Scholar] [CrossRef]
- Sun, H.; Xiao, Y.; Liu, J.; Wang, D.; Li, F.; Wang, C.; Li, C.; Zhu, J.; Song, J.; Sun, H.; et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl. Acad. Sci. USA 2020, 117, 17204–17210. [Google Scholar] [CrossRef]
- Husain, M. Avian influenza A (H7N9) virus infection in humans: Epidemiology, evolution, and pathogenesis. Infect. Genet. Evol. 2014, 28, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Hatta, M.; Gao, P.; Halfmann, P.; Kawaoka, Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science 2001, 293, 1840–1842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, D.B.; Mukatira, S.; Mehta, P.K.; Obenauer, J.C.; Su, X.; Webster, R.G.; Naeve, C.W. Persistent host markers in pandemic and H5N1 influenza viruses. J. Virol. 2007, 81, 10292–10299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taubenberger, J.K.; Reid, A.H.; Lourens, R.M.; Wang, R.; Jin, G.; Fanning, T.G. Characterization of the 1918 influenza virus polymerase genes. Nature 2005, 437, 889–893. [Google Scholar] [CrossRef] [PubMed]
- El-Shesheny, R.; Kandeil, A.; Bagato, O.; Maatouq, A.M.; Moatasim, Y.; Rubrum, A.; Song, M.S.; Webby, R.J.; Ali, M.A.; Kayali, G. Molecular characterization of avian influenza H5N1 virus in Egypt and the emergence of a novel endemic subclade. J. Gen. Virol. 2014, 95, 1444–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Li, Y.; Hu, Y.; Chang, G.; Sun, W.; Yang, Y.; Kang, X.; Wu, X.; Zhu, Q. PB1-mediated virulence attenuation of H5N1 influenza virus in mice is associated with PB2. J. Gen. Virol. 2011, 92, 1435–1444. [Google Scholar] [CrossRef]
- Li, J.; Ishaq, M.; Prudence, M.; Xi, X.; Hu, T.; Liu, Q.; Guo, D. Single mutation at the amino acid position 627 of PB2 that leads to increased virulence of an H5N1 avian influenza virus during adaptation in mice can be compensated by multiple mutations at other sites of PB2. Virus Res. 2009, 144, 123–129. [Google Scholar] [CrossRef]
- Shaw, M.; Cooper, L.; Xu, X.; Thompson, W.; Krauss, S.; Guan, Y.; Zhou, N.; Klimov, A.; Cox, N.; Webster, R. Molecular changes associated with the transmission of avian influenza a H5N1 and H9N2 viruses to humans. J. Med. Virol. 2002, 66, 107–114. [Google Scholar] [CrossRef]
- Salomon, R.; Franks, J.; Govorkova, E.A.; Ilyushina, N.A.; Yen, H.-L.; Hulse-Post, D.J.; Humberd, J.; Trichet, M.; Rehg, J.E.; Webby, R.J. The polymerase complex genes contribute to the high virulence of the human H5N1 influenza virus isolate A/Vietnam/1203/04. J. Exp. Med. 2006, 203, 689–697. [Google Scholar] [CrossRef] [Green Version]
- Mehle, A.; Doudna, J.A. Adaptive strategies of the influenza virus polymerase for replication in humans. Proc. Natl. Acad. Sci. USA 2009, 106, 21312–21316. [Google Scholar] [CrossRef] [Green Version]
- Yamada, S.; Hatta, M.; Staker, B.L.; Watanabe, S.; Imai, M.; Shinya, K.; Sakai-Tagawa, Y.; Ito, M.; Ozawa, M.; Watanabe, T. Biological and structural characterization of a host-adapting amino acid in influenza virus. PLoS Pathog. 2010, 6, e1001034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foeglein, A.; Loucaides, E.M.; Mura, M.; Wise, H.M.; Barclay, W.S.; Digard, P. Influence of PB2 host-range determinants on the intranuclear mobility of the influenza A virus polymerase. J. Gen. Virol. 2011, 92, 1650. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, G.; Abram, M.; Keiner, B.; Wagner, R.; Klenk, H.-D.; Stech, J. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J. Virol. 2007, 81, 9601–9604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elgendy, E.M.; Arai, Y.; Kawashita, N.; Daidoji, T.; Takagi, T.; Ibrahim, M.S.; Nakaya, T.; Watanabe, Y. Identification of polymerase gene mutations that affect viral replication in H5N1 influenza viruses isolated from pigeons. J. Gen. Virol. 2017, 98, 6–17. [Google Scholar] [CrossRef]
