Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond
Abstract
1. Introduction
2. History of the RNA Structure Epic: From Scratch to SHAPE
2.1. Birth of RNA Structure Probing
2.2. Childhood of RNA Probing and Investigation of Viral RNA Structures
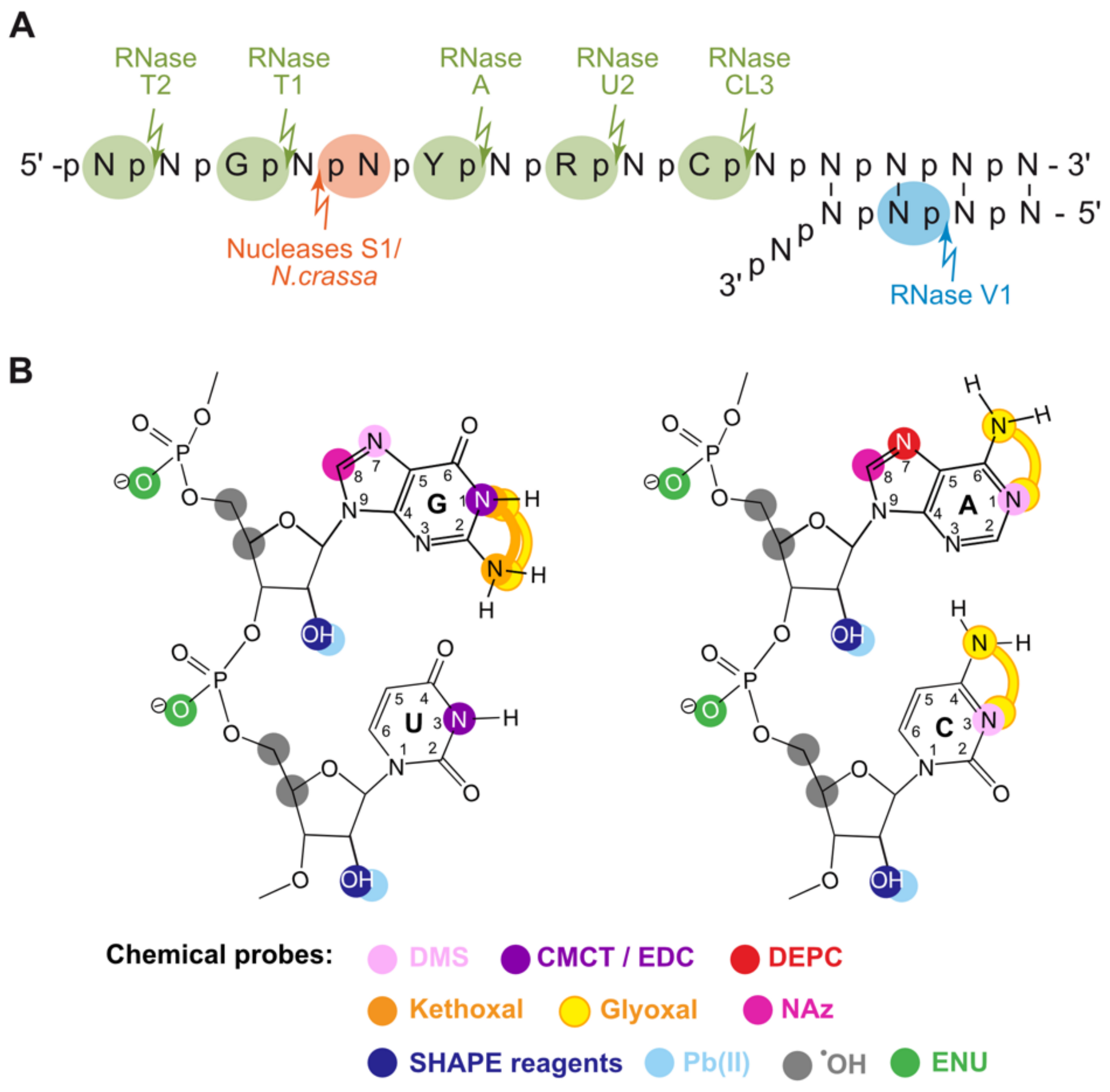
2.2.1. Characteristics and Specificities of the Pre-SHAPE Probes
2.2.2. Early Applications to Viral RNAs
3′-Terminal Region of Plant Viruses: tRNA Mimicry for Replication
3′-Terminal Region of Plant Viruses: Cap Independent Translation Enhancers
RNA Structures of Human Pathogenic Viruses
Enzymatic Viral RNAs: Self-Cleaving Ribozymes
3. The SHAPE (r)Evolution
4. A New Era for Enzymatic and Chemical Probing
4.1. The Deep-Sequencing Leaps
4.2. Old Probes Back in the Spotlight
4.3. Mutational Profiling
4.4. The Latest Developments
4.5. Applications of the MAP Strategies to Viral RNAs
5. Conclusions
6. Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Markham, R.; Smith, K.M. Studies on the Virus of Turnip Yellow Mosaic. Parasitology 1949, 39, 330–342. [Google Scholar] [CrossRef]
- Gierer, A.; Schramm, G. Infectivity of Ribonucleic Acid from Tobacco Mosaic Virus. Nature 1956, 177, 702–703. [Google Scholar] [CrossRef]
- Ennifar, E.; Walter, P.; Ehresmann, B.; Ehresmann, C.; Dumas, P. Crystal Structures of Coaxially Stacked Kissing Complexes of the HIV-1 RNA Dimerization Initiation Site. Nat. Struct. Biol. 2001, 8, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Suddala, K.C.; Zhang, J. High-Affinity Recognition of Specific TRNAs by an MRNA Anticodon-Binding Groove. Nat. Struct. Mol. Biol. 2019, 26, 1114–1122. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. Structure of the HIV-1 RNA Packaging Signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Li, S.; Kappel, K.; Pintilie, G.; Su, Z.; Mou, T.-C.; Schmid, M.F.; Das, R.; Chiu, W. Cryo-EM Structure of a 40 KDa SAM-IV Riboswitch RNA at 3.7 Å Resolution. Nat. Commun. 2019, 10, 5511. [Google Scholar] [CrossRef] [PubMed]
- Kappel, K.; Zhang, K.; Su, Z.; Watkins, A.M.; Kladwang, W.; Li, S.; Pintilie, G.; Topkar, V.V.; Rangan, R.; Zheludev, I.N.; et al. Accelerated Cryo-EM-Guided Determination of Three-Dimensional RNA-Only Structures. Nat. Methods 2020, 17, 699–707. [Google Scholar] [CrossRef]
- Mailler, E.; Paillart, J.-C.; Marquet, R.; Smyth, R.P.; Vivet-Boudou, V. The Evolution of RNA Structural Probing Methods: From Gels to next-Generation Sequencing. WIREs RNA 2019, 10, e1518. [Google Scholar] [CrossRef] [PubMed]
- Rich, A. The Era of RNA Awakening: Structural Biology of RNA in the Early Years. Q. Rev. Biophys. 2009, 42, 117–137. [Google Scholar] [CrossRef]
- Nguyen, T.C.; Zaleta-Rivera, K.; Huang, X.; Dai, X.; Zhong, S. RNA, Action through Interactions. Trends Genet. 2018, 34, 867–882. [Google Scholar] [CrossRef]
- Kudla, G.; Wan, Y.; Helwak, A. RNA Conformation Capture by Proximity Ligation. Annu. Rev. Genom. Hum. Genet. 2020, 21, 81–100. [Google Scholar] [CrossRef]
- Wang, X.-W.; Liu, C.-X.; Chen, L.-L.; Zhang, Q.C. RNA Structure Probing Uncovers RNA Structure-Dependent Biological Functions. Nat. Chem. Biol. 2021, 17, 755–766. [Google Scholar] [CrossRef]
- Brown, D.M.; Heppel, L.A.; Hilmoe, R.J. Nucleotides. Part XXIV. The Action of Some Nucleases on Simple Esters of Monoribonucleotides. J. Chem. Soc. 1954, 40–46. [Google Scholar] [CrossRef]
- Rich, A.; Davies, D.R. A New Two Stranded Helical Structure: Polyadenylic Acid and Polyuridylic Acid. J. Am. Chem. Soc. 1956, 78, 3548–3549. [Google Scholar] [CrossRef]
- Felsenfeld, G.; Davies, D.R.; Rich, A. Formation of a Three-Stranded Polynucleotide Molecule. J. Am. Chem. Soc. 1957, 79, 2023–2024. [Google Scholar] [CrossRef]
- Dubos, R.J. The Decomposition of Yeast Nucleic Acid by a Heat Resistant Enzyme. Science 1937, 85, 549. [Google Scholar] [CrossRef]
- Sato, K.; Egami, F. Studies on Ribonucleases in Takadiastase. I. J. Biochem. 1957, 44, 753–767. [Google Scholar] [CrossRef]
- Gomatos, P.J.; Tamm, I.; Dales, S.; Franklin, R.M. Reovirus Type 3: Physical Characteristics and Interaction with L Cells. Virology 1962, 17, 441–454. [Google Scholar] [CrossRef]
- Cantoni, G.L.; Gelboin, H.V.; Luborsky, S.W.; Richards, H.H.; Singer, M.F. Studies on Soluble Ribonucleic Acid of Rabbit Liver III. Preparation and Properties of Rabbit-Liver Soluble RNA. Biochim. Biophys. Acta 1962, 61, 354–367. [Google Scholar] [CrossRef]
- Geiduschek, E.P.; Moohr, J.W.; Weiss, S.B. The Secondary Structure of Complementary RNA. Proc. Natl. Acad. Sci. USA 1962, 48, 1078–1086. [Google Scholar] [CrossRef]
- Holley, R.W.; Everett, G.A.; Madison, J.T.; Zamir, A. Nucleotide Sequences in the Yeast Alanine Transfer Ribonucleic Acid. J. Biol. Chem. 1965, 240, 2122–2128. [Google Scholar] [CrossRef]
- Sato, K.; Egami, F. The specificity of T1 ribonuclease. Comptes Rendus Seances Soc. Biol. Fil. 1957, 151, 1792–1796. [Google Scholar]
- Doctor, B.P.; Connelly, C.M.; Rushizky, G.W.; Sober, H.A. Studies on the Chemical Structure of Yeast Amino Acid Acceptor Ribonucleic Acids. J. Biol. Chem. 1963, 238, 3985–3990. [Google Scholar] [CrossRef]
- Elmore, D.T.; Gulland, J.M.; Jordan, D.O.; Taylor, H.F.W. The Reaction of Nucleic Acids with Mustard Gas. Biochem. J. 1948, 42, 308–316. [Google Scholar] [CrossRef] [PubMed]
- Fraenkel-Conrat, H.; Singer, B.; Tsugita, A. Purification of Viral RNA by Means of Bentonite. Virology 1961, 14, 54–58. [Google Scholar] [CrossRef]
