Evaluation of the Presence of ASFV in Wolf Feces Collected from Areas in Poland with ASFV Persistence
Abstract
:1. Introduction
2. Materials and Methods
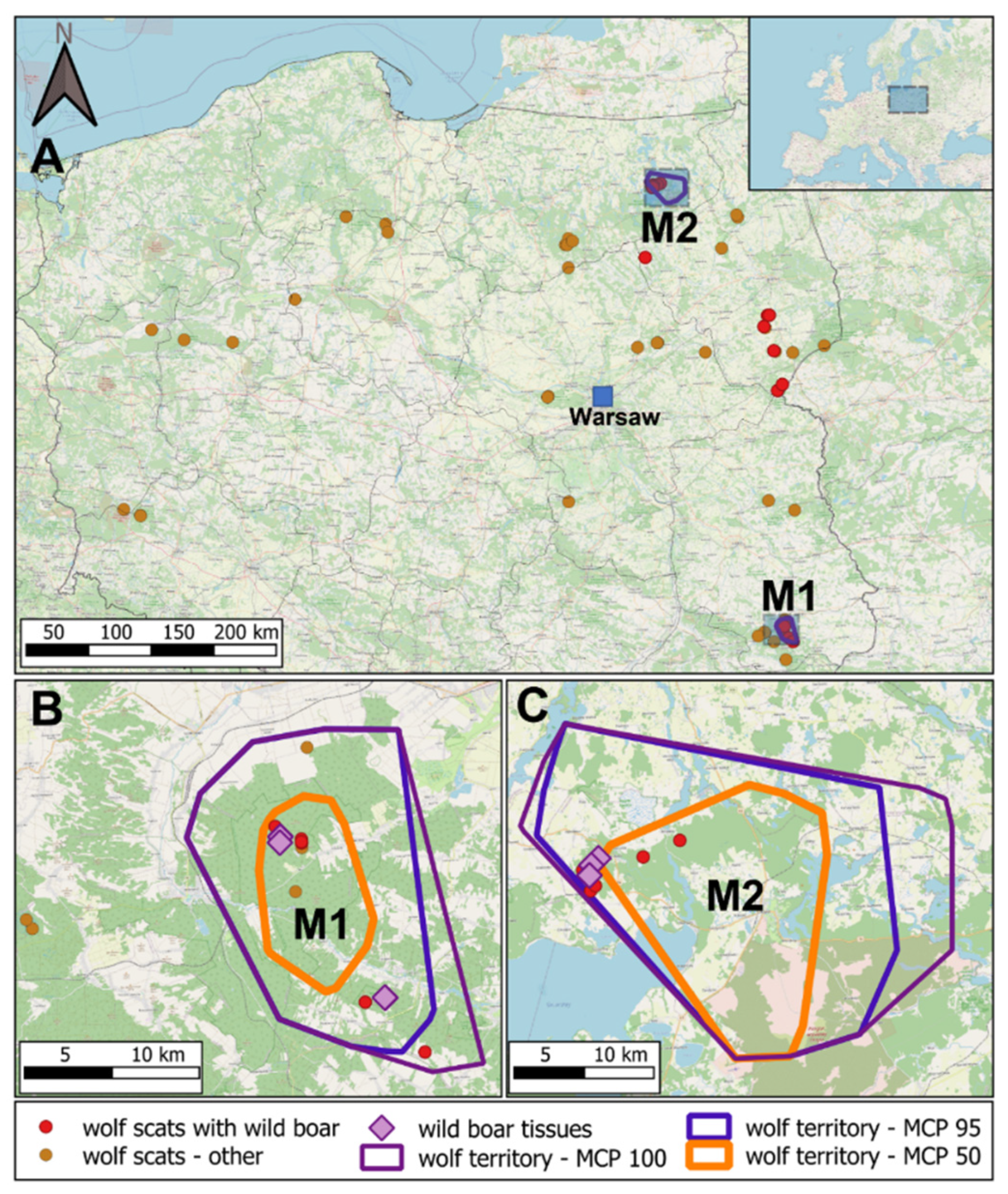
2.1. Wolf Telemetry and Sample Collection
2.2. Laboratory Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galindo, I.; Alonso, C. African Swine Fever Virus: A Review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [Green Version]
- Arias, M.; Jurado, C.; Gallardo, C.; Fernández-Pinero, J.; Sánchez-Vizcaíno, J.M. Gaps in African swine fever: Analysis and priorities. Transbound Emerg. Dis. 2018, 65, 235–247. [Google Scholar] [CrossRef]
- Jori, F.; Vial, L.; Penrith, M.L.; Pérez-Sánchez, R.; Etter, E.; Albina, E.; Michaud, V.; Roger, F. Review of the sylvatic cycle of African swine fever in sub-Saharan Africa and the Indian ocean. Virus Res. 2013, 173, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Śmietanka, K.; Woźniakowski, G.; Kozak, E.; Niemczuk, K.; Frączyk, M.; Bocian, Ł.; Kowalczyk, A.; Pejsak, Z. African Swine Fever Epidemic, Poland, 2014–2015. Emerg. Infect. Dis. 2016, 22, 1201–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- PAFF Committee (Standing Committee on Plants, Animals, Food and Feed of the European Commission). African Swine Fever in Germany [Presentation at PAFF Committee meeting, 13 July 2021]. Available online: https://ec.europa.eu/food/system/files/2021-07/reg-com_ahw_20210713_asf_deu.pdf (accessed on 22 July 2021).
- Oļševskis, E.; Guberti, V.; Seržants, M.; Westergaard, J.; Gallardo, C.; Rodze, I.; Depner, K. African swine fever virus introduction into the EU in 2014: Experience of Latvia. Res. Vet. Sci. 2016, 105, 28–30. [Google Scholar] [CrossRef] [PubMed]
- Podgórski, T.; Borowik, T.; Łyjak, M.; Woźniakowski, G. Spatial epidemiology of African swine fever: Host, landscape and anthropogenic drivers of disease occurrence in wild boar. Prev. Vet. Med. 2020, 177, 104691. [Google Scholar] [CrossRef]
- Podgórski, T.; Śmietanka, K. Do wild boar movements drive the spread of African Swine Fever? Transbound Emerg. Dis. 2018, 65, 1588–1596. [Google Scholar] [CrossRef]
- Pepin, K.M.; Golnar, A.; Podgórski, T. Social structure defines spatial transmission of African swine fever in wild boar. J. R. Soc. Interface 2021, 18, 20200761. [Google Scholar] [CrossRef] [PubMed]
