Evaluating RNA Structural Flexibility: Viruses Lead the Way
Abstract
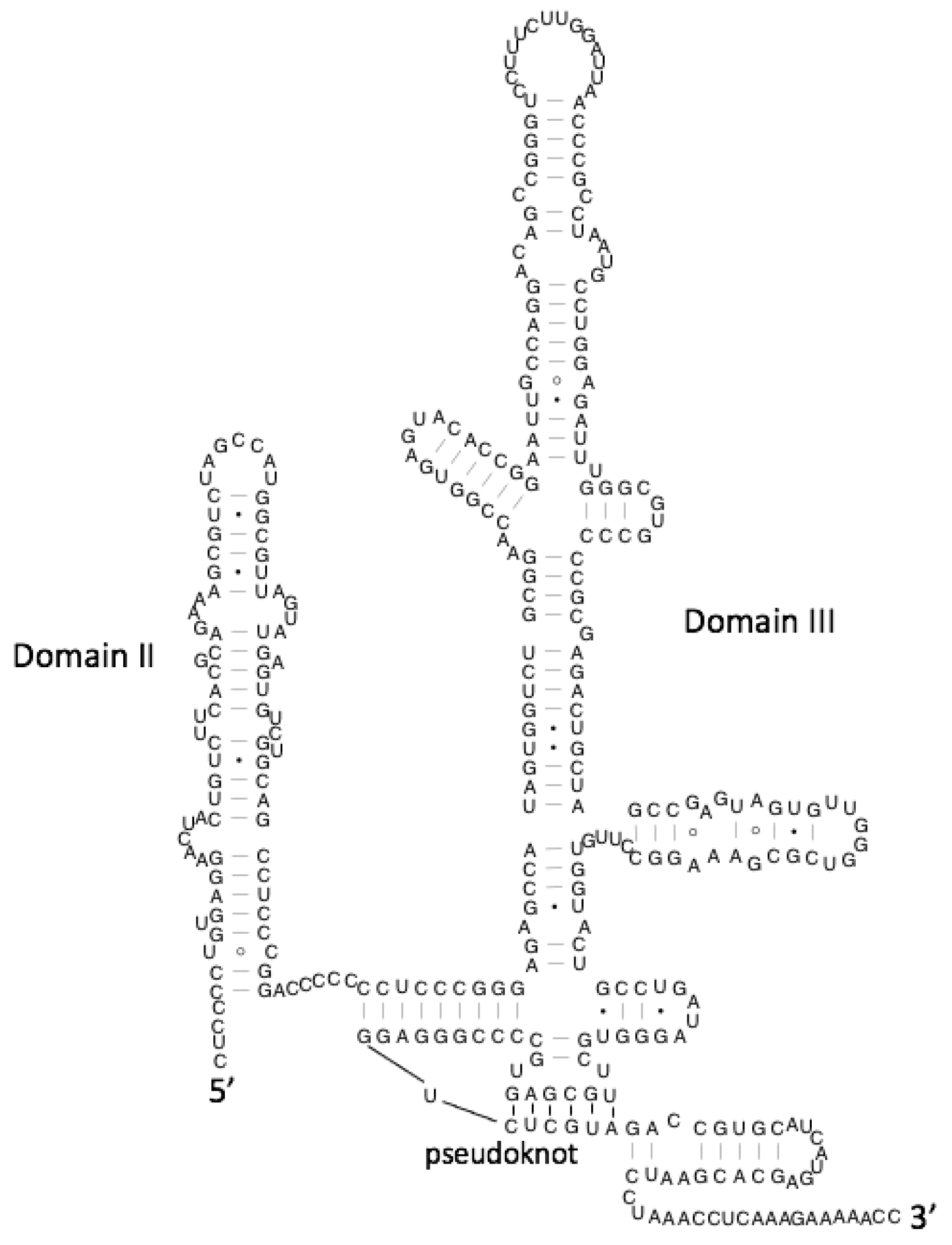
:1. Introduction
2. Analysis of RNA Structure and Flexibility
2.1. SHAPE and Other Secondary Structure Probing Techniques
2.2. Proximity Ligation
| Technique | Distinguishing Features | Reference |
|---|---|---|
| Psoralen cross-linked, ligated, and selected hybrids (PARIS) | Uses 2-dimensional gel electrophoresis to enrich cross linked fragments | [32] |
| Sequencing of psoralen cross-linked, ligated, and selected hybrids (SPLASH) | Biotin-streptavidin enrichment | [33] |
| Ligation of interacting RNA (LIGR) | RNase digestion enriches fragments | [34] |
| Dual crosslinking, immunoprecipitation, and proximity ligation (2CIMPL) | Enriches fragment using magnetic bead tethering via an anti-NP antibody, also contains an additional cross-linking step before introduction of AMT | [31] |
| Cross-linking of matched RNAs and deep sequencing (COMRADES) | Biotin-streptavidin enrichment | [35] |
| RNA proximity ligation (RPL) | Omits psoralen cross linking step, instead relying on spatial proximity of bases in RNA secondary structures to facilitate chimera formation | [36] |
2.3. NMR
2.4. Sm-FRET
2.5. SAXS
2.6. Other Techniques
3. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hagerman, P.J. FLEXIBILITY OF RNA. Annu. Rev. Biophys. Biomol. Struct. 1997, 26, 139–156. [Google Scholar] [CrossRef]
- Zwieb, C. The Principles of RNA Structure Architecture. Adv. Struct. Saf. Stud. 2013, 1097, 33–43. [Google Scholar] [CrossRef]
- Marin-Gonzalez, A.; Vilhena, J.G.; Moreno-Herrero, F.; Perez, R. Sequence-dependent mechanical properties of double-stranded RNA. Nanoscale 2019, 11, 21471–21478. [Google Scholar] [CrossRef] [Green Version]
- van der Kuyl, A.C.; Berkhout, B. The biased nucleotide composition of the HIV genome: A constant factor in a highly variable virus. Retrovirology 2012, 9, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Hemert, F.; van der Kuyl, A.C.; Berkhout, B. Impact of the biased nucleotide composition of viral RNA genomes on RNA structure and codon usage. J. Gen. Virol. 2016, 97, 2608–2619. [Google Scholar] [CrossRef] [PubMed]
- Olson, W.K.; Li, S.; Kaukonen, T.; Colasanti, A.V.; Xin, Y.; Lu, X.-J. Effects of Noncanonical Base Pairing on RNA Folding: Structural Context and Spatial Arrangements of G·A Pairs. Biochemistry 2019, 2019. 58, 2474–2487. [Google Scholar] [CrossRef]
- Holbrook, S.R. RNA structure: The long and the short of it. Curr. Opin. Struct. Biol. 2005, 15, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Reiter, N.J.; Chan, C.W.; Mondragón, A. Emerging structural themes in large RNA molecules. Curr. Opin. Struct. Biol. 2011, 21, 319–326. [Google Scholar] [CrossRef] [Green Version]