- Hulse-Post, D.; Franks, J.; Boyd, K.; Salomon, R.; Hoffmann, E.; Yen, H.; Webby, R.; Walker, D.; Nguyen, T.; Webster, R. Molecular changes in the polymerase genes (PA and PB1) associated with high pathogenicity of H5N1 influenza virus in mallard ducks. J. Virol. 2007, 81, 8515–8524. [Google Scholar] [CrossRef] [Green Version]
- Govorkova, E.A.; Rehg, J.E.; Krauss, S.; Yen, H.-L.; Guan, Y.; Peiris, M.; Nguyen, T.D.; Hanh, T.H.; Puthavathana, P.; Long, H.T. Lethality to ferrets of H5N1 influenza viruses isolated from humans and poultry in 2004. J. Virol. 2005, 79, 2191–2198. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Sun, H.; Sun, Z.; Sun, Y.; Kong, W.; Pu, J.; Ma, G.; Yin, Y.; Yang, H.; Guo, X. Influenza A virus acquires enhanced pathogenicity and transmissibility after serial passages in swine. J. Virol. 2014, 88, 11981–11994. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Hu, W.-B.; Xu, K.; He, Y.-X.; Wang, T.-Y.; Chen, Z.; Li, T.-X.; Liu, J.-H.; Buchy, P.; Sun, B. Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J. Gen. Virol. 2012, 93, 531–540. [Google Scholar] [CrossRef]
- Conenello, G.M.; Zamarin, D.; Perrone, L.A.; Tumpey, T.; Palese, P. A single mutation in the PB1-F2 of H5N1 (HK/97) and 1918 influenza A viruses contributes to increased virulence. PLoS Pathog. 2007, 3, e141. [Google Scholar] [CrossRef]
- Yamayoshi, S.; Yamada, S.; Fukuyama, S.; Murakami, S.; Zhao, D.; Uraki, R.; Watanabe, T.; Tomita, Y.; Macken, C.; Neumann, G. Virulence-affecting amino acid changes in the PA protein of H7N9 influenza A viruses. J. Virol. 2014, 88, 3127–3134. [Google Scholar] [CrossRef] [Green Version]
- Song, J.; Xu, J.; Shi, J.; Li, Y.; Chen, H. Synergistic effect of S224P and N383D substitutions in the PA of H5N1 avian influenza virus contributes to mammalian adaptation. Sci. Rep. 2015, 5, 10510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeo, S.J.; Than, D.D.; Park, H.S.; Sung, H.W.; Park, H. Molecular Characterization of a Novel Avian Influenza A (H2N9) Strain Isolated from Wild Duck in Korea in 2018. Viruses 2019, 11, 1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, B.W.; Chen, H.; Brownlee, G.G. Correlation between polymerase activity and pathogenicity in two duck H5N1 influenza viruses suggests that the polymerase contributes to pathogenicity. Virology 2010, 401, 96–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mei, K.; Liu, G.; Chen, Z.; Gao, Z.; Zhao, L.; Jin, T.; Yu, X.; Chen, Q. Deep sequencing reveals the viral adaptation process of environment-derived H10N8 in mice. Infect. Genet. Evol. 2016, 37, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Bussey, K.A.; Desmet, E.A.; Mattiacio, J.L.; Hamilton, A.; Bradel-Tretheway, B.; Bussey, H.E.; Kim, B.; Dewhurst, S.; Takimoto, T. PA residues in the 2009 H1N1 pandemic influenza virus enhance avian influenza virus polymerase activity in mammalian cells. J. Virol. 