- Probst, C.; Globig, A.; Knoll, B.; Conraths, F.J.; Depner, K. Behaviour of free ranging wild boar towards their dead fellows: Potential implications for the transmission of African swine fever. R. Soc. Open Sci. 2017, 4, 170054. [Google Scholar] [CrossRef] [Green Version]
- Morelle, K.; Jezek, M.; Licoppe, A.; Podgorski, T. Deathbed choice by ASF-infected wild boar can help find carcasses. Transbound Emerg. Dis. 2019, 66, 1821–1826. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Żmudzki, J.; Woźniakowski, G. African Swine Fever Virus—Persistence in Different Environmental Conditions and the Possibility of its Indirect Transmission. J. Vet. Res. 2019, 63, 303–310. [Google Scholar] [CrossRef] [Green Version]
- EFSA AHAW Panel. Scientific opinion on African swine fever. EFSA J. 2014, 12, 3628. [Google Scholar]
- Costard, S.; Mur, L.; Lubroth, J.; Sanchez-Vizcaino, J.M.; Pfeiffer, D.U. Epidemiology of African swine fever virus. Virus Res. 2013, 173, 191–197. [Google Scholar] [CrossRef] [PubMed]
- EFSA (European Food Safety Authority); Depner, K.; Gortazar, C.; Guberti, V.; Masiulis, M.; More, S.; Oļševskis, E.; Thulke, H.-H.; Viltrop, A.; Woźniakowski, G.; et al. Scientific report on the epidemiological analyses of African swine fever in the Baltic States and Poland. EFSA J. 2017, 15, 5068. [Google Scholar]
- Chenais, E.; Ståhl, K.; Guberti, V.; Depner, K. Identification of wild boar–habitat epidemiologic cycle in African swine fever epizootic. Emerg. Infect. Dis. 2018, 24, 810–812. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Liu, J.; Wang, L.; Fan, S.; Li, Z.; Li, Y.; Yi, L.; Ding, H.; Zhao, M.; Chen, J. Current state of global African Swine Fever vaccine development under the prevalence and transmission of ASF in China. Vaccines 2020, 8, 531. [Google Scholar] [CrossRef] [PubMed]
- Probst, C.; Gethmann, J.; Amler, S.; Globig, A.; Knoll, B.; Conraths, F.J. The potential role of scavengers in spreading African swine fever among wild boar. Sci. Rep. 2019, 9, 11450. [Google Scholar] [CrossRef]
- Parzybut, T. Czy Wilki Mogą Przenosić ASF? [In Polish] 2016. Available online: https://www.farmer.pl/fakty/czy-wilki-moga-przenosic-asf,67789.html (accessed on 15 July 2021).