- Lawley, P.D.; Brookes, P. Further Studies on the Alkylation of Nucleic Acids and Their Constituent Nucleotides. Biochem. J. 1963, 89, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Gilham, P.T. An Addition Reaction Specific for Uridine and Guanosine Nucleotides and Its Application to the Modification of Ribonuclease Action. J. Am. Chem. Soc. 1962, 84, 687–688. [Google Scholar] [CrossRef]
- Augusti-Tocco, G.; Brown, G.L. Reaction of N-Cyclohexyl, N’-Beta (4-Methylmorpholinium) Ethyl Carbodiimide Iodide with Nucleic Acids and Polynucleotides. Nature 1965, 206, 683–685. [Google Scholar] [CrossRef]
- Naylor, R.; Ho, N.W.Y.; Gilham, P.T. Selective Chemical Modifications of Uridine and Pseudouridine in Polynucleotides and Their Effect on the Specificities of Ribonuclease and Phosphodiesterases. J. Am. Chem. Soc. 1965, 87, 4209–4210. [Google Scholar] [CrossRef]
- Linn, S.; Lehman, I.R. An Endonuclease from Neurospora Crassa Specific for Polynucleotides Lacking an Ordered Structure. J. Biol. Chem. 1965, 240, 1294–1304. [Google Scholar] [CrossRef]
- Harada, F.; Dahlberg, J.E. Specific Cleavage of TRNA by Nuclease S1. Nucleic Acids Res. 1975, 2, 865–871. [Google Scholar] [CrossRef]
- Vary, C.P.; Vournakis, J.N. RNA Structure Analysis Using T2 Ribonuclease: Detection of PH and Metal Ion Induced Conformational Changes in Yeast TRNAPhe. Nucleic Acids Res. 1984, 12, 6763–6778. [Google Scholar] [CrossRef]
- Levy, C.C.; Karpetsky, T.P. The Purification and Properties of Chicken Liver RNase: An Enzyme Which Is Useful in Distinguishing between Cytidylic and Uridylic Acid Residues. J. Biol. Chem. 1980, 255, 2153–2159. [Google Scholar] [CrossRef]
- Uchida, T.; Arima, T.; Egami, F. Specificity of RNase U2. J. Biochem. 1970, 67, 91–102. [Google Scholar] [CrossRef]
- Holley, R.W.; Apgar, J.; Everett, G.A.; Madison, J.T.; Marquisee, M.; Merrill, S.H.; Penswick, J.R.; Zamir, A. Structure of a Ribonucleic Acid. Science 1965, 147, 1462–1465. [Google Scholar] [CrossRef]
- Vasilenko, S.K.; Babkina, G.T. Isolation and properties of ribonuclease isolated from cobra venom. Biokhimiia 1965, 30, 705–712. [Google Scholar]
- Vasilenko, S.K.; Ryte, V.C. Isolation of highly purified ribonuclease from cobra (Naja oxiana) venom. Biokhimiia 1975, 40, 578–583. [Google Scholar]
- Favorova, O.O.; Fasiolo, F.; Keith, G.; Vassilenko, S.K.; Ebel, J.P. Partial Digestion of TRNA—Aminoacyl-TRNA Synthetase Complexes with Cobra Venom Ribonuclease. Biochemistry 1981, 20, 1006–1011. [Google Scholar] [CrossRef]
- Desai, N.A.; Shankar, V. Single-Strand-Specific Nucleases. FEMS Microbiol. Rev. 2003, 26, 457–491. [Google Scholar] [CrossRef]
- Mitchell, D.; Ritchey, L.E.; Park, H.; Babitzke, P.; Assmann, S.M.; Bevilacqua, P.C. Glyoxals as in Vivo RNA Structural Probes of Guanine Base-Pairing. RNA 2018, 24, 114–124. [Google Scholar] [CrossRef]
- Staehelin, M. Inactivation of Virus Nucleic Acid with Glyoxal Derivatives. Biochim. Biophys. Acta 1959, 31, 448–454. [Google Scholar] [CrossRef]
- Litt, M. Structural Studies on Transfer Ribonucleic Acid. I. Labeling of Exposed Guanine Sites in Yeast Phenylalanine Transfer Ribonucleic Acid with Kethoxal. Biochemistry 1969, 8, 3249–3253. [Google Scholar] [CrossRef]
- Oberg, B. Biochemical and Biological Characteristics of Carbethoxylated Poliovirus and Viral RNA. Biochim. Biophys. Acta 1970, 204, 430–440. [Google Scholar] [CrossRef]
- Leonard, N.J.; McDonald, J.J.; Henderson, R.E.L.; Reichmann, M.E. Reaction of Diethyl Pyrocarbonate with Nucleic Acid Components. Adenosine. Biochemistry 1971, 10, 3335–3342. [Google Scholar] [CrossRef] [PubMed]
- Peattie, D.A.; Gilbert, W. Chemical Probes for Higher-Order Structure in RNA. Proc. Natl. Acad. Sci. USA 1980, 77, 4679–4682. [Google Scholar] [CrossRef]
- Stern, S.; Wilson, R.C.; Noller, H.F. Localization of the Binding Site for Protein S4 on 16 S Ribosomal RNA by Chemical and Enzymatic Probing and Primer Extension. J. Mol. Biol. 1986, 192, 101–110. [Google Scholar] [CrossRef]
- Inoue, T.; Cech, T.R. Secondary Structure of the Circular Form of the Tetrahymena RRNA Intervening Sequence: A Technique for RNA Structure Analysis Using Chemical Probes and Reverse Transcriptase. Proc. Natl. Acad. Sci. USA 1985, 82, 648–652. [Google Scholar] [CrossRef]
- Guo, J.U.; Bartel, D.P. RNA G-Quadruplexes Are Globally Unfolded in Eukaryotic Cells and Depleted in Bacteria. Science 2016, 353, aaf5371. [Google Scholar] [CrossRef]
- Singer, B. All Oxygens in Nucleic Acids React with Carcinogenic Ethylating Agents. Nature 1976, 264, 333–339. [Google Scholar] [CrossRef]
- Vlassov, V.V.; Giege, R.; Ebel, J.P. The Tertiary Structure of Yeast TRNAPhe in Solution Studied by Phosphodiester Bond Modification with Ethylnitrosourea. FEBS Lett. 1980, 120, 12–16. [Google Scholar] [CrossRef]
- Wang, X.D.; Padgett, R.A. Hydroxyl Radical “Footprinting” of RNA: Application to Pre-MRNA Splicing Complexes. Proc. Natl. Acad. Sci. USA 1989, 86, 7795–7799. [Google Scholar] [CrossRef]
- Vary, C.P.; Vournakis, J.N. RNA Structure Analysis Using Methidiumpropyl-EDTA.Fe(II): A Base-Pair-Specific RNA Structure Probe. Proc. Natl. Acad. Sci. USA 1984, 81, 6978–6982. [Google Scholar] [CrossRef]
- Zamir, A.; Holley, R.W.; Marquisee, M. Evidence for the Occurrence of a Common Pentanucleotide Sequence in the Structures of Transfer Ribonucleic Acids. J. Biol. Chem. 1965, 240, 1267–1273. [Google Scholar] [CrossRef]
- Wintermeyer, W.; Zachau, H.G. Tertiary Structure Interactions of 7-Methylguanosine in Yeast TRNA Phe as Studied by Borohydride Reduction. FEBS Lett. 1975, 58, 306–309. [Google Scholar] [CrossRef]