2011, 85, 7020–7028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, G.; Zhang, X.; Gao, W.; Wang, C.; Wang, J.; Sun, H.; Sun, Y.; Guo, L.; Zhang, R.; Chang, K.-C. Prevailing PA mutation K356R in avian influenza H9N2 virus increases mammalian replication and pathogenicity. J. Virol. 2016, 90, 8105–8114. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Hu, Z.; Zhang, X.; Chen, M.; Wang, Z.; Xu, G.; Bi, Y.; Tong, Q.; Wang, M.; Sun, H. An R195K Mutation in the PA-X Protein Increases the Virulence and Transmission of Influenza A Virus in Mammalian Hosts. J. Virol. 2020, 94, e01817-19. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Xiao, H.; Bi, Y.; Zhang, W.; Xu, Y.; Wang, F.; Shi, Y.; Gao, G.F. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep. 2017, 20, 1201–1214. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Wu, H.; Peng, X.; Wu, X.; Cheng, L.; Liu, F.; Ji, S.; Wu, N. Amino acid substitutions occurring during adaptation of an emergent H5N6 avian influenza virus to mammals. Arch. Virol. 2016, 161, 1665–1670. [Google Scholar] [CrossRef]
- Lee, M.; Deng, M.; Lin, Y.; Chang, C.; Shieh, H.K.; Shiau, J.; Huang, C. Characterization of an H5N1 avian influenza virus from Taiwan. Vet. Microbiol. 2007, 124, 193–201. [Google Scholar] [CrossRef]
- Katz, J.M.; Lu, X.; Tumpey, T.M.; Smith, C.B.; Shaw, M.W.; Subbarao, K. Molecular correlates of influenza A H5N1 virus pathogenesis in mice. J. Virol. 2000, 74, 10807–10810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeVries, A.; Wotton, J.; Lees, C.; Boxrud, D.; Uyeki, T.; Lynfield, R. Neuraminidase H275Y and hemagglutinin D222G mutations in a fatal case of 2009 pandemic influenza A (H1N1) virus infection. Influenza Other Respir. Viruses 2012, 6, e85–e88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karthick, V.; Ramanathan, K. Insight into the oseltamivir resistance R292K mutation in H5N1 influenza virus: A molecular docking and molecular dynamics approach. Cell Biochem. Biophys. 2014, 68, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Kiso, M.; Ozawa, M.; Imai, H.; Takahashi, K.; Kakugawa, S.; Noda, T.; Horimoto, T.; Kawaoka, Y. Effect of an asparagine-to-serine mutation at position 294 in neuraminidase on the pathogenicity of highly pathogenic H5N1 influenza A virus. J. Virol. 2011, 85, 4667–4672. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Sun, W.; Li, X.; Chen, Q.; Chai, H.; Gao, X.; Guo, J.; Zhang, K.; Wang, T.; Feng, N. Adaptive amino acid substitutions enhance the virulence of a reassortant H7N1 avian influenza virus isolated from wild waterfowl in mice. Virology 2015, 476, 233–239. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Zou, X.; Zhou, J.; Tang, J.; Shu, Y. Residues 41V and/or 210D in the NP protein enhance polymerase activities and potential replication of novel influenza (H7N9) viruses at low temperature. Virol. J. 2015, 12, 71. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Chen, H.; Huang, J.; Chen, Y.; Gu, M.; Wang, X.; Hu, S.; Liu, X.; Liu, X. A nonpathogenic duck-origin H9N2 influenza A virus adapts to high pathogenicity in mice. Arch. Virol. 2014, 159, 2243–2252. [Google Scholar] [CrossRef]
- Li, J.; Zheng, W.; Hou, L.; Chen, C.; Fan, W.; Qu, H.; Jiang, J.; Liu, J.; Gao, G.F.; Zhou, J. Differential nucleocytoplasmic shuttling of the nucleoprotein of influenza a viruses and association with host tropism. Cell. Microbiol. 2017, 19, e12692. [Google Scholar] [CrossRef] [Green Version]