- Chapron, G.; Kaczensky, P.; Linnell, J.D.C.; von Arx, M.; Huber, D.; Andrén, H.; López-Bao, J.V.; Adamec, M.; Álvares, F.; Anders, O.; et al. Recovery of large carnivores in Europe ‘ s modern human-dominated landscapes. Science 2014, 346, 1517–1519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, S.; Mysłajek, R.W. Wolf recovery and population dynamics in Western Poland, 2001–2012. Mammal Res. 2016, 61, 83–98. [Google Scholar] [CrossRef]
- Nowak, S.; Mysłajek, R.W.; Szewczyk, M.; Tomczak, P.; Borowik, T.; Jędrzejewska, B. Sedentary but not dispersing wolves Canis lupus recolonizing western Poland (2001–2016) conform to the predictions of a habitat suitability model. Divers. Distrib. 2017, 23, 1353–1364. [Google Scholar] [CrossRef] [Green Version]
- Szewczyk, M.; Nowak, S.; Niedźwiecka, N.; Hulva, P.; Špinkytė-Bačkaitienė, R.; Demjanovičová, K.; Bolfíková, B.Č.; Antal, V.; Fenchuk, V.; Figura, M.; et al. Dynamic range expansion leads to establishment of a new, genetically distinct wolf population in Central Europe. Sci. Rep. 2019, 9, 19003. [Google Scholar] [CrossRef]
- Szewczyk, M.; Nowak, C.; Hulva, P.; Mergeay, J.; Stronen, A.V.; Bolfíková, B.Č.; Czarnomska, S.D.; Diserens, T.A.; Fenchuk, V.; Figura, M.; et al. Genetic support for the current discrete conservation unit of the Central European wolf population. Wildl. Biol. 2021, 2, wlb.00809. [Google Scholar]
- Borowik, T.; Cornulier, T.; Jędrzejewska, B. Environmental factors shaping ungulate abundances in Poland. Acta Theriol. 2013, 58, 403–413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, S.; Mysłajek, R.W.; Kłosińska, A.; Gabryś, G. Diet and prey selection of wolves Canis lupus recolonising Western and Central Poland. Mammal Biol. 2011, 76, 709–715. [Google Scholar] [CrossRef]
- Klich, D.; Yanuta, G.; Sobczuk, M.; Balcerak, M. Indirect Effect of African Swine Fever on the Diet Composition of the Gray Wolf Canis lupus—A Case Study in Belarus. Animals 2021, 11, 1758. [Google Scholar] [CrossRef]
- Calenge, C. The package adehabitat for the R software: Tool for the analysis of space and habitat use by animals. Ecol. Model. 2006, 197, 516–519. [Google Scholar] [CrossRef]
- Mysłajek, R.W.; Tracz, M.; Tracz, M.; Tomczak, P.; Szewczyk, M.; Niedźwiecka, N.; Nowak, S. Spatial organization in wolves Canis lupus recolonizing north-west Poland: Large territories at low population density. Mammal Biol. 2019, 92, 37–44. [Google Scholar] [CrossRef]
- Teerink, B.J. Atlas and Identification Key. Hair of West European Mammals; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Agüero, M.; Fernández, J.; Romero, L.; Sánchez Mascaraque, C.; Arias, M.; Sánchez-Vizcaíno, J.M. Highly sensitive PCR assay for routine diagnosis of African swine fever virus in clinical samples. J. Clin. Microbiol. 2003, 41, 4431–4434. [Google Scholar] [CrossRef] [Green Version]
- Geospiza.com. FinchTV. 2015. Available online: http://www.geospiza.com/Products/finchtv.shtm (accessed on 12 July 2021).