- Ehresmann, C.; Baudin, F.; Mougel, M.; Romby, P.; Ebel, J.P.; Ehresmann, B. Probing the Structure of RNAs in Solution. Nucleic Acids Res. 1987, 15, 9109–9128. [Google Scholar] [CrossRef]
- Peattie, D.A. Direct Chemical Method for Sequencing RNA. Proc. Natl. Acad. Sci. USA 1979, 76, 1760–1764. [Google Scholar] [CrossRef]
- Kuśmierek, J.T.; Singer, B. Sites of Alkylation of Poly(U) by Agents of Varying Carcinogenicity and Stability of Products. Biochim. Biophys. Acta 1976, 442, 420–431. [Google Scholar] [CrossRef]
- Markham, R.; Smith, J.D. The Structure of Ribonucleic Acid. I. Cyclic Nucleotides Produced by Ribonuclease and by Alkaline Hydrolysis. Biochem. J. 1952, 552–557. [Google Scholar] [CrossRef]
- Rosenstein, S.P.; Been, M.D. Evidence That Genomic and Antigenomic RNA Self-Cleaving Elements from Hepatitis Delta Virus Have Similar Secondary Structures. Nucleic Acids Res. 1991, 19, 5409–5416. [Google Scholar] [CrossRef]
- Florentz, C.; Briand, J.P.; Romby, P.; Hirth, L.; Ebel, J.P.; Glegé, R. The TRNA-like Structure of Turnip Yellow Mosaic Virus RNA: Structural Organization of the Last 159 Nucleotides from the 3’ OH Terminus. EMBO J. 1982, 1, 269–276. [Google Scholar] [CrossRef]
- Rietveld, K.; Van Poelgeest, R.; Pleij, C.W.; Van Boom, J.H.; Bosch, L. The TRNA-like Structure at the 3’ Terminus of Turnip Yellow Mosaic Virus RNA. Differences and Similarities with Canonical TRNA. Nucleic Acids Res. 1982, 10, 1929–1946. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, K.; Takemura, S. Structure and Function of 5S Ribosomal Ribonucleic Acid from Torulopsis Utilis. J. Biochem. 1977, 81, 995–1003. [Google Scholar] [CrossRef]
- Ahlquist, P.; Dasgupta, R.; Kaesberg, P. Near Identity of 3′ RNA Secondary Structure in Bromoviruses and Cucumber Mosaic Virus. Cell 1981, 23, 183–189. [Google Scholar] [CrossRef]
- Troutt, A.; Savin, T.J.; Curtiss, W.C.; Celentano, J.; Vournakis, J.N. Secondary Structure of Bombyx Mori and Dictyostelium Discoideum 5S RRNA from S1 Nuclease and Cobra Venom Ribonuclease Susceptibility, and Computer Assisted Analysis. Nucleic Acids Res. 1982, 10, 653–664. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Skinner, M.A.; Racaniello, V.R.; Dunn, G.; Cooper, J.; Minor, P.D.; Almond, J.W. New Model for the Secondary Structure of the 5’ Non-Coding RNA of Poliovirus Is Supported by Biochemical and Genetic Data That Also Show That RNA Secondary Structure Is Important in Neurovirulence. J. Mol. Biol. 1989, 207, 379–392. [Google Scholar] [CrossRef]
- Garret, M.; Labouesse, B.; Litvak, S.; Romby, P.; Ebel, J.P.; Giegé, R. Tertiary Structure of Animal TRNATrp in Solution and Interaction of TRNATrp with Tryptophanyl-TRNA Synthetase. Eur. J. Biochem. 1984, 138, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Isel, C.; Marquet, R.; Keith, G.; Ehresmann, C.; Ehresmann, B. Modified Nucleotides of TRNA(3Lys) Modulate Primer/Template Loop-Loop Interaction in the Initiation Complex of HIV-1 Reverse Transcription. J. Biol. Chem. 1993, 268, 25269–25272. [Google Scholar] [CrossRef]
- Mougel, M.; Philippe, C.; Ebel, J.P.; Ehresmann, B.; Ehresmann, C. The E. Coli 16S RRNA Binding Site of Ribosomal Protein S15: Higher-Order Structure in the Absence and in the Presence of the Protein. Nucleic Acids Res. 1988, 16, 2825–2839. [Google Scholar] [CrossRef]
- Glickman, J.N.; Howe, J.G.; Steitz, J.A. Structural Analyses of EBER1 and EBER2 Ribonucleoprotein Particles Present in Epstein-Barr Virus-Infected Cells. J. Virol. 1988, 62, 902–911. [Google Scholar] [CrossRef]
- Hertzberg, R.P.; Dervan, P.B. Cleavage of Double Helical DNA by Methidium-Propyl-EDTA-Iron(II). J. Am. Chem. Soc. 1982, 104, 313–315. [Google Scholar] [CrossRef]
- Lusvarghi, S.; Sztuba-Solinska, J.; Purzycka, K.J.; Pauly, G.T.; Rausch, J.W.; Grice, S.F.J.L. The HIV-2 Rev-Response Element: Determining Secondary Structure and Defining Folding Intermediates. Nucleic Acids Res. 2013, 41, 6637–6649. [Google Scholar] [CrossRef]
- Tullius, T.D.; Dombroski, B.A. Hydroxyl Radical “Footprinting”: High-Resolution Information about DNA-Protein Contacts and Application to Lambda Repressor and Cro Protein. Proc. Natl. Acad. Sci. USA 1986, 83, 5469–5473. [Google Scholar] [CrossRef]
- Isel, C.; Westhof, E.; Massire, C.; Le Grice, S.F.J.; Ehresmann, B.; Ehresmann, C.; Marquet, R. Structural Basis for the Specificity of the Initiation of HIV-1 Reverse Transcription. EMBO J. 1999, 18, 1038–1048. [Google Scholar] [CrossRef]
- Werner, C.; Krebs, B.; Keith, G.; Dirheimer, G. Specific Cleavages of Pure TRNAs by Plumbous Ions. Biochim. Biophys. Acta 1976, 432, 161–175. [Google Scholar] [CrossRef]
- Gornicki, P.; Baudin, F.; Romby, P.; Wiewiorowski, M.; Kryzosiak, W.; Ebel, J.P.; Ehresmann, C.; Ehresmann, B. Use of Lead(II) to Probe the Structure of Large RNA’s. Conformation of the 3′ Terminal Domain of E. Coli 16S RRNA and Its Involvement in Building the TRNA Binding Sites. J. Biomol. Struct. Dynam. 1989, 6, 971–984. [Google Scholar] [CrossRef]
- Paillart, J.C.; Westhof, E.; Ehresmann, C.; Ehresmann, B.; Marquet, R. Non-Canonical Interactions in a Kissing Loop Complex: The Dimerization Initiation Site of HIV-1 Genomic RNA. J. Mol. Biol. 1997, 270, 36–49. [Google Scholar] [CrossRef]
- Qu, H.L.; Michot, B.; Bachellerie, J.P. Improved Methods for Structure Probing in Large RNAs: A Rapid “heterologous” Sequencing Approach Is Coupled to the Direct Mapping of Nuclease Accessible Sites. Application to the 5’ Terminal Domain of Eukaryotic 28S RRNA. Nucleic Acids Res. 1983, 11, 5903–5920. [Google Scholar] [CrossRef]