- Fan, S.; Deng, G.; Song, J.; Tian, G.; Suo, Y.; Jiang, Y.; Guan, Y.; Bu, Z.; Kawaoka, Y.; Chen, H. Two amino acid residues in the matrix protein M1 contribute to the virulence difference of H5N1 avian influenza viruses in mice. Virology 2009, 384, 28–32. [Google Scholar] [CrossRef] [Green Version]
- Center for Disease Control and Prevention. Update: Drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. Mmwr. Morb. Mortal. Wkly. Rep. 2009, 58, 433. [Google Scholar] [PubMed]
- Lycett, S.; Ward, M.; Lewis, F.; Poon, A.; Pond, S.K.; Brown, A.L. Detection of mammalian virulence determinants in highly pathogenic avian influenza H5N1 viruses: Multivariate analysis of published data. J. Virol. 2009, 83, 9901–9910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanrai, P.; Mostafa, A.; Madhugiri, R.; Lechner, M.; Wilk, E.; Schughart, K.; Ylösmäki, L.; Saksela, K.; Ziebuhr, J.; Pleschka, S. Identification of specific residues in avian influenza A virus NS1 that enhance viral replication and pathogenicity in mammalian systems. J. Gen. Virol. 2016, 97, 2135–2148. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Jiao, P.; Wang, A.; Zhao, F.; Tian, G.; Wang, X.; Yu, K.; Bu, Z.; Chen, H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 2006, 80, 11115–11123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thangavel, R.R.; Bouvier, N.M. Animal models for influenza virus pathogenesis, transmission, and immunology. J. Immunol. Methods 2014, 410, 60–79. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Reeves, A.B.; Ramey, A.M.; Poulson, R.L.; Stallknecht, D.E. The Genome Sequence of an H6N5 Influenza A Virus Strain Isolated from a Northern Pintail (Anas acuta) Sampled in Alaska, USA, Shares High Identity with That of a South Korean Wild Bird Strain. Microbiol. Resour. Announc. 2020, 9, 33. [Google Scholar] [CrossRef]
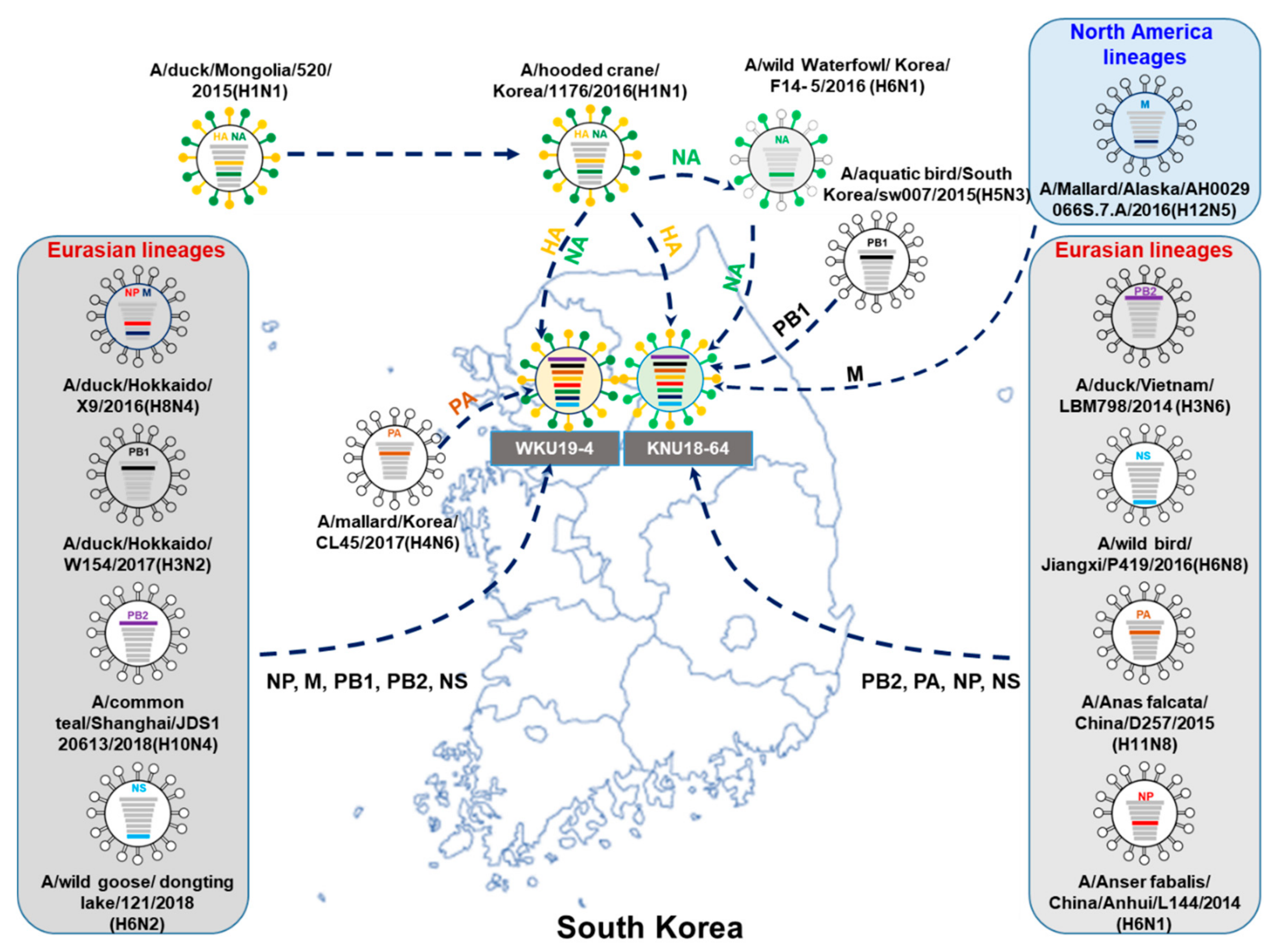
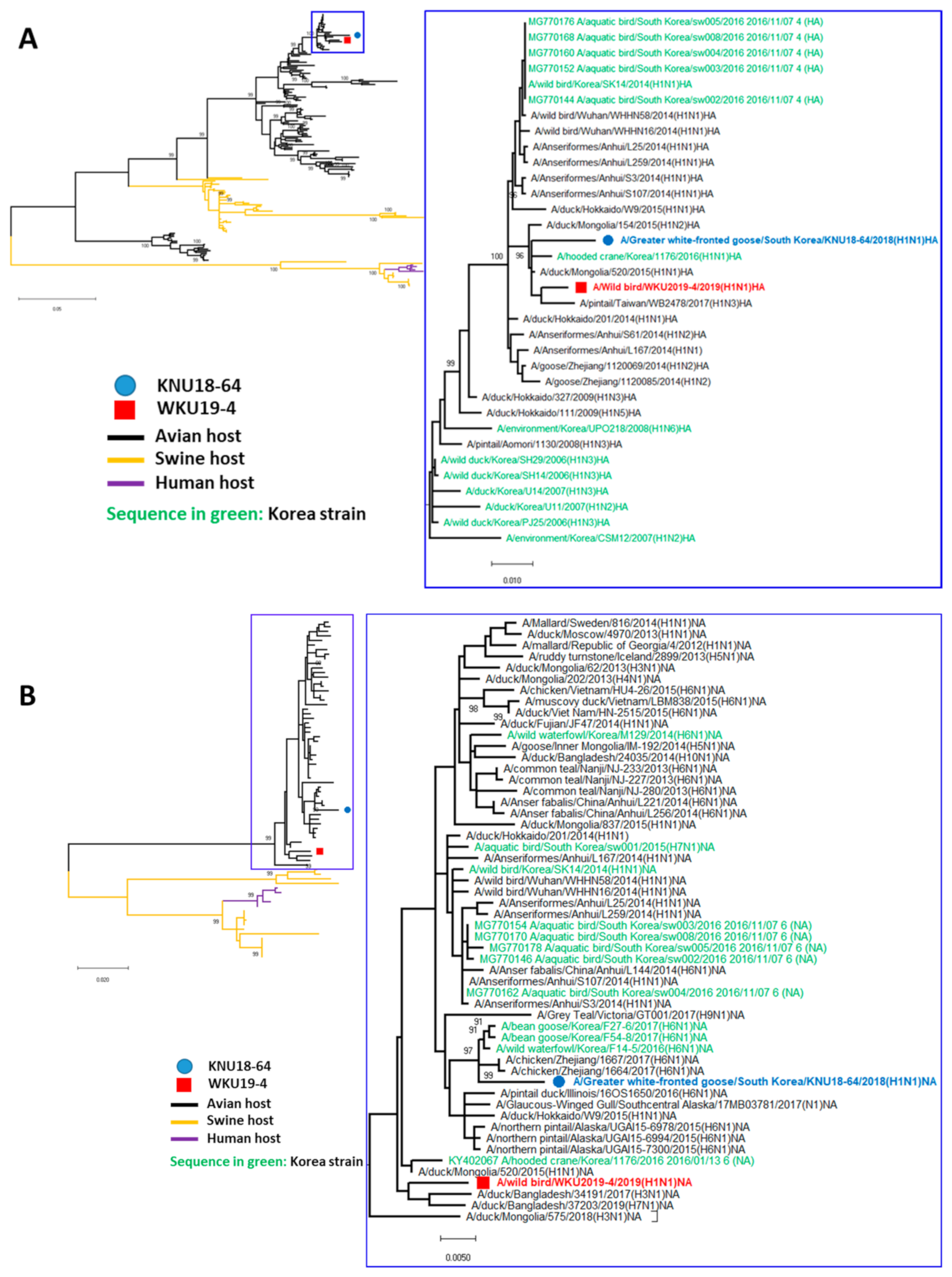
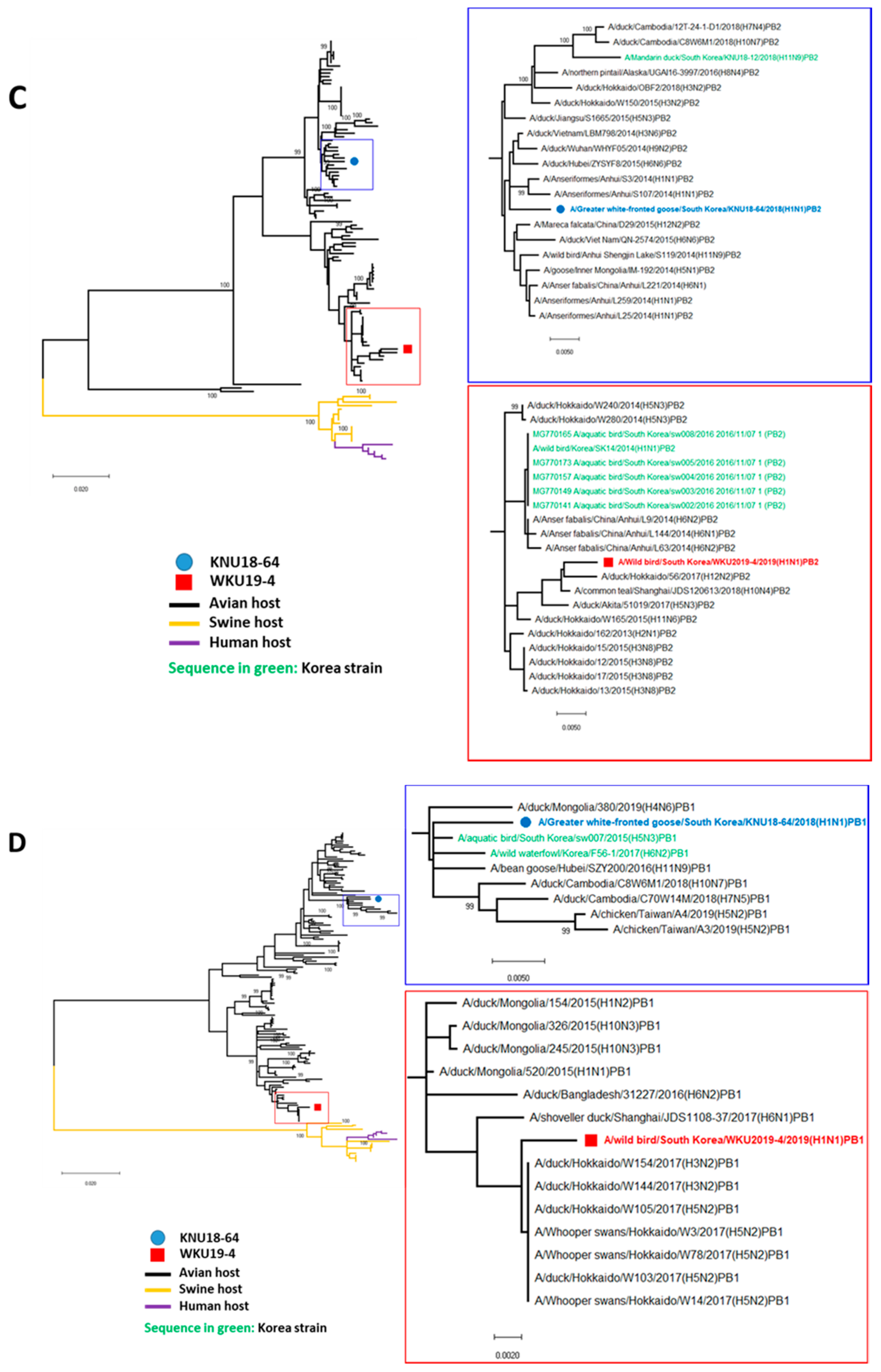
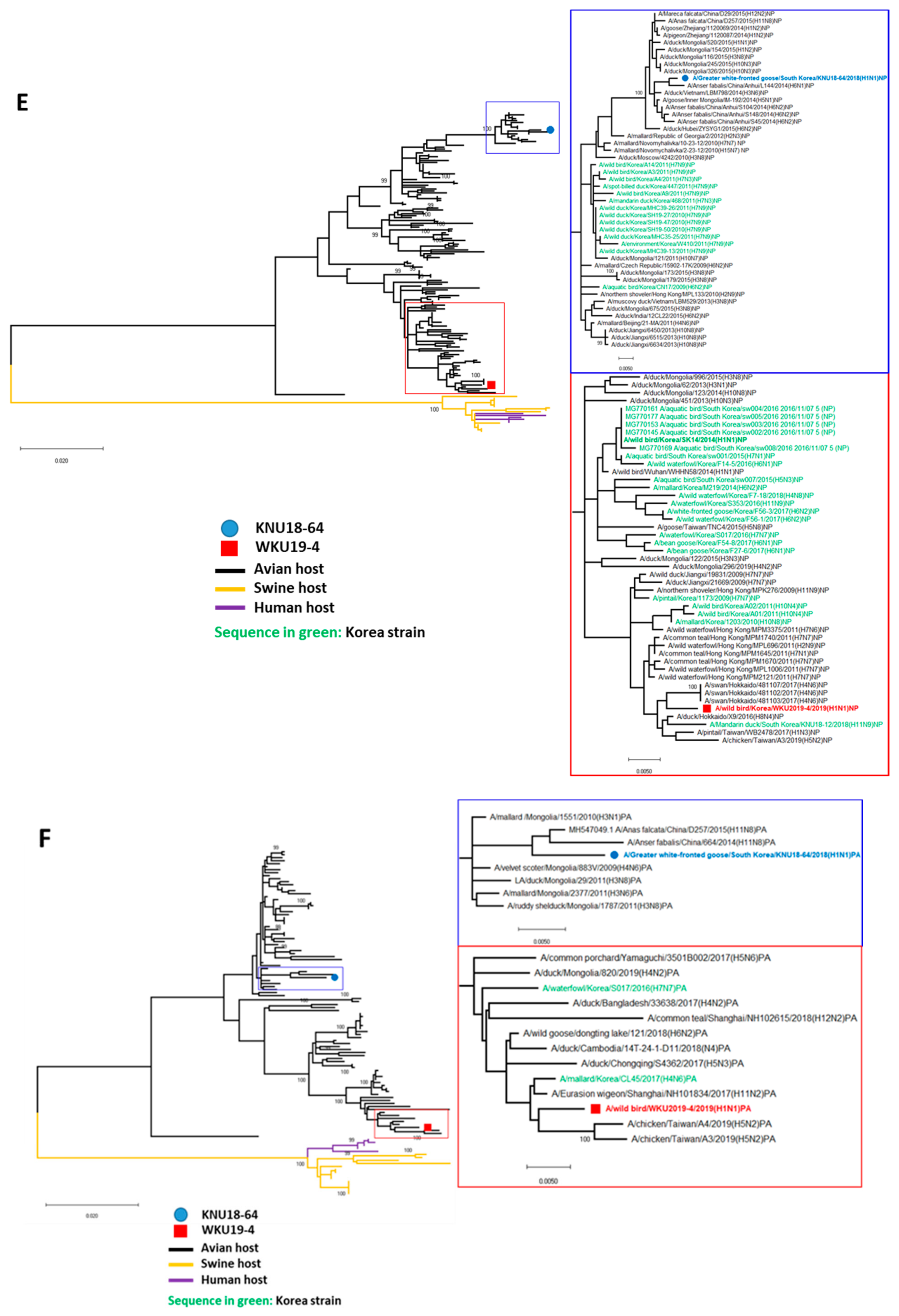
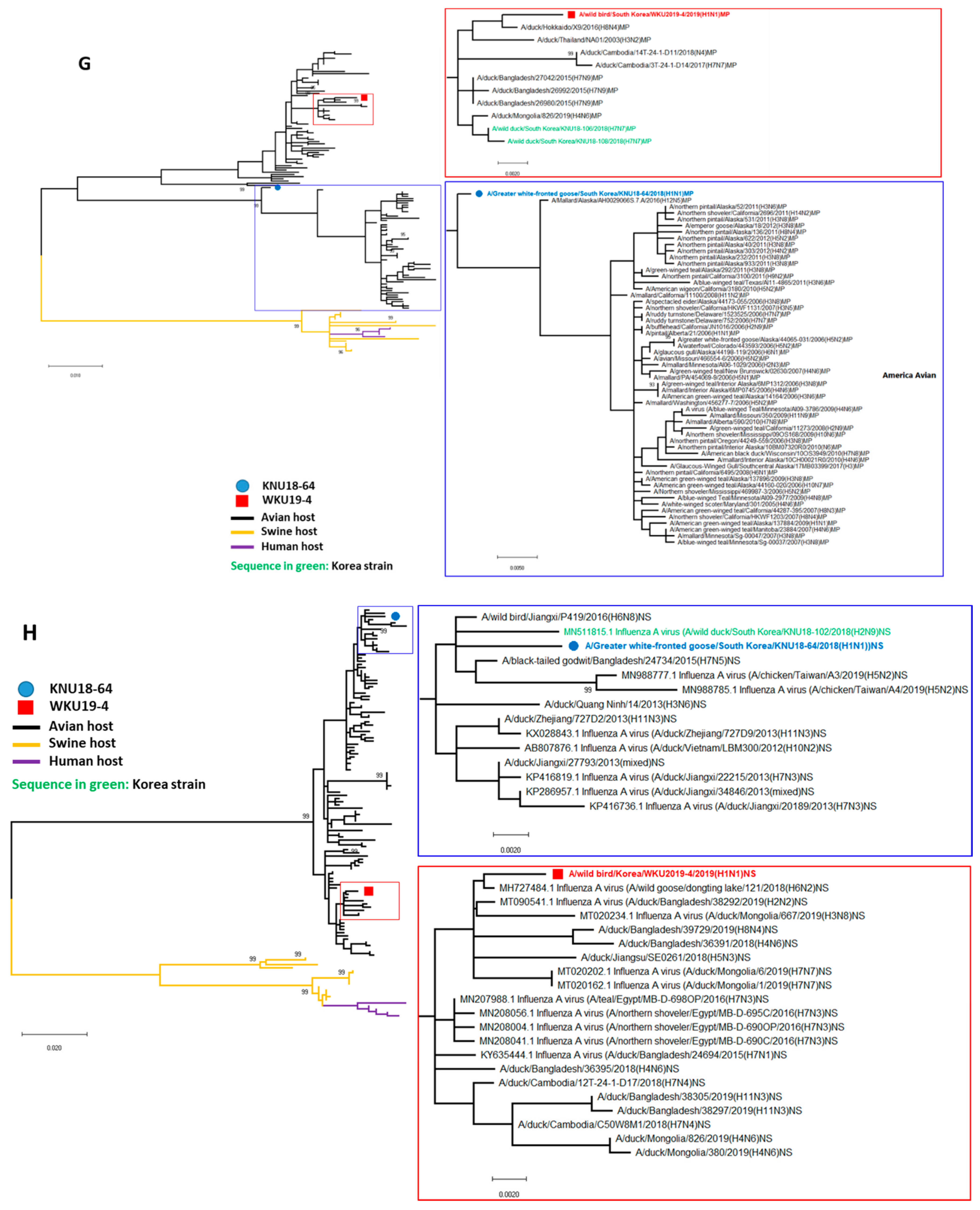
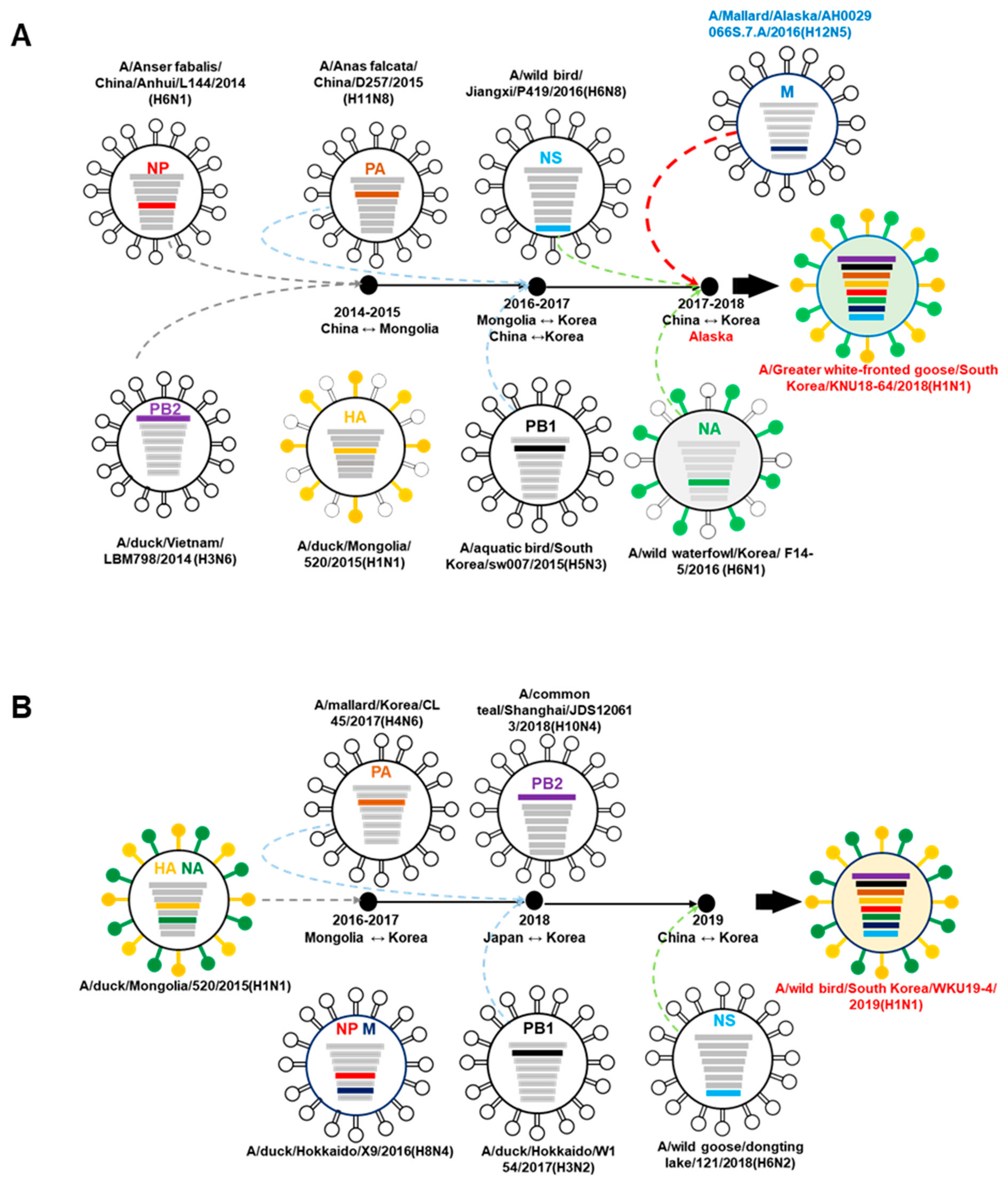



| A/Greater White-Fronted Goose/South Korea/KNU18-64/2018(H1N1) | A/Wildbird/South Korea/WKU19-4/2019(H1N1) | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene Segment | Genebank ID | Reference Strain Accession ID | Highest Similarly Strain Reference | Per. Ident (%) | Genebank ID | Reference Strain Accession ID | Highest Similarly Strain Reference | Per. Ident (%) |
| PB2 | MN584878.1 | LC053481.1 | A/duck/Vietnam/LBM798/2014(H3N6) | 99.08(2280/2280) | MT821115.1 | MN049531.1 | A/common teal/Shanghai/JDS120613/2018(H10N4) | 99.259(2259/2280) |
| MN171439.1 | A/duck/Jiangsu/S1665/2015(H5N3) | 98.86(2293/2280) | MK592490.1 | A/duck/Hokkaido/56/2017(H12N2) | 98.86(2280/2280) | |||
| KU881717.1 | A/Anseriformes/Anhui/L259/2014(H1N1) | 98.86(2280/2280) | MK592458.1 | A/duck/Akita/51019/2017(H5N3) | 98.64(2280/2280) | |||
| PB1, PB1F2 | MN584879.1 | MG386194.1 | A/aquatic bird/South Korea/sw007/2015(H5N3) | 99.08(2274/2274) | MT821116.1 | MK592547.1 | A/duck/Hokkaido/W154/2017(H3N2) | 99.56(2274/2274) |
| MH130136.1 | A/wild waterfowl/Korea/F56-1/2017(H6N2) | 98.77(2274/2274) | MK592531.1 | A/duck/Hokkaido/W144/2017(H3N2) | 99.56(2274/2274) | |||
