Research Advances on the Interactions between Rabies Virus Structural Proteins and Host Target Cells: Accrued Knowledge from the Application of Reverse Genetics Systems
Abstract
:1. Introduction
2. Interactions between RABV Structural Proteins and Host Target Cells
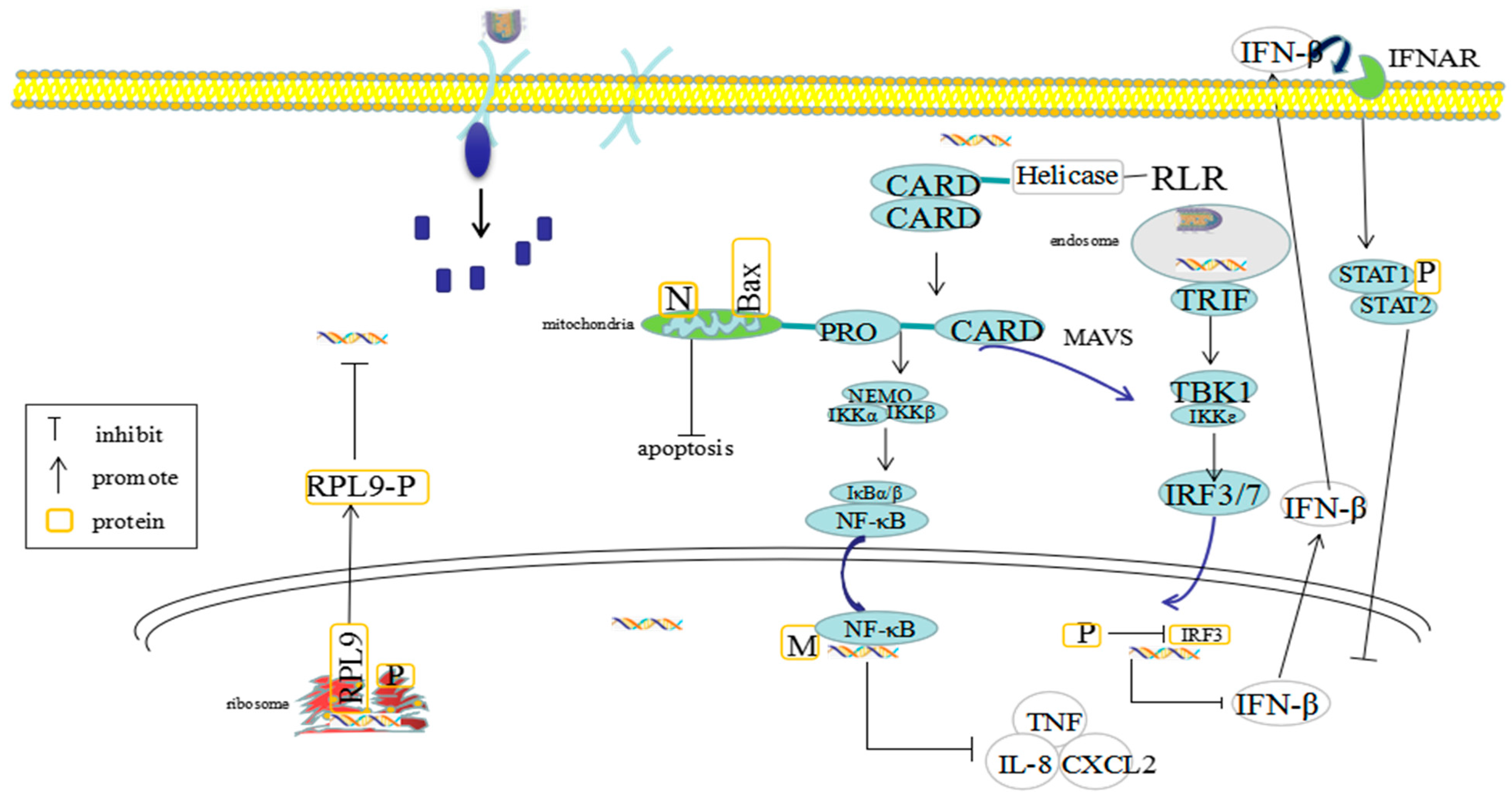
2.1. The Pathogenic Mechanisms of RABV N Protein
2.2. The Pathogenic Mechanisms of RABV P Protein
2.3. The Pathogenic Mechanisms of RABV M Protein
2.4. The Pathogeni Mechanisms of RABV G Protein
2.5. The Pathogenic Mechanisms of RABV L Protein
3. Research on the Manipulation of RABV by Using Reverse Genetics
3.1. The Establishment of Reverse Genetic Manipulation of RABV
3.2. The Application of Reverse Genetic Manipulation of RABV
3.2.1. The Development of Attenuated Rabies Vaccines
3.2.2. RABV-Based Vectors as Vaccines against Other Infectious Diseases
4. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BBB | blood-brain-barrier |
| BEFV | bovine ephemeral fever virus |
| CD | cytoplasmic domain |
| CXCL | CGXGC Motif Chemokine 13 |
| DCBp | DC binding peptide |
| DG | dentate gyrus |
| DLC1 | dynein light chain 1 |
| Flt 3L | Fms-like tyrosine kinase3 ligand |
| G | glycoprotein |
| GC | Germinal Center |
| HDAC6 | histone deacetylase 6 |
| HHrz | hammerhead ribozymes |
| HMGB 1 | high G mobility group Box 1 |
| IRF3 | IFN regulatory factor 3 |
| L | large RNA polymerase protein |
| M | matrix protein |
| MERS-CoV | Middle East Respiratory Syndrome coronavirus |
| MGluR2 | the metabolic glutamine receptor subtype II |
| N | nucleoprotein |
| nAChR | the acetylcholine receptor |
| NC | nucleocapsid |
| NCAM | the neural cell adhesion molecule |
| P | phosphoprotein |
| p75NTR | the low affinity neurotrophic receptor |
| PBM | PDZ binding motif |
| PCTD | the C-terminal domain of P protein |
| PEP | post-exposure prophylaxis |
| PRP | propagation replicon particle |
| PS | phosphatidylserine |
| RABV | rabies virus |
| RdRp | RNA-dependent RNA polymerase |
| RNP | ribonucleoprotein |
| RPL9 | ribosomal large subunit protein L9 |
| rRABV | recombinant rabies virus |
| SFTSV | severe fever with thrombocytopenia syndrome virus |
| VEEV | Venezuelan equine encephalitis virus |
| VNA | virus-neutralizing antibodies |
| VSV | vesicular stomatitis virus |
| ZIKV | Zika virus |
References
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Muller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Primers. 2017, 3, 17091. [Google Scholar] [CrossRef] [PubMed]
- Cleaveland, S.; Hampson, K. Rabies elimination research: Juxtaposing optimism, pragmatism and realism. Proc. R. Soc. B: Boil. Sci. 2017, 284, 20171880. [Google Scholar] [CrossRef] [PubMed]
- Finke, S.; Conzelmann, K.-K. Replication strategies of rabies virus. Virus Res. 2005, 111, 120–131. [Google Scholar] [CrossRef]
- Davis, B.M.; Rall, G.F.; Schnell, M.J. Everything You Always Wanted to Know About Rabies Virus (But Were Afraid to Ask). Annu. Rev. Virol. 2015, 2, 451–471. [Google Scholar] [CrossRef] [PubMed]