- Moazed, D.; Stern, S.; Noller, H.F. Rapid Chemical Probing of Conformation in 16 S Ribosomal RNA and 30 S Ribosomal Subunits Using Primer Extension. J. Mol. Biol. 1986, 187, 399–416. [Google Scholar] [CrossRef]
- Pinck, M.; Yot, P.; Chapeville, F.; Duranton, H.M. Enzymatic Binding of Valine to the 3′ End of TYMV-RNA. Nature 1970, 226, 954–956. [Google Scholar] [CrossRef]
- Yamakawa, M.; Shatkin, A.J.; Furuichi, Y. Chemical Methylation of RNA and DNA Viral Genomes as a Probe of in Situ Structure. J. Virol. 1981, 40, 482–490. [Google Scholar] [CrossRef]
- Joshi, R.L.; Joshi, S.; Chapeville, F.; Haenni, A.L. tRNA-like Structures of Plant Viral RNAs: Conformational Requirements for Adenylation and Aminoacylation. EMBO J. 1983, 2, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Langeberg, C.J.; Sherlock, M.E.; MacFadden, A.; Kieft, J.S. An Expanded Class of Histidine-Accepting Viral TRNA-like Structures. RNA 2021, 27, 653–664. [Google Scholar] [CrossRef] [PubMed]
- Hammond, J.A.; Rambo, R.P.; Filbin, M.E.; Kieft, J.S. Comparison and Functional Implications of the 3D Architectures of Viral TRNA-like Structures. RNA 2009, 15, 294–307. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.L.; White, K.A. Context-Influenced Cap-Independent Translation of Tombusvirus MRNAs in Vitro. Virology 2008, 380, 203–212. [Google Scholar] [CrossRef][Green Version]
- Wu, B.; White, K.A. A Primary Determinant of Cap-Independent Translation Is Located in the 3’-Proximal Region of the Tomato Bushy Stunt Virus Genome. J. Virol. 1999, 73, 8982–8988. [Google Scholar] [CrossRef]
- Danthinne, X.; Seurinck, J.; Meulewaeter, F.; Van Montagu, M.; Cornelissen, M. The 3’untranslated Region of Satellite Tobacco Necrosis Virus RNA Stimulates Translation in Vitro. Mol. Cell. Biol. 1993, 13, 3340–3349. [Google Scholar] [CrossRef]
- Miller, W.A.; Wang, Z.; Treder, K. The Amazing Diversity of Cap-Independent Translation Elements in the 3′-Untranslated Regions of Plant Viral RNAs. Biochem. Soc. Trans. 2007, 35, 1629–1633. [Google Scholar] [CrossRef]
- Fabian, M.R.; White, K.A. 5′-3′ RNA-RNA Interaction Facilitates Cap- and Poly(A) Tail-Independent Translation of Tomato Bushy Stunt Virus Mrna: A Potential Common Mechanism for Tombusviridae. J. Biol. Chem. 2004, 279, 28862–28872. [Google Scholar] [CrossRef]
- Pelletier, J.; Sonenberg, N. Internal Initiation of Translation of Eukaryotic MRNA Directed by a Sequence Derived from Poliovirus RNA. Nature 1988, 334, 320–325. [Google Scholar] [CrossRef]
- Jang, S.K.; Kräusslich, H.G.; Nicklin, M.J.; Duke, G.M.; Palmenberg, A.C.; Wimmer, E. A Segment of the 5’ Nontranslated Region of Encephalomyocarditis Virus RNA Directs Internal Entry of Ribosomes during in Vitro Translation. J. Virol. 1988, 62, 2636–2643. [Google Scholar] [CrossRef]
- Pilipenko, E.V.; Blinov, V.M.; Chernov, B.K.; Dmitrieva, T.M.; Agol, V.I. Conservation of the Secondary Structure Elements of the 5’-Untranslated Region of Cardio- and Aphthovirus RNAs. Nucleic Acids Res. 1989, 17, 5701–5711. [Google Scholar] [CrossRef]
- Mailliot, J.; Martin, F. Viral Internal Ribosomal Entry Sites: Four Classes for One Goal. WIREs RNA 2017, 9. [Google Scholar] [CrossRef]
- Evans, D.M.A.; Dunn, G.; Minor, P.D.; Schild, G.C.; Cann, A.J.; Stanway, G.; Almond, J.W.; Currey, K.; Maizel, J.V. Increased Neurovirulence Associated with a Single Nucleotide Change in a Noncoding Region of the Sabin Type 3 Poliovaccine Genome. Nature 1985, 314, 548–550. [Google Scholar] [CrossRef]
- Harrison, G.P.; Lever, A.M. The Human Immunodeficiency Virus Type 1 Packaging Signal and Major Splice Donor Region Have a Conserved Stable Secondary Structure. J. Virol. 1992, 66, 4144–4153. [Google Scholar] [CrossRef]
- Baudin, F.; Marquet, R.; Isel, C.; Darlix, J.-L.; Ehresmann, B.; Ehresmann, C. Functional Sites in the 5′ Region of Human Immunodeficiency Virus Type 1 RNA Form Defined Structural Domains. J. Mol. Biol. 1993, 229, 382–397. [Google Scholar] [CrossRef]
- Isel, C.; Ehresmann, C.; Keith, G.; Ehresmann, B.; Marquet, R. Initiation of Reverse Transcription of HIV-1: Secondary Structure of the HIV-1 RNA/TRNA(3Lys) (Template/Primer). J. Mol. Biol. 1995, 247, 236–250. [Google Scholar] [CrossRef]
- Ennifar, E.; Paillart, J.-C.; Bodlenner, A.; Walter, P.; Weibel, J.-M.; Aubertin, A.-M.; Pale, P.; Dumas, P.; Marquet, R. Targeting the Dimerization Initiation Site of HIV-1 RNA with Aminoglycosides: From Crystal to Cell. Nucleic Acids Res. 2006, 34, 2328–2339. [Google Scholar] [CrossRef]
- Abbink, T.E.M.; Berkhout, B. A Novel Long Distance Base-Pairing Interaction in Human Immunodeficiency Virus Type 1 RNA Occludes the Gag Start Codon*. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef]
- Huthoff, H.; Berkhout, B. Two Alternating Structures of the HIV-1 Leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef]
- Damgaard, C.K.; Andersen, E.S.; Knudsen, B.; Gorodkin, J.; Kjems, J. RNA Interactions in the 5′ Region of the HIV-1 Genome. J. Mol. Biol. 2004, 336, 369–379. [Google Scholar] [CrossRef]
- Symons, R.H. Self-Cleavage of RNA in the Replication of Small Pathogens of Plants and Animals. Trends Biochem. Sci. 1989, 14, 445–450. [Google Scholar] [CrossRef]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-Splicing RNA: Autoexcision and Autocyclization of the Ribosomal RNA Intervening Sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Wu, H.N.; Lin, Y.J.; Lin, F.P.; Makino, S.; Chang, M.F.; Lai, M.M. Human Hepatitis Delta Virus RNA Subfragments Contain an Autocleavage Activity. Proc. Natl. Acad. Sci. USA 1989, 86, 1831–1835. [Google Scholar] [CrossRef] [PubMed]