- Moore, P.B. Structural Motifs in RNA. Annu. Rev. Biochem. 1999, 68, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Haque, F.; Pi, F.; Zhao, Z.; Gu, S.; Hu, H.; Yu, H.; Guo, P. RNA versatility, flexibility, and thermostability for practice in RNA nanotechnology and biomedical applications. Wiley Interdiscip. Rev. RNA 2018, 9, e1452. [Google Scholar] [CrossRef]
- Tang, D.-J.; Du, X.; Shi, Q.; Zhang, J.-L.; He, Y.-P.; Chen, Y.-M.; Ming, Z.; Wang, D.; Zhong, W.-Y.; Liang, Y.-W.; et al. A SAM-I riboswitch with the ability to sense and respond to uncharged initiator tRNA. Nat. Commun. 2020, 11, 1–11. [Google Scholar] [CrossRef]
- Kim, H.D.; Nienhaus, G.U.; Ha, T.; Orr, J.W.; Williamson, J.; Chu, S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc. Natl. Acad. Sci. USA 2002, 99, 4284–4289. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.; Merriman, D.K.; Choi, S.H.; Schumacher, M.A.; Plangger, R.; Kreutz, C.; Horner, S.M.; Meyer, K.; Al-Hashimi, H.M. A potentially abundant junctional RNA motif stabilized by m6A and Mg2+. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Shajani, Z.; Deka, P.; Varani, G. Decoding RNA motional codes. Trends Biochem. Sci. 2006, 31, 421–424. [Google Scholar] [CrossRef]
- Kligun, E.; Mandel-Gutfreund, Y. The role of RNA conformation in RNA-protein recognition. RNA Biol. 2015, 12, 720–727. [Google Scholar] [CrossRef] [Green Version]
- Hill, A.; Bartley, L.E.; Schroeder, S.J. Prohead RNA: A noncoding viral RNA of novel structure and function. Wiley Interdiscip. Rev. RNA 2016, 7, 428–437. [Google Scholar] [CrossRef] [Green Version]
- Rangan, R.; Zheludev, I.N.; Hagey, R.J.; Pham, E.A.; Wayment-Steele, H.K.; Glenn, J.S.; Das, R. RNA genome conservation and secondary structure in SARS-CoV-2 and SARS-related viruses: A first look. RNA 2016, 26, 937–959. [Google Scholar] [CrossRef]
- Wilkinson, K.A.; Merino, E.J.; Weeks, K.M. Selective 2’-hydroxyl acylation analyzed by primer extension (SHAPE): Quanti-tative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006, 1, 1610–1616. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nat. Cell Biol. 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkinson, K.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-Throughput SHAPE Analysis Reveals Structures in HIV-1 Genomic RNA Strongly Conserved across Distinct Biological States. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenyon, J.C.; Prestwood, L.J.; Le Grice, S.F.J.; Lever, A.M.L. In-gel probing of individual RNA conformers within a mixed population reveals a dimerization structural switch in the HIV-1 leader. Nucleic Acids Res. 2013, 41, e174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spitale, R.C.; Crisalli, P.; Flynn, R.A.; Torre, E.A.; Kool, E.T.; Chang, H.Y. RNA SHAPE analysis in living cells. Nat. Chem. Biol. 2013, 9, 18–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grohman, J.K.; Gorelick, R.J.; Kottegoda, S.; Allbritton, N.L.; Rein, A.; Weeks, K.M. An Immature Retroviral RNA Genome Resembles a Kinetically Trapped Intermediate State. J. Virol. 