| KX121186.1 | A/bean goose/Hubei/SZY200/2016(H11N9) | 98.77(2274/2274) | MK592515.1 | A/duck/Hokkaido/W105/2017(H5N2) | 99.56(2274/2274) | |||
| PA | MN584880.1 | MH547049.1 | A/Anas falcata/China/D257/2015(H11N8) | 98.93(2151/2151) | MT821117.1 | MH579404.1 | A/mallard/Korea/CL45/2017(H4N6) | 99.12(2151/2151) |
| MK943289.1 | A/duck/Viet Nam/HN-2525/2015(mixed) | 98.47(2151/2151) | MH727479.1 | A/wild goose/dongting lake/121/2018(H6N2) | 98.98(2151/2151) | |||
| KF454809.1 | A/mallard/Mongolia/1551/2010(H3N1) | 98.47(2151/2151) | MN703036.1 | A/duck/Cambodia/10T-24-1-D14/2018(mixed) | 98.31(2151/2151) | |||
| HA | MN584881.1 | LC121396.1 | A/duck/Mongolia/520/2015(H1N1) | 98.53(1726/1701) | MT821118.1 | LC121396.1 | A/duck/Mongolia/520/2015(H1N1) | 99.06(1701/1701) |
| KY402065.1 | A/hooded crane/Korea/1176/2016(H1N1) | 98.12(1701/1701) | LC121276.1 | A/duck/Mongolia/154/2015(H1N2) | 98.88(1701/1701) | |||
| LC121276.1 | A/duck/Mongolia/154/2015(H1N2) | 98.12(1732/1701) | KY402065.1 | A/hooded crane/Korea/1176/2016(H1N1) | 98.65(1701/1701) | |||
| NP | MN584882.1 | KT717283.1 | A/Anser fabalis/China/Anhui/L144/2014(H6N1) | 99.53(1497/1497) | MT821119.1 | MK978905.1 | A/duck/Hokkaido/X9/2016(H8N4) | 99.2(1497/1497) |
| KR010412.1 | A/goose/Inner Mongolia/IM-192/2014(H5N1) | 99.4(1497/1497) | KF259820.1 | A/common teal/Hong Kong/MPM1670/2011(H7N7) | 99.13(1497/1497) | |||
| KT717243.1 | A/Anser fabalis/China/Anhui/S104/2014(H6N2) | 99.33(1497/1497) | KF259825.1 | A/wild waterfowl/Hong Kong/MPL1006/2011(H7N7) | 99.06(1497/1497) | |||
| NA | MN584883.1 | MH130116.1 | A/wild waterfowl/Korea/F14-5/2016(H6N1) | 98.94(1429/1410) | MT821120.1 | LC121398.1 | A/duck/Mongolia/520/2015(H1N1) | 98.94(1410/1410) |
| MH130132.1 | A/bean goose/Korea/F54-8/2017(H6N1) | 98.87(1424/1410) | MH791653.1 | A/duck/Bangladesh/34191/2017(H3N1) | 98.58(1410/1410) | |||
| MH130124.1 | A/bean goose/Korea/F27-6/2017(H6N1) | 98.87(1410/1410) | KY402067.1 | A/hooded crane/Korea/1176/2016(H1N1) | 98.51(1410/1410) | |||
| M2, M1 | MN584884.1 | MN253720.1 | A/Mallard/Alaska/AH0029066S.7.A/2016(H12N5) | 98.68(1007/982) | MT821121.1 | MK978907.1 | A/duck/Hokkaido/X9/2016(H8N4) | 99.49(982/982) |
| KY130619.1 | A/green-winged teal/Alaska/292/2011(H3N8) | 98.07(995/982) | KY635666.1 | A/duck/Bangladesh/26980/2015(H7N9) | 99.19(982/982) | |||
| CY120628.1 | A/American wigeon/California/3180/2010(H5N2) | 98.07(995/982) | KY635509.1 | A/duck/Bangladesh/27042/2015(H7N9) | 99.18(982/982) | |||
| NEP, NS1 | MN584885.1 | KX867861.1 | A/wild bird/Jiangxi/P419/2016(H6N8) | 99.16(864/838) | MT821122.1 | MN483241.1 | A/White-fronted Goose/South Korea/KNU18-119/2018(H7N7) | 99.64(855/838) |
| KY635798.1 | A/black-tailed godwit/Bangladesh/24734/2015(H7N5) | 99.05(849/838) | MH727484.1 | A/wild goose/dongting lake/121/2018(H6N2) | 99.64(838/838) | |||
| KT266946.1 | A/duck/Guangxi/113/2012(H6N8) | 99.28(864/838) | MT090541.1 | A/duck/Bangladesh/38292/2019(H2N2) | 99.52(852/838) | |||
| Virus Strain | HA Receptor-Binding Residues (H5 Numbering) | NA | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cleavage Sites | D/E94N | I116M | S121N | A134V | G139R | S142G | D221G/N | Q222L | Deleted Range from 50–70 | M26I | I106V | T223I | K/S373A/N | G394D | |