- Maillard, A.; Domanski, M.; Brunet, P.; Chaffotte, A.; Guittet, E.; Gaudin, Y. Spectroscopic characterization of two peptides derived from the stem of rabies virus glycoprotein. Virus Res. 2003, 93, 151–158. [Google Scholar] [CrossRef]
- Siler, C.A.; McGettigan, J.P.; Dietzschold, B.; Herrine, S.K.; Dubuisson, J.; Pomerantz, R.J.; Schnell, M.J. Live and Killed Rhabdovirus-Based Vectors as Potential Hepatitis C Vaccines. Virol. 2002, 292, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Banyard, A.; Tordo, N. Rabies pathogenesis and immunology. Rev. Sci. Tech. de l’OIE 2018, 37, 323–330. [Google Scholar] [CrossRef]
- Zhu, S.; Li, H.; Wang, C.; Luo, F.; Guo, C. Reverse genetics of rabies virus: New strategies to attenuate virus virulence for vaccine development. J. Neurovirol. 2015, 21, 335–345. [Google Scholar] [CrossRef]
- McGettigan, J.P.; Naper, K.; Orenstein, J.; Koser, M.; McKenna, P.M.; Schnell, M.J. Functional Human Immunodeficiency Virus Type 1 (HIV-1) Gag-Pol or HIV-1 Gag-Pol and Env Expressed from a Single Rhabdovirus-Based Vaccine Vector Genome. J. Virol. 2003, 77, 10889–10899. [Google Scholar] [CrossRef] [Green Version]
- Kurup, D.; Wirblich, C.; Feldmann, H.; Marzi, A.; Schnell, M.J. Rhabdovirus-Based Vaccine Platforms against Henipaviruses. J. Virol. 2015, 89, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Duan, M.; Wang, X.; Gao, J.; Guan, Z.; Zhang, M. Early events in rabies virus infection—Attachment, entry, and intracellular trafficking. Virus Res. 2019, 263, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Schnell, M.J.; Mcgettigan, J.P.; Wirblich, C.; Papaneri, A. The cell biology of rabies virus: Using stealth to reach the brain. Nat. Rev. Genet. 2009, 8, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Mebatsion, T.; Weiland, F.; Conzelmann, K.-K. Matrix Protein of Rabies Virus Is Responsible for the Assembly and Budding of Bullet-Shaped Particles and Interacts with the Transmembrane Spike Glycoprotein G. J. Virol. 1999, 73, 242–250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albertini, A.A.; Ruigrok, R.W.; Blondel, D. Rabies Virus Transcription and Replication. Int. Rev. Cytol. 2011, 79, 1–22. [Google Scholar]
- He, W.; Zhang, H.; Zhang, Y.; Wang, R.; Lu, S.; Ji, Y.; Liu, C.; Yuan, P.; Su, S. Codon usage bias in the N gene of rabies virus. Infect. Genet. Evol. 2017, 54, 458–465. [Google Scholar] [CrossRef]
- Shimizu, K.; Ito, N.; Mita, T.; Yamada, K.; Hosokawa-Muto, J.; Sugiyama, M.; Minamoto, N. Involvement of nucleoprotein, phosphoprotein, and matrix protein genes of rabies virus in virulence for adult mice. Virus Res. 2007, 123, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Schoehn, G.; Iseni, F.; Mavrakis, M.; Blondel, D.; Ruigrok, R.W.H. Structure of Recombinant Rabies Virus Nucleoprotein-RNA Complex and Identification of the Phosphoprotein Binding site. J. Virol. 2001, 75, 490–498. [Google Scholar] [CrossRef] [Green Version]