- Friedrich-Loeffler-Institut. FAQ: African Swine Fever in Wild Boar. Available online: https://www.openagrar.de/servlets/MCRFileNodeServlet/openagrar_derivate_00009659/FLI-Information_FAQ_ASP20180115_eng.pdf (accessed on 2 October 2021).
- Phillips, D.P.; Danilchuk, W.; Ryon, J.; Fentress, J.C. Food-caching in timber wolves, and the question of rules of action syntax. Behav. Brain Res. 1990, 38, 1–6. [Google Scholar] [CrossRef]
- Głowaciński, Z.; Profus, P. Potential impact of wolves Canis lupus on prey populations in eastern Poland. Biol. Cons. 1997, 80, 99–106. [Google Scholar] [CrossRef]
- Arias, M.; de la Torre, A.; Dixon, L.; Gallardo, C.; Jori, F.; Laddomada, A.; Martins, C.; Parkhouse, R.M.; Revilla, Y.; Rodriguez, F.A.J.; et al. Approaches and Perspectives for Development of African Swine Fever Virus Vaccines. Vaccines 2017, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Bartoń, K.A.; Zwijacz-Kozica, T.; Zięba, F.; Sergiel, A.; Selva, N. Bears without borders: Long-distance movement in human-dominated landscapes. Glob. Ecol. Conserv. 2019, 17, e00541. [Google Scholar] [CrossRef]
- Jędrzejewski, W.; Schmidt, K.; Theuerkauf, J.; Jędrzejewska, B.; Okarma, H. Daily movements and territory use by radio-collared wolves (Canis lupus) in Białowieża Primeval Forest in Poland. Can. J. Zool. 2001, 79, 1993–2004. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szewczyk, M.; Łepek, K.; Nowak, S.; Witek, M.; Bajcarczyk, A.; Kurek, K.; Stachyra, P.; Mysłajek, R.W.; Szewczyk, B. Evaluation of the Presence of ASFV in Wolf Feces Collected from Areas in Poland with ASFV Persistence. Viruses 2021, 13, 2062. https://doi.org/10.3390/v13102062
Szewczyk M, Łepek K, Nowak S, Witek M, Bajcarczyk A, Kurek K, Stachyra P, Mysłajek RW, Szewczyk B. Evaluation of the Presence of ASFV in Wolf Feces Collected from Areas in Poland with ASFV Persistence. Viruses. 2021; 13(10):2062. https://doi.org/10.3390/v13102062
Chicago/Turabian StyleSzewczyk, Maciej, Krzysztof Łepek, Sabina Nowak, Małgorzata Witek, Anna Bajcarczyk, Korneliusz Kurek, Przemysław Stachyra, Robert W. Mysłajek, and Bogusław Szewczyk. 2021. "Evaluation of the Presence of ASFV in Wolf Feces Collected from Areas in Poland with ASFV Persistence" Viruses 13, no. 10: 2062. https://doi.org/10.3390/v13102062
APA StyleSzewczyk, M., Łepek, K., Nowak, S., Witek, M., Bajcarczyk, A., Kurek, K., Stachyra, P., Mysłajek, R. W., & Szewczyk, B. (2021). Evaluation of the Presence of ASFV in Wolf Feces Collected from Areas in Poland with ASFV Persistence. Viruses, 13(10), 2062. https://doi.org/10.3390/v13102062






