In Vitro Infection with Hepatitis B Virus Using Differentiated Human Serum Culture of Huh7.5-NTCP Cells without Requiring Dimethyl Sulfoxide
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Lentiviral Vectors Expressing NTCP
2.2. Establishment of Stable NTCP-Expressing Huh7.5 Cell Line (Huh7.5-NTCP)
2.3. Conventional Culture of Huh7.5 Cells and Huh7.5-NTCP Cells
2.4. Differentiation of Huh7.5 and Huh7.5-NTCP Cells in Human Serum
2.5. Infection of Cells with HBV
2.6. Culture of PXB Cells
2.7. Analysis of Pregenomic RNA (pgRNA)
2.8. Analysis of Covalently Closed Circular DNA (cccDNA)
2.9. Enzyme-Linked Immunosorbent Assays (ELISAs)
2.10. Aptamer-Binding Assay for the E Antigen (HBeAg)
2.11. Immunofluorescence Staining of Huh7.5 Cells Overexpressing NTCP
2.12. Flow Cytometry Analysis of NTCP Expression
2.13. Western Blotting
2.14. Nanoluciferase Reporter Luminescence Assay
2.15. Statistical Analysis
3. Results
3.1. Huh7.5 Cell Line Overexpressing NTCP (Huh7.5-NTCP Cells)
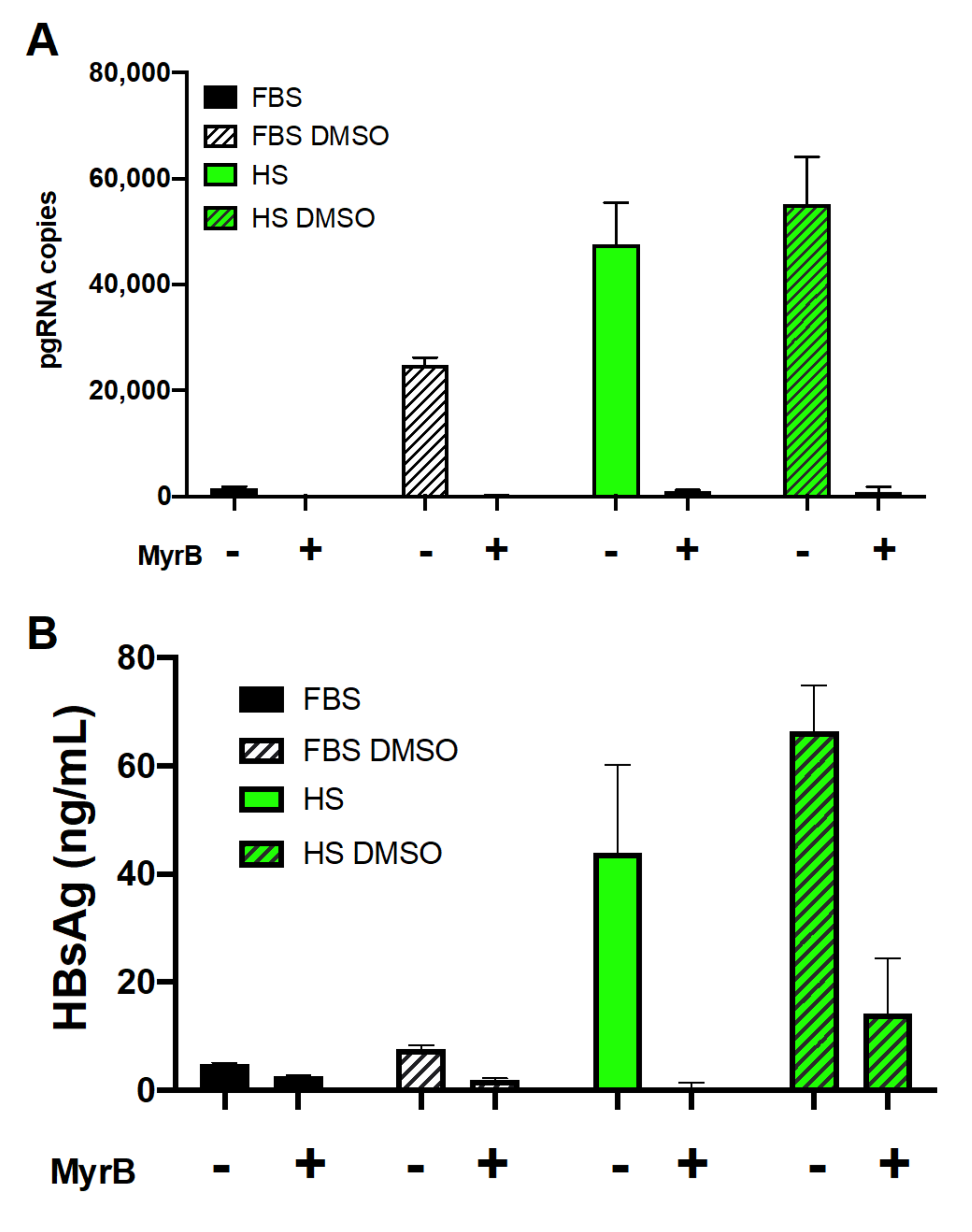
3.2. Human Serum Culture Enhanced Productive HBV Infection in Huh7.5-NTCP Cells
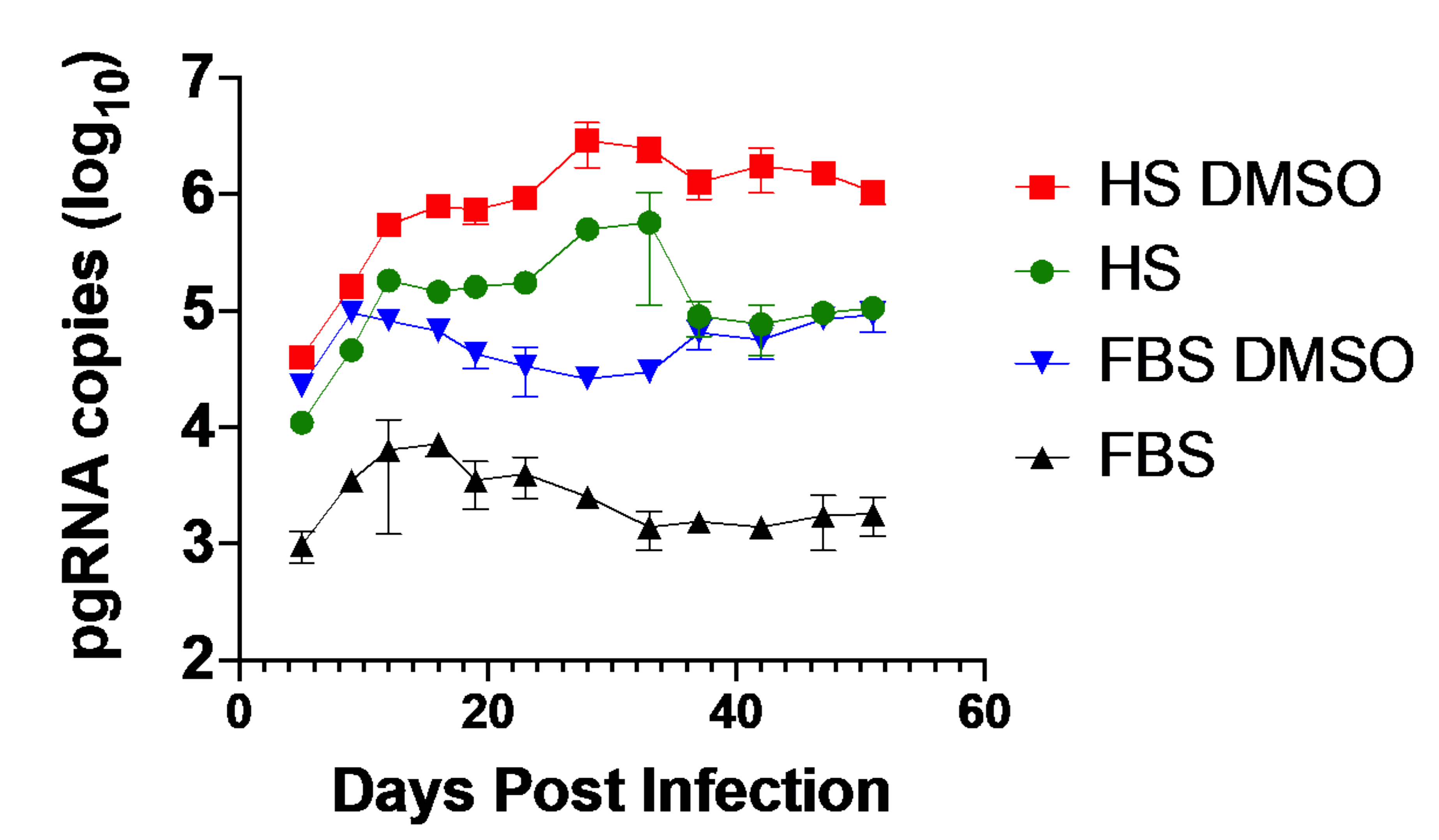
3.3. Huh7.5-NTCP Cells in a Human Serum Culture Serve as a Model for Long-Term HBV Infection
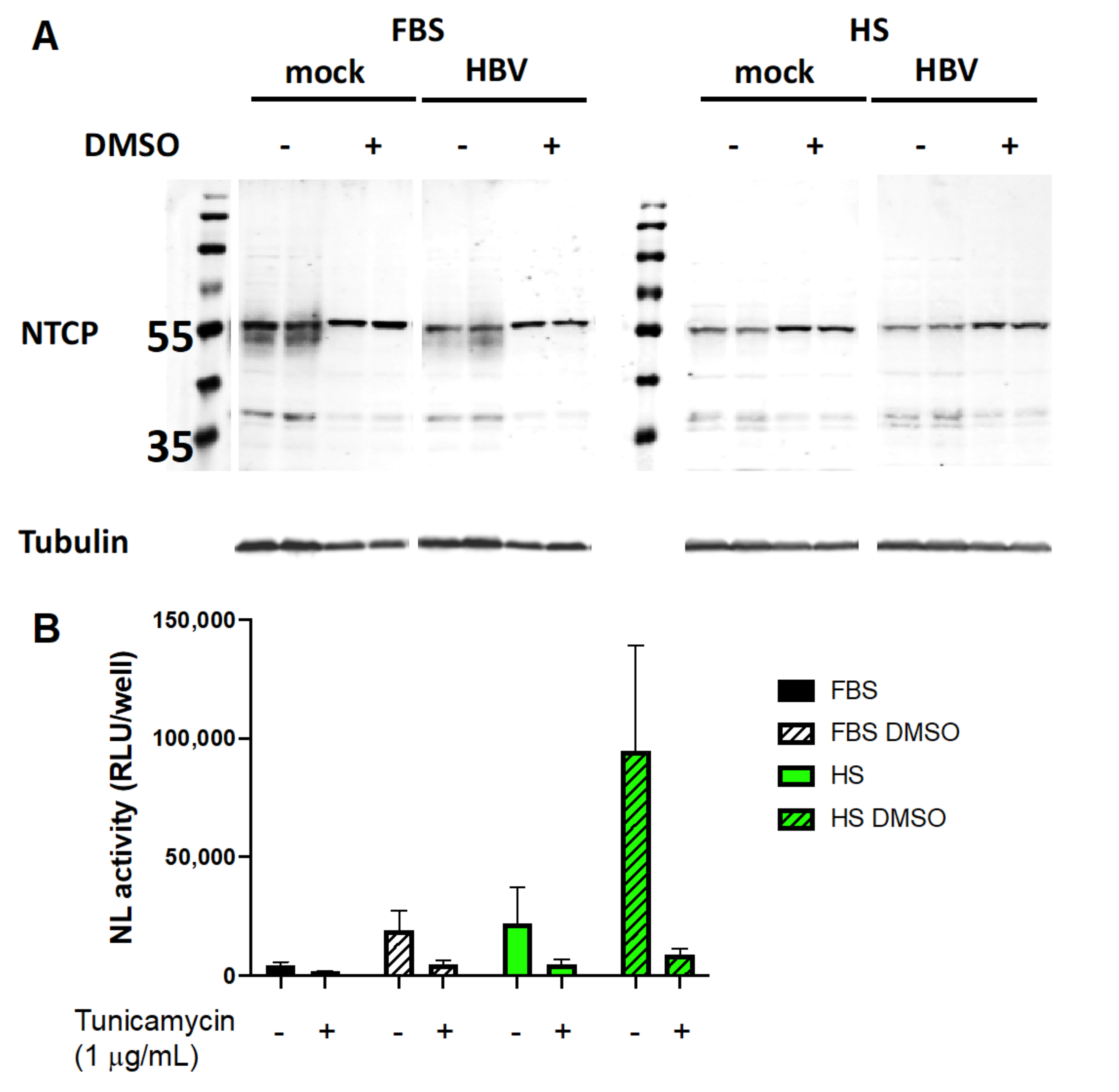
3.4. Human Serum Alters Hepatocyte Differentiation Markers in Huh7.5-NTCP
3.5. Involvement of NTCP and Possible Effect of Its N-Glycosylation on Viral Entry
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ott, J.; Stevens, G.; Groeger, J.; Wiersma, S. Global epidemiology of hepatitis B virus infection: New estimates of age-specific HBsAg seroprevalence and endemicity. Vaccine 2012, 30, 2212–2219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- Razavi-Shearer, D.; Gamkrelidze, I.; Nguyen, M.H.; Chen, D.S.; Van Damme, P.; Abbas, Z.; Abdulla, M.; Rached, A.A.; Adda, D.; Aho, I.; et al. Global prevalence, treatment, and prevention of hepatitis B virus infection in 2016: A modelling study. Lancet Gastroenterol. Hepatol. 2018, 3, 383–403. [Google Scholar] [CrossRef]
- Tang, L.S.Y.; Covert, E.; Wilson, E.; Kottilil, S. Chronic hepatitis B infection. JAMA 2018, 319, 1802–1813. [Google Scholar] [CrossRef]