2014, 88, 6061–6068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.; Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Huston, N.C.; Wan, H.; Strine, M.S.; de Cesaris Araujo Tavares, R.; Wilen, C.B.; Pyle, A.M. Comprehensive in vivo secondary structure of the SARS-CoV-2 genome reveals novel regulatory motifs and mechanisms. Mol. Cell 2021, 81, 584–598.e5. [Google Scholar] [CrossRef]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513–11518. [Google Scholar] [CrossRef] [Green Version]
- Huber, R.G.; Ni Lim, X.; Ng, W.C.; Sim, A.Y.L.; Poh, H.X.; Shen, Y.; Lim, S.Y.; Sundstrom, K.B.; Sun, X.; Aw, J.G.; et al. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kenyon, J.C.; Prestwood, L.J.; Lever, A.M.L. A novel combined RNA-protein interaction analysis distinguishes HIV-1 Gag protein binding sites from structural change in the viral RNA leader. Sci. Rep. 2015, 5, srep14369. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wei, Y.; Mei, M.; Tang, L.; Sun, L.; Huang, W.; Zhou, J.; Zou, C.; Zhang, S.; Qin, C.-F.; et al. Integrative Analysis of Zika Virus Genome RNA Structure Reveals Critical Determinants of Viral Infectivity. Cell Host Microbe 2018, 24, 875–886.e5. [Google Scholar] [CrossRef] [Green Version]
- Granneman, S.; Kudla, G.; Petfalski, E.; Tollervey, D. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 9613–9618. [Google Scholar] [CrossRef] [Green Version]
- Le Sage, V.; Kanarek, J.P.; Snyder, D.J.; Cooper, V.S.; Lakdawala, S.S.; Lee, N. Mapping of Influenza Virus RNA-RNA Interactions Reveals a Flexible Network. Cell Rep. 2020, 31, 107823. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, Q.C.; Lee, B.; Flynn, R.; Smith, M.; Robinson, J.T.; Davidovich, C.; Gooding, A.R.; Goodrich, K.J.; Mattick, J.; et al. RNA Duplex Map in Living Cells Reveals Higher-Order Transcriptome Structure. Cell 2016, 165, 1267–1279. [Google Scholar] [CrossRef] [Green Version]
- Aw, J.G.; Shen, Y.; Wilm, A.; Sun, M.; Lim, X.N.; Boon, K.L.; Tapsin, S.; Chan, Y.S.; Tan, C.P.; Sim, A.Y.; et al. In Vivo Mapping of Eukaryotic RNA Interactomes Reveals Principles of Higher-Order Organization and Regulation. Mol. Cell 2016, 62, 603–617. [Google Scholar] [CrossRef] [Green Version]
- Sharma, E.; Sterne-Weiler, T.; O’Hanlon, D.; Blencowe, B.J. Global Mapping of Human RNA-RNA Interactions. Mol. Cell 2016, 62, 618–626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziv, O.; Gabryelska, M.M.; Lun, A.T.L.; Gebert, L.F.; Sheu-Gruttadauria, J.; Meredith, L.W.; Liu, Z.-Y.; Kwok, C.K.; Qin, C.-F.; MacRae5, I.J.; et al. COMRADES determines in vivo RNA structures and interactions. Nat. Methods. 2018, 15, 785–788. [Google Scholar] [CrossRef] [PubMed]
- Ramani, V.; Qiu, R.; Shendure, J. High-throughput determination of RNA structure by proximity ligation. Nat. Biotechnol. 2015, 33, 980–984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dadonaite, B.; Gilbertson, B.; Knight, M.L.; Trifkovic, S.; Rockman, S.; Laederach, A.; Brown, L.E.; Fodor, E.; Bauer, D.L.V. The structure of the influenza A virus genome. Nat. Microbiol. 2019, 4, 1781–1789. [Google Scholar] [CrossRef]