- Strobel, B.; Spöring, M.; Klein, H.; Blazevic, D.; Rust, W.; Sayols, S.; Hartig, J.S.; Kreuz, S. High-Throughput Identification of Synthetic Riboswitches by Barcode-Free Amplicon-Sequencing in Human Cells. Nat. Commun. 2020, 11, 714. [Google Scholar] [CrossRef]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA Structure Analysis at Single Nucleotide Resolution by Selective 2’-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef]
- Chamberlin, S.I.; Weeks, K.M. Differential Helix Stabilities and Sites Pre-Organized for Tertiary Interactions Revealed by Monitoring Local Nucleotide Flexibility in the BI5 Group I Intron RNA. Biochemistry 2003, 42, 901–909. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension (SHAPE): Quantitative RNA Structure Analysis at Single Nucleotide Resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. RNA SHAPE Chemistry Reveals Nonhierarchical Interactions Dominate Equilibrium Structural Transitions in TRNA(Asp) Transcripts. J. Am. Chem. Soc. 2005, 127, 4659–4667. [Google Scholar] [CrossRef]
- Mortimer, S.A.; Weeks, K.M. A Fast-Acting Reagent for Accurate Analysis of RNA Secondary and Tertiary Structure by SHAPE Chemistry. J. Am. Chem. Soc. 2007, 129, 4144–4145. [Google Scholar] [CrossRef]
- Gherghe, C.M.; Mortimer, S.A.; Krahn, J.M.; Thompson, N.L.; Weeks, K.M. Slow Conformational Dynamics at C2′-Endo Nucleotides in RNA. J. Am. Chem. Soc. 2008, 130, 8884–8885. [Google Scholar] [CrossRef]
- Mortimer, S.A.; Weeks, K.M. Time-Resolved RNA SHAPE Chemistry. J. Am. Chem. Soc. 2008, 130, 16178–16180. [Google Scholar] [CrossRef]
- Spitale, R.C.; Crisalli, P.; Flynn, R.A.; Torre, E.A.; Kool, E.T.; Chang, H.Y. RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol. 2013, 9, 18–20. [Google Scholar] [CrossRef]
- Tyrrell, J.; McGinnis, J.L.; Weeks, K.M.; Pielak, G.J. The Cellular Environment Stabilizes Adenine Riboswitch RNA Structure. Biochemistry 2013, 52, 8777–8785. [Google Scholar] [CrossRef]
- Vasa, S.M.; Guex, N.; Wilkinson, K.A.; Weeks, K.M.; Giddings, M.C. ShapeFinder: A Software System for High-Throughput Quantitative Analysis of Nucleic Acid Reactivity Information Resolved by Capillary Electrophoresis. RNA 2008, 14, 1979–1990. [Google Scholar] [CrossRef]
- Mitra, S.; Shcherbakova, I.V.; Altman, R.B.; Brenowitz, M.; Laederach, A. High-Throughput Single-Nucleotide Structural Mapping by Capillary Automated Footprinting Analysis. Nucleic Acids Res. 2008, 36, e63. [Google Scholar] [CrossRef]
- Yoon, S.; Kim, J.; Hum, J.; Kim, H.; Park, S.; Kladwang, W.; Das, R. HiTRACE: High-Throughput Robust Analysis for Capillary Electrophoresis. Bioinformatics 2011, 27, 1798–1805. [Google Scholar] [CrossRef]
- Pang, P.S.; Elazar, M.; Pham, E.A.; Glenn, J.S. Simplified RNA Secondary Structure Mapping by Automation of SHAPE Data Analysis. Nucleic Acids Res. 2011, 39, e151. [Google Scholar] [CrossRef]
- Karabiber, F.; McGinnis, J.L.; Favorov, O.V.; Weeks, K.M. QuShape: Rapid, Accurate, and Best-Practices Quantification of Nucleic Acid Probing Information, Resolved by Capillary Electrophoresis. RNA 2013, 19, 63–73. [Google Scholar] [CrossRef]
- Cantara, W.A.; Hatterschide, J.; Wu, W.; Musier-Forsyth, K. RiboCAT: A New Capillary Electrophoresis Data Analysis Tool for Nucleic Acid Probing. RNA 2017, 23, 240–249. [Google Scholar] [CrossRef]
- Gumna, J.; Zok, T.; Figurski, K.; Pachulska-Wieczorek, K.; Szachniuk, M. RNAthor—Fast, Accurate Normalization, Visualization and Statistical Analysis of RNA Probing Data Resolved by Capillary Electrophoresis. PLoS ONE 2020, 15, e0239287. [Google Scholar] [CrossRef]
- Wirecki, T.K.; Merdas, K.; Bernat, A.; Boniecki, M.J.; Bujnicki, J.M.; Stefaniak, F. RNAProbe: A Web Server for Normalization and Analysis of RNA Structure Probing Data. Nucleic Acids Res. 2020, 48, W292–W299. [Google Scholar] [CrossRef]
- Poulsen, L.D.; Kielpinski, L.J.; Salama, S.R.; Krogh, A.; Vinther, J. SHAPE Selection (SHAPES) Enrich for RNA Structure Signal in SHAPE Sequencing-Based Probing Data. RNA 2015, 21, 1042–1052. [Google Scholar] [CrossRef]
- Spitale, R.C.; Flynn, R.A.; Zhang, Q.C.; Crisalli, P.; Lee, B.; Jung, J.-W.; Kuchelmeister, H.Y.; Batista, P.J.; Torre, E.A.; Kool, E.T.; et al. Structural Imprints in Vivo Decode RNA Regulatory Mechanisms. Nature 2015, 519, 486–490. [Google Scholar] [CrossRef]
- Wang, P.Y.; Sexton, A.N.; Culligan, W.J.; Simon, M.D. Carbodiimide Reagents for the Chemical Probing of RNA Structure in Cells. RNA 2019, 25, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Feng, C.; Chan, D.; Joseph, J.; Muuronen, M.; Coldren, W.H.; Dai, N.; Corrêa, I.R.; Furche, F.; Hadad, C.M.; Spitale, R.C. Light-Activated Chemical Probing of Nucleobase Solvent Accessibility Inside Cells. Nat. Chem. Biol. 2018, 14, 276–283. [Google Scholar] [CrossRef]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and Secondary Structure of an Entire HIV-1 RNA Genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Burrill, C.P.; Westesson, O.; Schulte, M.B.; Strings, V.R.; Segal, M.; Andino, R. Global RNA Structure Analysis of Poliovirus Identifies a Conserved RNA Structure Involved in Viral Replication and Infectivity. J. Virol. 2013, 87, 11670–11683. [Google Scholar] [CrossRef]
- Pirakitikulr, N.; Kohlway, A.; Lindenbach, B.D.; Pyle, A.M. The Coding Region of the HCV Genome Contains a Network of Regulatory RNA Structures. Mol. Cell 2016, 62, 111–120. [Google Scholar] [CrossRef]