- Yuen, M.F.; Chen, D.S.; Dusheiko, G.M.; Janssen, H.L.A.; Lau, D.T.Y.; Locarnini, S.A.; Peters, M.G.; Lai, C.L. Hepatitis B virus infection. Nat. Rev. Dis. Prim. 2018, 4, 18035. [Google Scholar] [CrossRef]
- Hall, S.; Howell, J.; Visvanathan, K.; Thompson, A.J. The Yin and the Yang of treatment for chronic hepatitis B—When to start, when to stop nucleos(t)ide analogue therapy. Viruses 2020, 12, 934. [Google Scholar] [CrossRef]
- De Clercq, E.; Férir, G.; Kaptein, S.J.F.; Neyts, J. Antiviral treatment of chronic hepatitis B virus (HBV) infections. Viruses 2010, 2, 1279–1305. [Google Scholar] [CrossRef] [Green Version]
- Chevaliez, S.; Hezode, C.; Bahrami, S.; Grare, M.; Pawlotsky, J.-M. Long-term hepatitis B surface antigen (HBsAg) kinetics during nucleoside/nucleotide analogue therapy: Finite treatment duration unlikely. J. Hepatol. 2013, 58, 676–683. [Google Scholar] [CrossRef] [Green Version]
- Perrillo, R. Benefits and risks of interferon therapy for hepatitis B. Hepatology 2009, 49, S103–S111. [Google Scholar] [CrossRef]
- Janssen, H.L.A.; Van Zonneveld, M.; Senturk, H.; Zeuzem, S.; Akarca, U.S.; Cakaloglu, Y.; Simon, C.; So, T.M.K.; Gerken, G.; A De Man, R.; et al. Pegylated interferon alfa-2b alone or in combination with lamivudine for HBeAg-positive chronic hepatitis B: A randomised trial. Lancet 2005, 365, 123–129. [Google Scholar] [CrossRef]
- Marcellin, P.; Lau, G.; Bonino, F.; Farci, P.; Hadziyannis, S.; Jin, R.; Lu, Z.-M.; Piratvisuth, T.; Germanidis, G.; Yurdaydin, C.; et al. Peginterferon alfa-2a alone, Lamivudine alone, and the two in combination in patients with HBeAg-negative chronic hepatitis B. N. Engl. J. Med. 2004, 351, 1206–1217. [Google Scholar] [CrossRef] [Green Version]
- Lampertico, P.; Viganò, M.; Di Costanzo, G.G.; Sagnelli, E.; Fasano, M.; Di Marco, V.; Boninsegna, S.; Farci, P.; Fargion, S.; Giuberti, T.; et al. Randomised study comparing 48 and 96 weeks peginterferon α-2a therapy in genotype D HBeAg-negative chronic hepatitis B. Gut 2013, 62, 290–298. [Google Scholar] [CrossRef]
- Duraisamy, G.S.; Bhosale, D.; Lipenská, I.; Huvarova, I.; Ruzek, D.; Windisch, M.P.; Miller, A.D. Advanced therapeutics, vaccinations, and precision medicine in the treatment and management of chronic hepatitis B viral infections; Where are we and where are we going? Viruses 2020, 12, 998. [Google Scholar] [CrossRef]