- Mei, M.; Long, T.; Zhang, Q.; Zhao, J.; Tian, Q.; Peng, J.; Luo, J.; Jiang, H.; Lin, Y.; Lin, Z.; et al. Phenotypic Consequence of Rearranging the N Gene of RABV HEP-Flury. Viruses 2019, 11, 402. [Google Scholar] [CrossRef] [Green Version]
- Masatani, T.; Ito, N.; Ito, Y.; Nakagawa, K.; Abe, M.; Yamaoka, S.; Okadera, K.; Sugiyama, M. Importance of rabies virus nucleoprotein in viral evasion of interferon response in the brain. Microbiol. Immunol. 2013, 57, 511–517. [Google Scholar] [CrossRef]
- Masatani, T.; Ito, N.; Shimizu, K.; Ito, Y.; Nakagawa, K.; Abe, M.; Yamaoka, S.; Sugiyama, M. Amino acids at positions 273 and 394 in rabies virus nucleoprotein are important for both evasion of host RIG-I-mediated antiviral response and pathogenicity. Virus Res. 2011, 155, 168–174. [Google Scholar] [CrossRef]
- Masatani, T.; Ito, N.; Shimizu, K.; Ito, Y.; Nakagawa, K.; Sawaki, Y.; Koyama, H.; Sugiyama, M. Rabies Virus Nucleoprotein Functions To Evade Activation of the RIG-I-Mediated Antiviral Response. J. Virol. 2010, 84, 4002–4012. [Google Scholar] [CrossRef] [Green Version]
- Zan, J.; Liu, J.; Zhou, J.-W.; Wang, H.-L.; Mo, K.-K.; Yan, Y.; Xu, Y.-B.; Liao, M.; Su, S.; Hu, R.-L.; et al. Rabies virus matrix protein induces apoptosis by targeting mitochondria. Exp. Cell Res. 2016, 347, 83–94. [Google Scholar] [CrossRef]
- Sugiyama, A.; Nomai, T.; Jiang, X.; Minami, M.; Yao, M.; Maenaka, K.; Ito, N.; Gooley, P.R.; Moseley, G.W.; Ose, T. Structural comparison of the C-terminal domain of functionally divergent lyssavirus P proteins. Biochem. Biophys. Res. Commun. 2020, 529, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Mavrakis, M.; Méhouas, S.; Réal, E.; Iseni, F.; Blondel, D.; Tordo, N.; Ruigrok, R.W. Rabies virus chaperone: Identification of the phosphoprotein peptide that keeps nucleoprotein soluble and free from non-specific RNA. Virology 2006, 349, 422–429. [Google Scholar] [CrossRef] [Green Version]
- Chenik, M.; Schnell, M.; Conzelmann, K.-K.; Blondel, D. Mapping the Interacting Domains between the Rabies Virus Polymerase and Phosphoprotein. J. Virol. 1998, 72, 1925–1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morin, B.; Liang, B.; Gardner, E.; Ross, R.A.; Whelan, S.P.J. An In Vitro RNA Synthesis Assay for Rabies Virus Defines Ribonucleoprotein Interactions Critical for Polymerase Activity. J. Virol. 2017, 91, e01508–e01516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyrat, C.; Schneider, R.; Ribeiro, E.A.; Yabukarski, F.; Yao, M.; Gérard, F.C.; Jensen, M.R.; Ruigrok, R.W.; Blackledge, M.; Jamin, M. Ensemble Structure of the Modular and Flexible Full-Length Vesicular Stomatitis Virus Phosphoprotein. J. Mol. Biol. 2012, 423, 182–197. [Google Scholar] [CrossRef]
- Zhan, J.; Harrison, A.R.; Portelli, S.; Nguyen, T.B.; Kojima, I.; Zheng, S.; Yan, F.; Masatani, T.; Rawlinson, S.M.; Sethi, A.; et al. Definition of the immune evasion-replication interface of rabies virus P protein. PLoS Pathog. 2021, 17, e1009729. [Google Scholar] [CrossRef]