| A/wild bird/South Korea/WKU2019-4/2019(H1N1) | PSIQRS↓GLF | E | I | S | A | G | S | G | Q | No deletion | I | I | I | N | G |
| A/Greater white-fronted goose/South Korea/KNU18-64/2018 | PSIQRS↓GLF | E | I | S | A | G | S | G | Q | No deletion | I | I | I | N | G |
| A/wild bird/Korea/SK14/2014 a | PSIQRS↓GLF | E | I | S | A | G | S | G | Q | No deletion | I | I | I | N | G |
| A/duck/Mongolia/520/2015 b | PSIQRS↓GLF | E | I | S | A | G | S | G | Q | No deletion | I | I | I | N | G |
| A/California/04/2009 c | PSIQRS↓GLF | D | I | S | A | G | S | D | Q | No deletion | I | V | I | N | G |
| A/swine/Shandong/1207/2016 d | PSIQRS↓GLF | E | I | S | A | G | S | E | Q | No deletion | I | V | I | N | G |
| Viral Protein | Amino Acid | W4 | K64 | SK14 | M520 | CA/04/09 | SW/1207 | Comments | Reference |
|---|---|---|---|---|---|---|---|---|---|
| PB2 | E627K | E | E | E | E | E | E | Mammalian host adaptation, Human host marker, Enhanced polymerase activity, Increased virulence in mammals | [46,47,48,49,50,51] |
| T63I (with PB1 M677T) | I | I | I | I | I | I | Enhanced polymerase activity, Increased virulence in mice | [51] | |
| L89V | V | V | V | V | V | V | Enhanced polymerase activity, Increased virulence in mice | [52] | |
| K251R | R | R | R | R | R | R | Increased virulence in mice | [41] | |
| T271A | T | T | T | T | A | A | Human host marker (host-specific polymerase activity) | [53] | |
| G309D | D | D | D | D | D | D | Enhanced polymerase activity, Increased virulence in mice | [52] | |
| T339K | K | K | K | K | K | K | Enhanced polymerase activity, Increased virulence in mice | [52] | |
| R/Q355K | R | R | R | R | R | K | Increased virulence in mammals | [42] | |
| Q368R | R | R | R | R | R | R | Increased polymerase activity, Increased virulence in mammals | [54] | |
| H447Q | Q | Q | Q | Q | Q | Q | Increased polymerase activity, Increased virulence in mammals | [54] | |
| R477G | G | G | G | G | G | G | Enhanced polymerase activity, Increased virulence in mice | [52] | |
| I495V | V | V | V | V | V | V | Enhanced polymerase activity, Increased virulence in mice | [52] | |
| A588I/V | A | A | A | A | T | I | Human host marker (host-specific polymerase activity) | [53] | |
| GQ590/591SR/K | GQ | GQ | GQ | GQ | SR | SR | Increased polymerase activity, Human host adaptation | [55] | |
| Q591K | Q | Q | Q | Q | R | R | Increased virulence in mammals | [56] | |
| A/S674T | A | A | A | A | A | A | Human host marker | [22] | |
| A676T | T | T | T | T | T | T | Enhanced polymerase active, Increased virulence in mice | [52] | |
| D701N | D | D | D | D | D | D | Increased polymerase activity, Increased virulence in mammals, Mammalian host marker | [55,57,58] | |
| K702R | K | K | K | K | K | K | Human host marker | [48,49,57] | |
| PB1 | D3V | V | V | V | V | V | V | Increased polymerase activity, Increased virulence in mammals | [59] |
| L13P | P | P | P | P | P | P | Increased polymerase activity, Increased virulence in mammals, Mammalian host marker | [58] | |
| R207K | K | K | K | K | K | K | Increased polymerase activity in mammalian cells | [60] | |
| K328N | N | N | N | N | N | N | Increased polymerase activity, Increased virulence in mammals | [61] | |
| S375N | N | N | N | N | N | N | Increased polymerase activity, Increased virulence in mammals, Human host marker | [49] | |