- Berg, F.V.D.; Limani, S.W.; Mnyandu, N.; Maepa, M.B.; Ely, A.; Arbuthnot, P. Advances with RNAi-based therapy for hepatitis B virus infection. Viruses 2020, 12, 851. [Google Scholar] [CrossRef] [PubMed]
- Schulze, A.; Mills, K.; Weiss, T.S.; Urban, S. Hepatocyte polarization is essential for the productive entry of the hepatitis B virus. Hepatology 2011, 55, 373–383. [Google Scholar] [CrossRef]
- Thomas, E.; Liang, T.J. Experimental models of hepatitis B and C—new insights and progress. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 362–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allweiss, L.; Dandri, M. Experimental in vitro and in vivo models for the study of human hepatitis B virus infection. J. Hepatol. 2016, 64, S17–S31. [Google Scholar] [CrossRef]
- Hu, J.; Lin, Y.; Chen, P.; Watashi, K.; Wakita, T. Cell and animal models for studying hepatitis B virus infection and drug development. Gastroenterology 2019, 156, 338–354. [Google Scholar] [CrossRef]
- Baumert, T.F.; Colpitts, C.C.; Schuster, C.; Zeisel, M.B.; Baumert, T.F. Cell culture models for the investigation of hepatitis B and D virus infection. Viruses 2016, 8, 261. [Google Scholar] [CrossRef]
- Witt-Kehati, D.; Alaluf, M.B.; Shlomai, A. Advances and challenges in studying hepatitis B virus in vitro. Viruses 2016, 8, 21. [Google Scholar] [CrossRef]
- Wettengel, J.; Burwitz, B.J. Innovative HBV animal models based on the entry receptor NTCP. Viruses 2020, 12, 828. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Diot, C.; Thézé, N.; Fourel, I.; Loreal, O.; Brechot, C.; Guguen-Guillouzo, C. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol. 1988, 62, 4136–4143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gripon, P.; Diot, C.; Guillouzo, C.G. Reproducible high level infection of cultured adult human hepatocytes by hepatitis B virus: Effect of polyethylene glycol on adsorption and penetration. Virology 1993, 192, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, N.J.; Lechón, M.J.G.; Houston, J.B.; Hallifax, D.; Brown, H.S.; Maurel, P.; Kenna, J.G.; Gustavsson, L.; Lohmann, C.; Skonberg, C.; et al. Primary hepatocytes: Current understanding of the regulation of metabolic enzymes and transporter proteins, and pharmaceutical practice for the use of hepatocytes in metabolism, enzyme induction, transporter, clearance, and hepatotoxicity studies. Drug Metab. Rev. 2007, 39, 159–234. [Google Scholar] [CrossRef] [PubMed]