- Wacker, A.; Weigand, J.E.; Akabayov, S.R.; Altincekic, N.; Bains, J.K.; Banijamali, E.; Binas, O.; Castillo-Martinez, J.; Cetiner, E.; Ceylan, B.; et al. Secondary structure determination of conserved SARS-CoV-2 RNA elements by NMR spectroscopy. Nucleic Acids Res. 2020, 48, 12415–12435. [Google Scholar] [CrossRef]
- D’Souza, A.R.; Buckingham, A.B.; Salasc, F.; Ingemarsdotter, C.K.; Iaconis, G.; Jarvis, I.; Groom, H.C.T.; Kenyon, J.C.; Lever, A.M.L. Duplex formation between the template and the nascent strand in the transcription-regulating sequences is associated with the site of template switching in SARS-CoV-2. RNA Biol. 2021, 18, 1–9. [Google Scholar] [CrossRef]
- Takahashi, K.-I.; Baba, S.; Hayashi, Y.; Koyanagi, Y.; Yamamoto, N.; Takaku, H.; Kawai, G. NMR analysis of intra- and inter-molecular stems in the dimerization initiation site of the HIV-1 genome. J. Biochem. 2000, 127, 681–686. [Google Scholar] [CrossRef]
- Greatorex, J.; Gallego, J.; Varani, G.; Lever, A. Structure and Stability of Wild-type and Mutant RNA Internal Loops from the SL-1 Domain of the HIV-1 Packaging Signal. J. Mol. Biol. 2002, 322, 543–557. [Google Scholar] [CrossRef]
- Keane, S.C.; Van, V.; Frank, H.; Sciandra, C.A.; McCowin, S.; Santos, J.; Heng, X.; Summers, M.F. NMR detection of intermolecular interaction sites in the dimeric 5′-leader of the HIV-1 genome. Proc. Natl. Acad. Sci. USA 2016, 113, 13033–13038. [Google Scholar] [CrossRef] [Green Version]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR Detection of Structures in the HIV-1 5’-Leader RNA That Regulate Genome Packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gavory, G.; Symmons, M.F.; Ghosh, Y.K.; Klenerman, D.; Balasubramanian, S. Structural Analysis of the Catalytic Core of Human Telomerase RNA by FRET and Molecular Modeling. Biochemistry 2006, 45, 13304–13311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boerneke, M.A.; Hermann, T. Conformational flexibility of viral RNA switches studied by FRET. Methods 2015, 91, 35–39. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, B.M.; E Graham, M.; O′donoghue, Z.; Beckham, J.D.; Kieft, J.S. Three-dimensional structure of a flavivirus dumbbell RNA reveals molecular details of an RNA regulator of replication. Nucleic Acids Res. 2021, 49, 7122–7138. [Google Scholar] [CrossRef] [PubMed]
- Meyer, A.; Freier, M.; Schmidt, T.; Rostowski, K.; Zwoch, J.; Lilie, H.; Behrens, S.-E.; Friedrich, S. An RNA Thermometer Activity of the West Nile Virus Genomic 3′-Terminal Stem-Loop Element Modulates Viral Replication Efficiency during Host Switching. Viruses 2020, 12, 104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, A.; Grosely, R.; Petrov, A.N.; Puglisi, J.D. Dynamics of IRES-mediated translation. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160177. [Google Scholar] [CrossRef]