- Cordero, P.; Kladwang, W.; VanLang, C.C.; Das, R. Quantitative DMS Mapping for Automated RNA Secondary Structure Inference. Biochemistry 2012, 51, 7037–7039. [Google Scholar] [CrossRef]
- Maurin, T.; Melko, M.; Abekhoukh, S.; Khalfallah, O.; Davidovic, L.; Jarjat, M.; D’Antoni, S.; Catania, M.V.; Moine, H.; Bechara, E.; et al. The FMRP/GRK4 MRNA Interaction Uncovers a New Mode of Binding of the Fragile X Mental Retardation Protein in Cerebellum. Nucleic Acids Res. 2015, 43, 8540–8550. [Google Scholar] [CrossRef] [PubMed]
- Alghoul, F.; Eriani, G.; Martin, F. RNA Secondary Structure Study by Chemical Probing Methods Using DMS and CMCT. In Small Non-Coding RNAs: Methods and Protocols; Methods in Molecular Biology; Rederstorff, M., Ed.; Springer: New York, NY, USA, 2021; pp. 241–250. ISBN 978-1-07-161386-3. [Google Scholar]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-Wide Measurement of RNA Secondary Structure in Yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef]
- Underwood, J.G.; Uzilov, A.V.; Katzman, S.; Onodera, C.S.; Mainzer, J.E.; Mathews, D.H.; Lowe, T.M.; Salama, S.R.; Haussler, D. FragSeq: Transcriptome-Wide RNA Structure Probing Using High-Throughput Sequencing. Nat. Methods 2010, 7, 995–1001. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Chang, H.Y. Genome-Wide Mapping of RNA Structure Using Nuclease Digestion and High-Throughput Sequencing. Nat. Protoc. 2013, 8, 849–869. [Google Scholar] [CrossRef]
- Wan, Y.; Qu, K.; Ouyang, Z.; Kertesz, M.; Li, J.; Tibshirani, R.; Makino, D.L.; Nutter, R.C.; Segal, E.; Chang, H.Y. Genome-Wide Measurement of RNA Folding Energies. Mol. Cell 2012, 48, 169–181. [Google Scholar] [CrossRef]
- Saus, E.; Willis, J.R.; Pryszcz, L.P.; Hafez, A.; Llorens, C.; Himmelbauer, H.; Gabaldón, T. NextPARS: Parallel Probing of RNA Structures in Illumina. RNA 2018, 24, 609–619. [Google Scholar] [CrossRef]
- Lucks, J.B.; Mortimer, S.A.; Trapnell, C.; Luo, S.; Aviran, S.; Schroth, G.P.; Pachter, L.; Doudna, J.A.; Arkin, A.P. Multiplexed RNA Structure Characterization with Selective 2′-Hydroxyl Acylation Analyzed by Primer Extension Sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci. USA 2011, 108, 11063–11068. [Google Scholar] [CrossRef]
- Loughrey, D.; Watters, K.E.; Settle, A.H.; Lucks, J.B. SHAPE-Seq 2.0: Systematic Optimization and Extension of High-Throughput Chemical Probing of RNA Secondary Structure with next Generation Sequencing. Nucleic Acids Res. 2014, 42, e165. [Google Scholar] [CrossRef]
- Li, P.; Zhou, X.; Xu, K.; Zhang, Q.C. RASP: An Atlas of Transcriptome-Wide RNA Secondary Structure Probing Data. Nucleic Acids Res. 2021, 49, D183–D191. [Google Scholar] [CrossRef]
- Kielpinski, L.J.; Vinther, J. Massive Parallel-Sequencing-Based Hydroxyl Radical Probing of RNA Accessibility. Nucleic Acids Res. 2014, 42, e70. [Google Scholar] [CrossRef][Green Version]
- Ding, Y.; Tang, Y.; Kwok, C.K.; Zhang, Y.; Bevilacqua, P.C.; Assmann, S.M. In Vivo Genome-Wide Profiling of RNA Secondary Structure Reveals Novel Regulatory Features. Nature 2014, 505, 696–700. [Google Scholar] [CrossRef]
- Rouskin, S.; Zubradt, M.; Washietl, S.; Kellis, M.; Weissman, J.S. Genome-Wide Probing of RNA Structure Reveals Active Unfolding of MRNA Structures in Vivo. Nature 2014, 505, 701–705. [Google Scholar] [CrossRef] [PubMed]
- Talkish, J.; May, G.; Lin, Y.; Woolford, J.L.; McManus, C.J. Mod-Seq: High-Throughput Sequencing for Chemical Probing of RNA Structure. RNA 2014, 20, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Incarnato, D.; Neri, F.; Anselmi, F.; Oliviero, S. Genome-Wide Profiling of Mouse RNA Secondary Structures Reveals Key Features of the Mammalian Transcriptome. Genome Biol. 2014, 15, 491. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.I.; Clark, W.C.; Pan, D.W.; Eckwahl, M.J.; Dai, Q.; Pan, T. Pseudouridines Have Context-Dependent Mutation and Stop Rates in High-Throughput Sequencing. RNA Biol. 2018, 15, 892–900. [Google Scholar] [CrossRef]
- Mitchell, D.; Renda, A.J.; Douds, C.A.; Babitzke, P.; Assmann, S.M.; Bevilacqua, P.C. In Vivo RNA Structural Probing of Uracil and Guanine Base-Pairing by 1-Ethyl-3-(3-Dimethylaminopropyl)Carbodiimide (EDC). RNA 2019, 25, 147–157. [Google Scholar] [CrossRef]
- Twittenhoff, C.; Brandenburg, V.B.; Righetti, F.; Nuss, A.M.; Mosig, A.; Dersch, P.; Narberhaus, F. Lead-Seq: Transcriptome-Wide Structure Probing in Vivo Using Lead(II) Ions. Nucleic Acids Res. 2020, 48, e71. [Google Scholar] [CrossRef]
- Métifiot, M.; Amrane, S.; Litvak, S.; Andreola, M.-L. G-Quadruplexes in Viruses: Function and Potential Therapeutic Applications. Nucleic Acids Res. 2014, 42, 12352–12366. [Google Scholar] [CrossRef]
- Lavezzo, E.; Berselli, M.; Frasson, I.; Perrone, R.; Palù, G.; Brazzale, A.R.; Richter, S.N.; Toppo, S. G-Quadruplex Forming Sequences in the Genome of All Known Human Viruses: A Comprehensive Guide. PLoS Comput. Biol. 2018, 14, e1006675. [Google Scholar] [CrossRef]
- Lyu, K.; Chow, E.Y.-C.; Mou, X.; Chan, T.-F.; Kwok, C.K. RNA G-Quadruplexes (RG4s): Genomics and Biological Functions. Nucleic Acids Res. 2021, 49, 5426–5450. [Google Scholar] [CrossRef]
- Kwok, C.K.; Balasubramanian, S. Targeted Detection of G-Quadruplexes in Cellular RNAs. Angew Chem. Int. Ed. 2015, 54, 6751–6754. [Google Scholar] [CrossRef]
- Kwok, C.K.; Marsico, G.; Sahakyan, A.B.; Chambers, V.S.; Balasubramanian, S. RG4-Seq Reveals Widespread Formation of G-Quadruplex Structures in the Human Transcriptome. Nat. Methods 2016, 13, 841–844. [Google Scholar] [CrossRef]