- Gripon, P.; Rumin, S.; Urban, S.; Le Seyec, J.; Glaise, D.; Cannie, I.; Guyomard, C.; Lucas, J.; Trepo, C.; Guguen-Guillouzo, C. Nonlinear partial differential equations and applications: Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA 2002, 99, 15655–15660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hantz, O.; Parent, R.; Durantel, D.; Gripon, P.; Guguen-Guillouzo, C.; Zoulim, F. Persistence of the hepatitis B virus covalently closed circular DNA in HepaRG human hepatocyte-like cells. J. Gen. Virol. 2009, 90, 127–135. [Google Scholar] [CrossRef]
- Ning, X.; Nguyen, D.; Mentzer, L.; Adams, C.; Lee, H.; Ashley, R.; Hafenstein, S.; Hu, J. Secretion of genome-free hepatitis B virus–single strand blocking model for virion morphogenesis of para-retrovirus. PLoS Pathog. 2011, 7, e1002255. [Google Scholar] [CrossRef]
- Sureau, C.; Romet-Lemonne, J.L.; Mullins, J.I.; Essex, M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 1986, 47, 37–47. [Google Scholar] [CrossRef]
- Sells, M.A.; Chen, M.L.; Acs, G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 1005–1009. [Google Scholar] [CrossRef] [Green Version]
- Ladner, S.K.; Otto, M.J.; Barker, C.S.; Zaifert, K.; Wang, G.H.; Guo, J.T.; Seeger, C.; King, R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997, 41, 1715–1720. [Google Scholar] [CrossRef] [Green Version]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, 00049. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Watashi, K.; Tsukuda, S.; Aly, H.H.; Fukasawa, M.; Fujimoto, A.; Suzuki, R.; Aizaki, H.; Ito, T.; Koiwai, O.; et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 2014, 443, 808–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seeger, C.; Sohn, J.A. Targeting hepatitis B virus with CRISPR/Cas9. Mol. Ther.-Nucleic Acids 2014, 3, e216. [Google Scholar] [CrossRef]