- Stephenson, J.D.; Li, H.; Kenyon, J.C.; Symmons, M.; Klenerman, D.; Lever, A.M. Three-Dimensional RNA Structure of the Major HIV-1 Packaging Signal Region. Structure 2013, 21, 951–962. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; Hohng, S.; Ha, T. A practical guide to single-molecule FRET. Nat. Methods 2008, 5, 507–516. [Google Scholar] [CrossRef]
- Zhao, R.; Rueda, D. RNA folding dynamics by single-molecule fluorescence resonance energy transfer. Methods 2009, 49, 112–117. [Google Scholar] [CrossRef]
- Fuchs, G.; Petrov, A.N.; Marceau, C.D.; Popov, L.M.; Chen, J.; O’Leary, S.E.; Wang, R.; Carette, J.E.; Sarnow, P.; Puglisi, J.D. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc. Natl. Acad. Sci. USA 2015, 112, 319–325. [Google Scholar] [CrossRef] [Green Version]
- Théobald-Dietrich, A.; De Wijn, R.; Rollet, K.; Bluhm, A.; Rudinger-Thirion, J.; Paulus, C.; Lorber, B.; Thureau, A.; Frugier, M.; Sauter, C. Structural Analysis of RNA by Small-Angle X-ray Scattering. Methods Mol. Biol. 2020, 2113, 189–215. [Google Scholar] [CrossRef]
- Tuukkanen, A.T.; Kleywegt, G.J.; Svergun, D.I. Resolution of ab initio shapes determined from small-angle scattering. IUCrJ 2016, 3, 440–447. [Google Scholar] [CrossRef] [Green Version]
- Gräwert, T.W.; Svergun, D.I. Structural Modeling Using Solution Small-Angle X-ray Scattering (SAXS). J. Mol. Biol. 2020, 432, 3078–3092. [Google Scholar] [CrossRef] [PubMed]
- Bernadó, P.; Mylonas, E.; Petoukhov, M.V.; Blackledge, M.; Svergun, D.I. Structural Characterization of Flexible Proteins Using Small-Angle X-ray Scattering. J. Am. Chem. Soc. 2007, 129, 5656–5664. [Google Scholar] [CrossRef] [PubMed]
- Warden, M.S.; Cai, K.; Cornilescu, G.; Burke, J.E.; Ponniah, K.; Butcher, S.E.; Pascal, S.M. Conformational flexibility in the enterovirus RNA replication platform. RNA 2018, 25, 376–387. [Google Scholar] [CrossRef] [PubMed]
- A Beckham, S.; Matak, M.Y.; Belousoff, M.J.; Venugopal, H.; Shah, N.; Vankadari, N.; Elmlund, H.; Nguyen, J.H.C.; Semler, B.L.; Wilce, M.C.J.; et al. Structure of the PCBP2/stem–loop IV complex underlying translation initiation mediated by the poliovirus type I IRES. Nucleic Acids Res. 2020, 48, 8006–8021. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, R.M.; Kasprzak, W.K.; Longhini, A.P.; Olenginski, L.T.; Abulwerdi, F.; Ginocchio, S.; Shields, B.; Nyman, J.; Svirydava, M.; Del Vecchio, C.; et al. Structural insights of the conserved “priming loop” of hepatitis B virus pre-genomic RNA. J. Bio-Mol. Struct. Dyn. 2021, 1–13. [Google Scholar] [CrossRef]
- Cantero-Camacho, Á.; Fan, L.; Wang, Y.-X.; Gallego, J. Three-dimensional structure of the 3′X-tail of hepatitis C virus RNA in monomeric and dimeric states. RNA 2017, 23, 1465–1476. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Zhang, Y.; Liu, Z.Y. Long non-coding subgenomic flavivirus RNAs have extended 3D structures and are flexible in solution. EMBO Rep. 2019, 20, e47016. [Google Scholar] [CrossRef]