| H436Y | Y | Y | Y | Y | Y | Y | Increased polymerase activity and virulence in mallards, ferrets and mice | [60] | |
| A469T | T | T | T | T | T | T | Conferred in contact transmissibility in guinea pigs. | [62] | |
| L473V | V | V | V | V | V | V | Increased polymerase activity and replication efficiency | [63] | |
| V652A | A | A | A | A | A | A | Increased virulence in mice | [41] | |
| M677T | T | T | T | T | T | T | Pathogenic in mice | [51] | |
| PB1-F2 | N66S | S | N | N | S | - | - | Increased virulence in mammals | [64] |
| PA | S37A | A | A | A | A | A | A | Significantly increased viral growth and polymerase activity in mammalian cells | [65] |
| S224P (with N383D) | S | S | S | S | P | S | Enhanced the pathogenicity and viral replication of H5N1 virus in mice | [66] | |
| H266R | R | R | R | R | R | R | Increased polymerase activity. Increased virulence in mammals and birds | [67,68] | |
| F277S | F | S | S | S | H | H | Contributed to the virulence and mammalian adaptation | [67] | |
| C278Q | Q | Q | Q | Q | Q | Q | Adapt to mammalian hosts | [69] | |
| N321K | I * | N | N | N | N | V | Increased polymerase activity | [37] | |
| K328R | K | R | K | K | K | K | Increased virulence in mice | [40] | |
| L336M | L | L | L | L | M | M | Increased virulence in mammals | [70] | |
| K356R | K | K | K | K | R | R | Increased virulence in mammals and mice | [71] | |
| N383D (with S224P) | D | D | D | D | D | D | Enhanced the pathogenicity and viral replication of H5N1 virus in mice | [66] | |
| S409N | S | S | S | S | N | N | Enhanced Transmission, Human host marker | [70] | |
| S/A515T | T | T | T | T | T | T | Increased polymerase activity. Increased virulence in mammals and birds | [60] | |
| PA-X | R195K | R | R | R | R | - | K | Increased virulence in mammals | [72] |
| HA | Cleavage site | PSIQRS↓GLF | PSIQRS↓GLF | PSIQRS↓GLF | PSIQRS↓GLF | PSIQRS↓GLF | PSIQRS↓GLF | [39] | |
| D/E94N | E | E | E | E | D | E | Increased virus binding to 2,6, Enhanced virus fusion | [33] | |
| I116M | I | I | I | I | I | I | Potential to alter the virulence of H1N1pdm09 in swine | [34] | |
| S121N | S | S | S | S | S | S | Increased virus binding to 2,6, Increased replication in mammals | [35] | |
| A134V | A | A | A | A | A | A | Increased virus binding to 2,6 | [36] | |
| G139R | G | G | G | G | G | G | Increased virus binding to 2,6 | [37] | |
| S142G | S | S | S | S | S | S | Potential to alter the virulence of H1N1pdm09 in swine | [34] | |
| D221G/N | G | G | G | G | D | E | Change in receptor binding | [35] | |
| Q222L | Q | Q | Q | Q | Q | Q | Increased virus binding to 2,6 | [73] | |
| NA | M26I | I | I | I | I | I | I | Increased virulence in mice | [40] |
| DELETED Range from aa 50–70 | NO DELETION | NO DELETION | NO DELETION | NO DELETION | NO DELETION | NO DELETION | the adaptation of influenza viruses from wild aquatic birds to domestic chickens | [39] | |
| I106V | I | I | I | I | V | V | Increased virulence in mice | [41] | |
| R143K | K | K | K | K | K | K | Increased virulence in mammals and mice | [74] | |
| T223I | I | I | I | I | I | I | Increased virulence in mammals | [42,75,76] | |
| V241I | V | V | V | V | V | V | Contributed to the significantly higher baseline IC50 value obtained to oseltamivir for 2010/2011 viruses | [43] | |
| N248D | N | N | N | N | N | N | Increased virulence in mice | [41] | |