- Hossain, A.; Larrous, F.; Rawlinson, S.M.; Zhan, J.; Sethi, A.; Ibrahim, Y.; Aloi, M.; Lieu, K.G.; Mok, Y.-F.; Griffin, M.D.; et al. Structural Elucidation of Viral Antagonism of Innate Immunity at the STAT1 Interface. Cell Rep. 2019, 29, 1934–1945.e8. [Google Scholar] [CrossRef] [Green Version]
- Brzózka, K.; Finke, S.; Conzelmann, K.-K. Identification of the Rabies Virus Alpha/Beta Interferon Antagonist: Phosphoprotein P Interferes with Phosphorylation of Interferon Regulatory Factor 3. J. Virol. 2005, 79, 7673–7681. [Google Scholar] [CrossRef] [Green Version]
- Rieder, M.; Conzelmann, K.-K. Interferon in Rabies Virus Infection. Adv. Virus Res. 2011, 79, 91–114. [Google Scholar] [PubMed]
- Rieder, M.; Brzózka, K.; Pfaller, C.K.; Cox, J.H.; Stitz, L.; Conzelmann, K.-K. Genetic Dissection of Interferon-Antagonistic Functions of Rabies Virus Phosphoprotein: Inhibition of Interferon Regulatory Factor 3 Activation Is Important for Pathogenicity. J. Virol. 2010, 85, 842–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidy, A.; Chelbi-Alix, M.; Blondel, D. Rabies Virus P Protein Interacts with STAT1 and Inhibits Interferon Signal Transduction Pathways. J. Virol. 2005, 79, 14411–14420. [Google Scholar] [CrossRef] [Green Version]
- Chelbi-Alix, M.K.; Vidy, A.; El Bougrini, J.; Blondel, D. Rabies Viral Mechanisms to Escape the IFN System: The Viral Protein P Interferes with IRF-3, Stat1, and PML Nuclear Bodies. J. Interf. Cytokine Res. 2006, 26, 271–280. [Google Scholar] [CrossRef]
- Yamaoka, S.; Okada, K.; Ito, N.; Okadera, K.; Mitake, H.; Nakagawa, K.; Sugiyama, M. Defect of rabies virus phosphoprotein in its interferon-antagonist activity negatively affects viral replication in muscle cells. J. Veter. Med. Sci. 2017, 79, 1394–1397. [Google Scholar] [CrossRef] [Green Version]
- Yamaoka, S.; Ito, N.; Ohka, S.; Kaneda, S.; Nakamura, H.; Agari, T.; Masatani, T.; Nakagawa, K.; Okada, K.; Okadera, K.; et al. Involvement of the Rabies Virus Phosphoprotein Gene in Neuroinvasiveness. J. Virol. 2013, 87, 12327–12338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fouquet, B.; Nikolic, J.; Larrous, F.; Bourhy, H.; Wirblich, C.; Lagaudrière-Gesbert, C.; Blondel, D. Focal Adhesion Kinase Is Involved in Rabies Virus Infection through Its Interaction with Viral Phosphoprotein P. J. Virol. 2015, 89, 1640–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Dong, W.; Shi, Y.; Deng, F.; Chen, X.; Wan, C.; Zhou, M.; Zhao, L.; Fu, Z.F.; Peng, G. Rabies virus phosphoprotein interacts with ribosomal protein L9 and affects rabies virus replication. Virology 2016, 488, 216–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raux, H.; Flamand, A.; Blondel, D. Interaction of the Rabies Virus P Protein with the LC8 Dynein Light Chain. J. Virol. 2000, 74, 10212–10216. [Google Scholar] [CrossRef] [Green Version]