- Yang, X.; Cheema, J.; Zhang, Y.; Deng, H.; Duncan, S.; Umar, M.I.; Zhao, J.; Liu, Q.; Cao, X.; Kwok, C.K.; et al. RNA G-Quadruplex Structures Exist and Function in Vivo in Plants. Genome Biol. 2020, 21, 226. [Google Scholar] [CrossRef]
- Yang, S.Y.; Lejault, P.; Chevrier, S.; Boidot, R.; Robertson, A.G.; Wong, J.M.Y.; Monchaud, D. Transcriptome-Wide Identification of Transient RNA G-Quadruplexes in Human Cells. Nat. Commun. 2018, 9, 4730. [Google Scholar] [CrossRef]
- Weng, X.; Gong, J.; Chen, Y.; Wu, T.; Wang, F.; Yang, S.; Yuan, Y.; Luo, G.; Chen, K.; Hu, L.; et al. Keth-Seq for Transcriptome-Wide RNA Structure Mapping. Nat. Chem. Biol. 2020, 16, 489–492. [Google Scholar] [CrossRef]
- Sun, L.; Li, P.; Ju, X.; Rao, J.; Huang, W.; Ren, L.; Zhang, S.; Xiong, T.; Xu, K.; Zhou, X.; et al. In Vivo Structural Characterization of the SARS-CoV-2 RNA Genome Identifies Host Proteins Vulnerable to Repurposed Drugs. Cell 2021, 184, 1865–1883.e20. [Google Scholar] [CrossRef]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.E.; Weeks, K.M. RNA Motif Discovery by SHAPE and Mutational Profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [Google Scholar] [CrossRef]
- Smola, M.J.; Rice, G.M.; Busan, S.; Siegfried, N.A.; Weeks, K.M. Selective 2’-Hydroxyl Acylation Analyzed by Primer Extension and Mutational Profiling (SHAPE-MaP) for Direct, Versatile and Accurate RNA Structure Analysis. Nat. Protoc. 2015, 10, 1643–1669. [Google Scholar] [CrossRef]
- Zubradt, M.; Gupta, P.; Persad, S.; Lambowitz, A.M.; Weissman, J.S.; Rouskin, S. DMS-MaPseq for Genome-Wide or Targeted RNA Structure Probing in Vivo. Nat. Methods 2017, 14, 75–82. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA Methylomes at Base Resolution Show Widespread Epigenomic Differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef]
- Zinshteyn, B.; Chan, D.; England, W.; Feng, C.; Green, R.; Spitale, R.C. Assaying RNA Structure with LASER-Seq. Nucleic Acids Res. 2019, 47, 43–55. [Google Scholar] [CrossRef]
- Marinus, T.; Fessler, A.B.; Ogle, C.A.; Incarnato, D. A Novel SHAPE Reagent Enables the Analysis of RNA Structure in Living Cells with Unprecedented Accuracy. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Homan, P.J.; Favorov, O.V.; Lavender, C.A.; Kursun, O.; Ge, X.; Busan, S.; Dokholyan, N.V.; Weeks, K.M. Single-Molecule Correlated Chemical Probing of RNA. Proc. Natl. Acad. Sci. USA 2014, 111, 13858–13863. [Google Scholar] [CrossRef] [PubMed]
- Krokhotin, A.; Mustoe, A.M.; Weeks, K.M.; Dokholyan, N.V. Direct Identification of Base-Paired RNA Nucleotides by Correlated Chemical Probing. RNA 2017, 23, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Mustoe, A.M.; Lama, N.N.; Irving, P.S.; Olson, S.W.; Weeks, K.M. RNA Base-Pairing Complexity in Living Cells Visualized by Correlated Chemical Probing. Proc. Natl. Acad. Sci. USA 2019, 116, 24574–24582. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive Tertiary Structure in the Dengue Virus RNA Genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513–11518. [Google Scholar] [CrossRef]
- Sengupta, A.; Rice, G.M.; Weeks, K.M. Single-Molecule Correlated Chemical Probing Reveals Large-Scale Structural Communication in the Ribosome and the Mechanism of the Antibiotic Spectinomycin in Living Cells. PLoS Biol. 2019, 17, e3000393. [Google Scholar] [CrossRef]
- Ehrhardt, J.E.; Weeks, K.M. Time-Resolved, Single-Molecule, Correlated Chemical Probing of RNA. J. Am. Chem. Soc. 2020, 142, 18735–18740. [Google Scholar] [CrossRef]
- Tomezsko, P.J.; Corbin, V.D.A.; Gupta, P.; Swaminathan, H.; Glasgow, M.; Persad, S.; Edwards, M.D.; Mcintosh, L.; Papenfuss, A.T.; Emery, A.; et al. Determination of RNA Structural Diversity and Its Role in HIV-1 RNA Splicing. Nature 2020, 582, 438–442. [Google Scholar] [CrossRef]
- Kladwang, W.; Cordero, P.; Das, R. A Mutate-and-Map Strategy Accurately Infers the Base Pairs of a 35-Nucleotide Model RNA. RNA 2011, 17, 522–534. [Google Scholar] [CrossRef]
- Kladwang, W.; VanLang, C.C.; Cordero, P.; Das, R. A Two-Dimensional Mutate-and-Map Strategy for Non-Coding RNA Structure. Nat. Chem. 2011, 3, 954–962. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Kladwang, W.; Yesselman, J.D.; Das, R. RNA Structure Inference through Chemical Mapping after Accidental or Intentional Mutations. Proc. Natl. Acad. Sci. USA 2017, 114, 9876–9881. [Google Scholar] [CrossRef]
- Weidmann, C.A.; Mustoe, A.M.; Jariwala, P.B.; Calabrese, J.M.; Weeks, K.M. Analysis of RNA-Protein Networks with RNP-MaP Defines Functional Hubs on RNA. Nat. Biotechnol. 2021, 39, 347–356. [Google Scholar] [CrossRef]
- Dadonaite, B.; Gilbertson, B.; Knight, M.L.; Trifkovic, S.; Rockman, S.; Laederach, A.; Brown, L.E.; Fodor, E.; Bauer, D.L.V. The Structure of the Influenza A Virus Genome. Nat. Microbiol. 2019, 4, 1781–1789. [Google Scholar] [CrossRef]
- Simon, L.M.; Morandi, E.; Luganini, A.; Gribaudo, G.; Martinez-Sobrido, L.; Turner, D.H.; Oliviero, S.; Incarnato, D. In Vivo Analysis of Influenza A MRNA Secondary Structures Identifies Critical Regulatory Motifs. Nucleic Acids Res. 2019, 47, 7003–7017. [Google Scholar] [CrossRef]
- Madden, E.A.; Plante, K.S.; Morrison, C.R.; Kutchko, K.M.; Sanders, W.; Long, K.M.; Taft-Benz, S.; Cruz Cisneros, M.C.; White, A.M.; Sarkar, S.; et al. Using SHAPE-MaP To Model RNA Secondary Structure and Identify 3′UTR Variation in Chikungunya Virus. J. Virol. 2020, 94, e00701-20. [Google Scholar] [CrossRef]