- Yan, R.; Zhang, Y.; Cai, D.; Liu, Y.; Cuconati, A.; Guo, H. Spinoculation enhances HBV infection in NTCP-reconstituted hepatocytes. PLoS ONE 2015, 10, e0129889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W. The hepatitis B virus receptor. Annu. Rev. Cell Dev. Biol. 2015, 31, 125–147. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Lempp, F.A.; Mehrle, S.; Nkongolo, S.; Kaufman, C.; Fälth, M.; Stindt, J.; Königer, C.; Nassal, M.; Kubitz, R.; et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology 2014, 146, 1070–1083.e6. [Google Scholar] [CrossRef]
- Lempp, F.A.; Qu, B.; Wang, Y.X.; Urban, S. Hepatitis B virus infection of a mouse hepatic cell line reconstituted with human sodium taurocholate cotransporting polypeptide. J. Virol. 2016, 90, 4827–4831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Zong, L.; Sureau, C.; Barker, L.; Wands, J.R.; Tong, S. Unusual features of sodium taurocholate cotransporting polypeptide as a hepatitis B virus receptor. J. Virol. 2016, 90, 8302–8313. [Google Scholar] [CrossRef] [Green Version]
- Michailidis, E.; Pabon, J.; Xiang, K.; Park, P.; Ramanan, V.; Hoffmann, H.H.; Schneider, W.M.; Bhatia, S.N.; De Jong, Y.P.; Shlomai, A.; et al. A robust cell culture system supporting the complete life cycle of hepatitis B virus. Sci. Rep. 2017, 7, 16616. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Zhao, K.; Yao, Y.; Yuan, Y.; Pei, R.; Wang, Y.; Chen, J.; Hu, X.; Zhou, Y.; Chen, X.; et al. Productive HBV infection of well-differentiated, hNTCP-expressing human hepatoma-derived (Huh7) cells. Virol. Sin. 2017, 32, 465–475. [Google Scholar] [CrossRef]
- Qiao, L.; Sui, J.; Luo, G. Robust human and murine hepatocyte culture models of hepatitis B virus infection and replication. J. Virol. 2018, 92, e01255-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asabe, S.; Wieland, S.F.; Chattopadhyay, P.K.; Roederer, M.; Engle, R.E.; Purcell, R.H.; Chisari, F.V. The size of the viral inoculum contributes to the outcome of hepatitis B virus infection. J. Virol. 2009, 83, 9652–9662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steenbergen, R.H.; Joyce, M.A.; Thomas, B.; Jones, D.; Law, J.; Russell, R.; Houghton, M.; Tyrrell, D.L. Human serum leads to differentiation of human hepatoma cells, restoration of very-low-density lipoprotein secretion, and a 1000-fold increase in HCV Japanese fulminant