- Sherpa, C.; Grice, S.F.J.L. Structural Fluidity of the Human Immunodeficiency Virus Rev Response Element. Viruses 2020, 12, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bai, Y.; Tambe, A.; Zhou, K.; A Doudna, J. RNA-guided assembly of Rev-RRE nuclear export complexes. eLife 2014, 3, e03656. [Google Scholar] [CrossRef]
- Sherpa, C.; Rausch, J.W.; Le Grice, S.F.; Hammarskjold, M.-L.; Rekosh, D. The HIV-1 Rev response element (RRE) adopts alternative conformations that promote different rates of virus replication. Nucleic Acids Res. 2015, 43, 4676–4686. [Google Scholar] [CrossRef] [PubMed]
- Herzik, M.A.; Wu, M.; Lander, G.C. High-resolution structure determination of sub-100 kDa complexes using conventional cryo-EM. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, T.; Machida, K.; Iwasaki, W.; Shigeta, T.; Nishimoto, M.; Takahashi, M.; Sakamoto, A.; Yonemochi, M.; Harada, Y.; Shigematsu, H.; et al. HCV IRES Captures an Actively Translating 80S Ribosome. Mol. Cell 2019, 74, 1205–1214.e8. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, S.J. Perspectives on Viral RNA Genomes and the RNA Folding Problem. Viruses 2020, 12, 1126. [Google Scholar] [CrossRef] [PubMed]
- Larson, S.B.; McPherson, A. Satellite tobacco mosaic virus RNA: Structure and implications for assembly. Curr. Opin. Struct. Biol. 2001, 11, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Geng, G.; Yu, C.; Li, X.; Yuan, X. A unique internal ribosome entry site representing a dynamic equilibrium state of RNA tertiary structure in the 5′-UTR of Wheat yellow mosaic virus RNA1. Nucleic Acids Res. 2019, 48, 390–404. [Google Scholar] [CrossRef] [PubMed]
- Badmalia, M.D.; Siddiqui, M.Q.; Mrozowich, T.; Gemmill, D.L.; Patel, T.R. Analytical ultracentrifuge: An ideal tool for characterization of non-coding RNAs. Eur. Biophys. J. 2020, 49, 809–818. [Google Scholar] [CrossRef] [PubMed]
- Mrozowich, T.; Henrickson, A.; Demeler, B.; Patel, T.R. Nanoscale Structure Determination of Murray Valley Encephalitis and Powassan Virus Non-Coding RNAs. Viruses 2020, 12, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Brun, E.; Arluison, V.; Wien, F. Application of Synchrotron Radiation Circular Dichroism for RNA Structural Analysis. Methods Mol. Biol. 2020, 2113, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Gaston, H.B.H. Application of NIR Raman Spectroscopy to Probe the Flexibility of RNA Structure. Methods Mol. Biol. 2020, 2113, 149–164. [Google Scholar] [CrossRef] [PubMed]
- Assi, H.A.; Rangadurai, A.K.; Shi, H.; Liu, B.; Clay, M.C.; Erharter, K.; Kreutz, C.; Holley, C.L.; Al-Hashimi, H.M. 2′-O-Methylation can increase the abundance and lifetime of alternative RNA conformational states. Nucleic Acids Res. 2020, 48, 12365–12379. [Google Scholar] [CrossRef] [PubMed]