| H275Y | H | H | H | H | H | H | Increased virulence in mammals | [77] | |
| R293K | R | R | R | R | R | R | Highly resistant to the oseltamivir | [78] | |
| S354N | N | N | N | N | N | N | Increased virulence in mice | [79] | |
| K/S373A/N | N | N | N | N | N | N | Increased virulence in mammals | [80] | |
| NP | V41I | I | I | I | I | I | I | Might contribute to viral transmissibility | [81] |
| V105M | M | M | M | M | M | M | Contribute to the increased virulence of the H9N2 | [82] | |
| D210E | E | E | E | E | E | E | Might contribute to viral transmissibility | [81] | |
| F253I | I | I | I | I | I | I | Results in attenuated pathogenicity of the virus in mice | [83] | |
| R305K | R | R | R | R | K | K | Human host marker | [48] | |
| F313V | F | F | F | F | V | V | Human host marker | [53] | |
| I353V | I | I | I | I | V | V | Increased virulence in mice | [41] | |
| Q357L/K (WITH PB2 I27K) | Q | Q | Q | Q | K | K | Increased virulence in mammals, Human host marker | [48] | |
| M1 | V15I/T | V | V | V | V | I | I | Increased virulence in mammals | [42] |
| N30D | D | D | D | D | S | S | Increased virulence in mammals | [84] | |
| A166V | V | V | V | V | A | A | Contribute to the increased virulence of the H9N2. | [82] | |
| M2 | S31N L55F | S L | S L | S L | S L | N F | N F | resistance to the adamantane antivirals and are sensitive to oseltamivir and zanamivir Enhanced Transmission | [53] [85] |
| NS1 | A/P42S | S | S | S | S | S | S | Increased virulence in mammals, Antagonism of IFNinduction | [86] |
| D74N | D | D | D | D | S | S | Increased virulence in mammals | [87] | |
| T/D/V/R/A127N | N | N | N | N | N | N | Increased virulence in mammals | [86] | |
| V149A | A | A | A | A | A | A | Pathogenicity in mice, Antagonism of IFN induction | [88] | |
| N/G205R/K (with NEP/NS2 M51I, T48N; PB2 A469T) | S | S | S | S | N | K | Decreased IFN antagonism, Conferred enhanced in-contact transmissibility in guinea pigs | [62] | |
| NEP/ NS2 | M31I | M | M | M | M | M | I | Increased virulence in mammals | [86] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trinh, T.-T.T.; Duong, B.T.; Nguyen, A.T.V.; Tuong, H.T.; Hoang, V.T.; Than, D.D.; Nam, S.; Sung, H.W.; Yun, K.-J.; Yeo, S.-J.; et al. Emergence of Novel Reassortant H1N1 Avian Influenza Viruses in Korean Wild Ducks in 2018 and 2019. Viruses 2021, 13, 30. https://doi.org/10.3390/v13010030
Trinh T-TT, Duong BT, Nguyen ATV, Tuong HT, Hoang VT, Than DD, Nam S, Sung HW, Yun K-J, Yeo S-J, et al. Emergence of Novel Reassortant H1N1 Avian Influenza Viruses in Korean Wild Ducks in 2018 and 2019. Viruses. 2021; 13(1):30. https://doi.org/10.3390/v13010030
Chicago/Turabian StyleTrinh, Thuy-Tien Thi, Bao Tuan Duong, Anh Thi Viet Nguyen, Hien Thi Tuong, Vui Thi Hoang, Duong Duc Than, SunJeong Nam, Haan Woo Sung, Ki-Jung Yun, Seon-Ju Yeo, and et al. 2021. "Emergence of Novel Reassortant H1N1 Avian Influenza Viruses in Korean Wild Ducks in 2018 and 2019" Viruses 13, no. 1: 30. https://doi.org/10.3390/v13010030
APA StyleTrinh, T.-T. T., Duong, B. T., Nguyen, A. T. V., Tuong, H. T., Hoang, V. T., Than, D. D., Nam, S., Sung, H. W., Yun, K.-J., Yeo, S.-J., & Park, H. (2021). Emergence of Novel Reassortant H1N1 Avian Influenza Viruses in Korean Wild Ducks in 2018 and 2019. Viruses, 13(1), 30. https://doi.org/10.3390/v13010030