hepatitis type 1 titers. Hepatology 2013, 58, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, R.; Oti, M.; Ter Horst, R.; Tat, W.; Neufeldt, C.; Belovodskiy, A.; Chua, T.T.; Cho, W.J.; Joyce, M.; Dutilh, B.E.; et al. Establishing normal metabolism and differentiation in hepatocellular carcinoma cells by culturing in adult human serum. Sci. Rep. 2018, 8, 11685. [Google Scholar] [CrossRef] [PubMed]
- Hart, S.N.; Li, Y.; Nakamoto, K.; Subileau, E.-A.; Steen, D.; Zhong, X.-B. A comparison of whole genome gene expression profiles of HepaRG cells and HepG2 cells to primary human hepatocytes and human liver tissues. Drug Metab. Dispos. 2010, 38, 988–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, L.; Dial, S.; Shi, L.; Branham, W.; Liu, J.; Fang, J.L.; Green, B.; Deng, H.; Kaput, J.; Ning, B. Similarities and differences in the expression of drug-metabolizing enzymes between human hepatic cell lines and primary human hepatocytes. Drug Metab. Dispos. 2011, 39, 528–538. [Google Scholar] [CrossRef] [Green Version]
- Vondráček, J.; Souček, K.; Sheard, M.A.; Chramostová, K.; Andrysík, Z.; Hofmanová, J.; Kozubík, A. Dimethyl sulfoxide potentiates death receptor-mediated apoptosis in the human myeloid leukemia U937 cell line through enhancement of mitochondrial membrane depolarization. Leuk. Res. 2006, 30, 81–89. [Google Scholar] [CrossRef]
- Song, Y.M.; Song, S.O.; Jung, Y.K.; Kang, E.S.; Cha, B.S.; Lee, H.C.; Lee, B. Dimethyl sulfoxide reduces hepatocellular lipid accumulation through autophagy induction. Autophagy 2012, 8, 1085–1097. [Google Scholar] [CrossRef] [Green Version]
- Galvao, J.; Davis, B.; Tilley, M.; Normando, E.; Duchen, M.R.; Cordeiro, M.F. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J. 2014, 28, 1317–1330. [Google Scholar] [CrossRef]
- Pal, R.; Mamidi, M.K.; Das, A.; Bhonde, R. Diverse effects of dimethyl sulfoxide (DMSO) on the differentiation potential of human embryonic stem cells. Arch. Toxicol. 2012, 86, 651–661. [Google Scholar] [CrossRef]