- Manfredonia, I.; Nithin, C.; Ponce-Salvatierra, A.; Ghosh, P.; Wirecki, T.K.; Marinus, T.; Ogando, N.S.; Snijder, E.J.; van Hemert, M.J.; Bujnicki, J.M.; et al. Genome-Wide Mapping of SARS-CoV-2 RNA Structures Identifies Therapeutically-Relevant Elements. Nucleic Acids Res. 2020, 48, 12436–12452. [Google Scholar] [CrossRef]
- Huston, N.C.; Wan, H.; Strine, M.S.; de Cesaris Araujo Tavares, R.; Wilen, C.B.; Pyle, A.M. Comprehensive in Vivo Secondary Structure of the SARS-CoV-2 Genome Reveals Novel Regulatory Motifs and Mechanisms. Mol. Cell 2021, 81, 584–598.e5. [Google Scholar] [CrossRef]
- Lan, T.C.; Allan, M.F.; Malsick, L.; Khandwala, S.; Nyeo, S.S.; Sun, Y.; Guo, J.U.; Bathe, M.; Griffiths, A.; Rouskin, S. Insights into the Secondary Structural Ensembles of the Full SARS-CoV-2 RNA Genome in Infected Cells. Biorxiv 2021. [Google Scholar] [CrossRef]
- Zhao, J.; Qiu, J.; Aryal, S.; Hackett, J.L.; Wang, J. The RNA Architecture of the SARS-CoV-2 3’-Untranslated Region. Viruses 2020, 12, 1473. [Google Scholar] [CrossRef]
- Christy, T.W.; Giannetti, C.A.; Houlihan, G.; Smola, M.J.; Rice, G.M.; Wang, J.; Dokholyan, N.V.; Laederach, A.; Holliger, P.; Weeks, K.M. Direct Mapping of Higher-Order RNA Interactions by SHAPE-JuMP. Biochemistry 2021, 60, 1971–1982. [Google Scholar] [CrossRef]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Gong, J.; Zhang, Q.C. PARIS: Psoralen Analysis of RNA Interactions and Structures with High Throughput and Resolution. Methods Mol. Biol. 2018, 1649, 59–84. [Google Scholar] [CrossRef] [PubMed]
- Ziv, O.; Gabryelska, M.M.; Lun, A.T.L.; Gebert, L.F.R.; Sheu-Gruttadauria, J.; Meredith, L.W.; Liu, Z.-Y.; Kwok, C.K.; Qin, C.-F.; MacRae, I.J.; et al. COMRADES Determines in Vivo RNA Structures and Interactions. Nat. Methods 2018, 15, 785–788. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Chou, F.-C.; Kladwang, W.; Tian, S.; Cordero, P.; Das, R. Consistent Global Structures of Complex RNA States through Multidimensional Chemical Mapping. eLife 2015, 4, e07600. [Google Scholar] [CrossRef]
- Aw, J.G.A.; Lim, S.W.; Wang, J.X.; Lambert, F.R.P.; Tan, W.T.; Shen, Y.; Zhang, Y.; Kaewsapsak, P.; Li, C.; Ng, S.B.; et al. Determination of Isoform-Specific RNA Structure with Nanopore Long Reads. Nat. Biotechnol. 2021, 39, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Cubi, R.; Bouhedda, F.; Collot, M.; Klymchenko, A.S.; Ryckelynck, M. ΜIVC-Useq: A Microfluidic-Assisted High-Throughput Functional Screening in Tandem with next-Generation Sequencing and Artificial Neural Network to Rapidly Characterize RNA Molecules. RNA 2021, 27, 841–853. [Google Scholar] [CrossRef]


| Probe | Target | In Cell-In Viro | Original Publication (* Related to Nucleic Acid) | Early Use as Structural Probe | Application to Viral RNA |
|---|---|---|---|---|---|
| Enzymatic | |||||
| RNase A | ssC and U | No | Markham and Smith (1952) * [58] | tRNAAla (1965) [21] | HDV (1991) [59] |
| RNase T1 | ssG | No | Sato and Egami (1957) * [22] | tRNAAla (1965) [21] | TYMV (1982) [60,61] |
| Nuclease S1 | ssN | No | Harada and Dahlberg (1975) * [31] | 5S rRNA (1977) [62] | Bromoviruses (1981) [63] |
| RNase CL3 | ssC>>A>U | No | Levy and Karpetsky (1980) * [33] | TYMV RNA (1982) [60] | TYMV (1982) [60] |
| RNase V1 | 4–6 nts in helices or stacked nts | No | Favorova et al. (1981) * [38] | 5S rRNA (1982) [64] | TYMV (1982) [60,61] |
| RNase T2 | ssA>ssN | No | Sato and Egami (1957) * [17] | yeast tRNA (1984) [32] | Poliovirus (1989) [65] |
| N. crassa endonuclease | ssN | No | Linn and Lehman (1965) [30] | tRNATrp (1984) [66] | HIV-1 (1993) [67] |
| RNase U2 | ssA>G>>C>U | No | Uchida et al. (1970) * [34] | 16S rRNA (1987) [68] | HDV (1991) [59] |
| Chemical | |||||
| Kethoxal | N1 and N2- ssG | No | Staehelin (1959) * [41] | Yeast tRNA (1969) [42] | EBV (1988) [69] |
| DMS | N7-G/N1-ssA N3-ssC | Yes | Lawley and Brookes (1963) * [26] | Yeast tRNAPhe (1980) [45] | TYMV (1982) [61] |
| DEPC | N7-A | No | Oberg (1970) * [43] | Yeast tRNAPhe (1980) [45] | TYMV (1982) [61] |
| ENU | Phosphates of ssN and dsN | No | Singer (1976) * [49] | tRNAPhe (1980) [50] | TYMV (1982) [60] |
| CMCT | N3-U and N1-G ss | No | Augusti and Brown (1965) * [28] | 16S rRNA (1986) [46] | EBV (1988) [69] |
| MPE-Fe(II) (•OH source) | Phosphate/sugar backbone dsN | No | Hertzberg and Dervan (1982) * [70] | tRNAPhe (1984) [52] | HIV-2 (2013) [71] |
| Fe-EDTA (•OH source) | Solvent accessibility | Yes | Tullius and Dombroski (1986) * [72] | pre-mRNA (1989) [51] | HIV-1 (1999) [73] |
| Lead (II) | Phosphate/sugar backbone ssN | Yes | Werner et al. (1976) * [74] | 16S rRNA (1989) [75] | HIV-1 (1997) [76] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gilmer, O.; Quignon, E.; Jousset, A.-C.; Paillart, J.-C.; Marquet, R.; Vivet-Boudou, V. Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond. Viruses 2021, 13, 1894. https://doi.org/10.3390/v13101894
Gilmer O, Quignon E, Jousset A-C, Paillart J-C, Marquet R, Vivet-Boudou V. Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond. Viruses. 2021; 13(10):1894. https://doi.org/10.3390/v13101894
Chicago/Turabian StyleGilmer, Orian, Erwan Quignon, Anne-Caroline Jousset, Jean-Christophe Paillart, Roland Marquet, and Valérie Vivet-Boudou. 2021. "Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond" Viruses 13, no. 10: 1894. https://doi.org/10.3390/v13101894
APA StyleGilmer, O., Quignon, E., Jousset, A.-C., Paillart, J.-C., Marquet, R., & Vivet-Boudou, V. (2021). Chemical and Enzymatic Probing of Viral RNAs: From Infancy to Maturity and Beyond. Viruses, 13(10), 1894. https://doi.org/10.3390/v13101894