- Takitou, S.; Taneda, A. Ant colony optimization for predicting RNA folding pathways. Comput. Biol. Chem. 2019, 83, 107118. [Google Scholar] [CrossRef] [PubMed]
- Sieradzan, A.K.; Golon, Ł.; Liwo, A. Prediction of DNA and RNA structure with the NARES-2P force field and conformational space annealing. Phys. Chem. Chem. Phys. 2018, 20, 19656–19663. [Google Scholar] [CrossRef]
- Puton, T.; Kozlowski, L.; Rother, K.M.; Bujnicki, J.M. CompaRNA: A server for continuous benchmarking of automated methods for RNA secondary structure prediction. Nucleic Acids Res. 2013, 41, 4307–4323. [Google Scholar] [CrossRef] [PubMed]
- Boniecki, M.J.; Lach, G.; Dawson, W.; Tomala, K.; Lukasz, P.; Soltysinski, T.; Rother, K.M.; Bujnicki, J.M. SimRNA: A coarse-grained method for RNA folding simulations and 3D structure prediction. Nucleic Acids Res. 2016, 44, e63. [Google Scholar] [CrossRef]
- Miao, Z.; Adamiak, R.W.; Antczak, M.; Boniecki, M.J.; Bujnicki, J.M.; Chen, S.-J.; Cheng, C.Y.; Cheng, Y.; Chou, F.-C.; Das, R.; et al. RNA-Puzzles Round IV: 3D structure predictions of four ribozymes and two aptamers. RNA 2020, 26, 982–995. [Google Scholar] [CrossRef]




| Motif | Characteristics |
|---|---|
| Three-way junction (3WJ) | The junction of three helices. |
| Four-way junction (4WJ) | The junction of four helices. |
| Kink-turn | Helix–internal loop–helix motif with a 50° bend in the helical axis. Two classes, N1 and N3. |
| C-loop | Increased helical twist. |
| Right angle motif | Internal helical angle of 90°. |
| Stem loop | Intramolecular helix with a terminal loop of three to eight nucleotides connecting the two sides of the stem. Tetraloops, where four nucleotides are in the loop, are particularly common, with UNCG and GNRA being especially stable. |
| Paranemic motif | Crossover motif of stacked helices |
| Kissing loops | Base pairing between two helix loops. |
| Ribose zippers | Hydrogen bonding interaction between the 2′OH groups on ribose sugars of unpaired, anti parallel bases. |
| Cross strand purine stacks | Internal loop motif where the six membered rings of two purines stack across two strands, as opposed to classical same strand stacking |
| Bulge-helix-bulge | 4 bp A-form helix with a 3 bp bulge either side. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fairman, C.W.; Lever, A.M.L.; Kenyon, J.C. Evaluating RNA Structural Flexibility: Viruses Lead the Way. Viruses 2021, 13, 2130. https://doi.org/10.3390/v13112130
Fairman CW, Lever AML, Kenyon JC. Evaluating RNA Structural Flexibility: Viruses Lead the Way. Viruses. 2021; 13(11):2130. https://doi.org/10.3390/v13112130
Chicago/Turabian StyleFairman, Connor W., Andrew M. L. Lever, and Julia C. Kenyon. 2021. "Evaluating RNA Structural Flexibility: Viruses Lead the Way" Viruses 13, no. 11: 2130. https://doi.org/10.3390/v13112130
APA StyleFairman, C. W., Lever, A. M. L., & Kenyon, J. C. (2021). Evaluating RNA Structural Flexibility: Viruses Lead the Way. Viruses, 13(11), 2130. https://doi.org/10.3390/v13112130