- Verheijen, M.; Lienhard, M.; Schrooders, Y.; Clayton, O.; Nudischer, R.; Boerno, S.; Timmermann, B.; Selevsek, N.; Schlapbach, R.; Gmuender, H.; et al. DMSO induces drastic changes in human cellular processes and epigenetic landscape in vitro. Sci. Rep. 2019, 9, 4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, C.; Du, Y.; Meng, G.; Yi, L.S.; Sun, S.; Song, N.; Zhang, X.; Xiao, Y.; Wang, J.; Yi, Z.; et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science 2019, 364, 399–402. [Google Scholar] [CrossRef] [PubMed]
- Schoggins, J.W.; MacDuff, D.A.; Imanaka, N.; Gainey, M.D.; Shrestha, B.; Eitson, J.L.; Mar, K.B.; Richardson, R.B.; Ratushny, A.V.; Litvak, V.; et al. Pan-viral specificity of IFN-induced genes reveals new roles for cGAS in innate immunity. Nature 2014, 505, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Yamasaki, C.; Yanagi, A.; Yoshizane, Y.; Fujikawa, K.; Watashi, K.; Abe, H.; Wakita, T.; Hayes, C.N.; Chayama, K.; et al. Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. Am. J. Pathol. 2015, 185, 1275–1285. [Google Scholar] [CrossRef]
- Qu, B.; Ni, Y.; Lempp, F.A.; Vondran, F.W.R.; Urban, S. T5 exonuclease hydrolysis of hepatitis B virus replicative intermediates allows reliable quantification and fast drug efficacy testing of covalently closed circular DNA by PCR. J. Virol. 2018, 92, e01117-18. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Le, C.; Tyrrell, D.L.; Le, X.C.; Li, X.-F. Aptamer binding assay for the E antigen of hepatitis B using modified aptamers with G-quadruplex structures. Anal. Chem. 2020, 92, 6495–6501. [Google Scholar] [CrossRef] [Green Version]
- Nishitsuji, H.; Ujino, S.; Shimizu, Y.; Harada, K.; Zhang, J.; Sugiyama, M.; Mizokami, M.; Shimotohno, K. Novel reporter system to monitor early stages of the hepatitis B virus life cycle. Cancer Sci. 2015, 106, 1616–1624. [Google Scholar] [CrossRef]
- Shlomai, A.; Schwartz, R.E.; Ramanan, V.; Bhatta, A.; De Jong, Y.P.; Bhatia, S.N.; Rice, C.M. Modeling host interactions with hepatitis B virus using primary and induced pluripotent stem cell-derived hepatocellular systems. Proc. Natl. Acad. Sci. USA 2014, 111, 12193–12198. [Google Scholar] [CrossRef] [Green Version]
- Kidambi, S.; Yarmush, R.S.; Novik, E.; Chao, P.; Yarmush, M.L.; Nahmias, Y. Oxygen-mediated enhancement of primary hepatocyte metabolism, functional polarization, gene expression, and drug clearance. Proc. Natl. Acad. Sci. USA 2009, 106, 15714–15719. [Google Scholar] [CrossRef] [Green Version]
- Meier, A.; Mehrle, S.; Weiss, T.S.; Mier, W.; Urban, S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 2013, 58, 31–42. [Google Scholar] [CrossRef]
- Appelman, M.D.; Chakraborty, A.; Protzer, U.; McKeating, J.A.; Van De Graaf, S.F. N-Glycosylation of the Na+-taurocholate cotransporting polypeptide (NTCP) determines its trafficking and stability and is required for hepatitis B virus infection. PLoS ONE 2017, 12, e0170419. [Google Scholar] [CrossRef] [PubMed]
- Sargiacomo, C.; El-Kehdy, H.; Pourcher, G.; Stieger, B.; Najimi, M.; Sokal, E. Age-dependent glycosylation of the sodium taurocholate cotransporter polypeptide: From fetal to adult human livers. Hepatol. Commun. 2018, 2, 693–702. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Zong, L.; Krotow, A.; Qin, Y.; Jia, L.; Zhang, J.; Tong, S.; Li, J. N-Linked glycosylation is not essential for sodium taurocholate cotransporting polypeptide to mediate hepatitis B virus infection in vitro. J. Virol. 2018, 92, e00732-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le, C.; Sirajee, R.; Steenbergen, R.; Joyce, M.A.; Addison, W.R.; Tyrrell, D.L. In Vitro Infection with Hepatitis B Virus Using Differentiated Human Serum Culture of Huh7.5-NTCP Cells without Requiring Dimethyl Sulfoxide. Viruses 2021, 13, 97. https://doi.org/10.3390/v13010097
Le C, Sirajee R, Steenbergen R, Joyce MA, Addison WR, Tyrrell DL. In Vitro Infection with Hepatitis B Virus Using Differentiated Human Serum Culture of Huh7.5-NTCP Cells without Requiring Dimethyl Sulfoxide. Viruses. 2021; 13(1):97. https://doi.org/10.3390/v13010097
Chicago/Turabian StyleLe, Connie, Reshma Sirajee, Rineke Steenbergen, Michael A. Joyce, William R. Addison, and D. Lorne Tyrrell. 2021. "In Vitro Infection with Hepatitis B Virus Using Differentiated Human Serum Culture of Huh7.5-NTCP Cells without Requiring Dimethyl Sulfoxide" Viruses 13, no. 1: 97. https://doi.org/10.3390/v13010097
APA StyleLe, C., Sirajee, R., Steenbergen, R., Joyce, M. A., Addison, W. R., & Tyrrell, D. L. (2021). In Vitro Infection with Hepatitis B Virus Using Differentiated Human Serum Culture of Huh7.5-NTCP Cells without Requiring Dimethyl Sulfoxide. Viruses, 13(1), 97. https://doi.org/10.3390/v13010097