- Finke, S.; Granzow, H.; Hurst, J.; Pollin, R.; Mettenleiter, T.C. Intergenotypic Replacement of Lyssavirus Matrix Proteins Demonstrates the Role of Lyssavirus M Proteins in Intracellular Virus Accumulation. J. Virol. 2010, 84, 1816–1827. [Google Scholar] [CrossRef] [Green Version]
- Finke, S.; Conzelmann, K.-K. Dissociation of Rabies Virus Matrix Protein Functions in Regulation of Viral RNA Synthesis and Virus Assembly. J. Virol. 2003, 77, 12074–12082. [Google Scholar] [CrossRef] [Green Version]
- Mita, T.; Shimizu, K.; Ito, N.; Yamada, K.; Ito, Y.; Sugiyama, M.; Minamoto, N. Amino acid at position 95 of the matrix protein is a cytopathic determinant of rabies virus. Virus Res. 2008, 137, 33–39. [Google Scholar] [CrossRef]
- Kojima, I.; Izumi, F.; Ozawa, M.; Fujimoto, Y.; Okajima, M.; Ito, N.; Sugiyama, M.; Masatani, T. Analyses of cell death mechanisms related to amino acid substitution at position 95 in the rabies virus matrix protein. J. Gen. Virol. 2021, 102, 001594. [Google Scholar] [CrossRef]
- Mebatsion, T.; König, M.; Conzelmann, K.-K. Budding of Rabies Virus Particles in the Absence of the Spike Glycoprotein. Cell 1996, 84, 941–951. [Google Scholar] [CrossRef] [Green Version]
- Sonthonnax, F.; Besson, B.; Bonnaud, E.; Jouvion, G.; Merino, D.; Larrous, F.; Bourhy, H. Lyssavirus matrix protein cooperates with phosphoprotein to modulate the Jak-Stat pathway. Sci. Rep. 2019, 9, 12171. [Google Scholar] [CrossRef] [PubMed]
- Pulmanausahakul, R.; Li, J.; Schnell, M.J.; Dietzschold, B. The Glycoprotein and the Matrix Protein of Rabies Virus Affect Pathogenicity by Regulating Viral Replication and Facilitating Cell-to-Cell Spread. J. Virol. 2008, 82, 2330–2338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finke, S.; Mueller-Waldeck, R.; Conzelmann, K.-K. Rabies virus matrix protein regulates the balance of virus transcription and replication. J. Gen. Virol. 2003, 84, 1613–1621. [Google Scholar] [CrossRef]
- Zan, J.; Liu, S.; Sun, D.-N.; Mo, K.-K.; Yan, Y.; Liu, J.; Hu, B.-L.; Gu, J.-Y.; Liao, M.; Zhou, J.-Y. Rabies Virus Infection Induces Microtubule Depolymerization to Facilitate Viral RNA Synthesis by Upregulating HDAC6. Front. Cell. Infect. Microbiol. 2017, 7, 146. [Google Scholar] [CrossRef]
- Ben Khalifa, Y.; Luco, S.; Besson, B.; Sonthonnax, F.; Archambaud, M.; Grimes, J.M.; Larrous, F.; Bourhy, H. The matrix protein of rabies virus binds to RelAp43 to modulate NF-kappaB-dependent gene expression related to innate immunity. Sci. Rep. 2016, 6, 39420. [Google Scholar] [CrossRef] [Green Version]
- Real, L.A.; Henderson, J.C.; Biek, R.; Snaman, J.; Jack, T.L.; Childs, J.E.; Stahl, E.; Waller, L.; Tinline, R.; Nadin-Davis, S. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc. Natl. Acad. Sci. USA 2005, 102, 12107–12111. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.; Wang, H.; Mahmood, F.; Fu, Z.F. Rabies virus glycoprotein is an important determinant for the induction of innate immune responses and the pathogenic mechanisms. Veter. Microbiol. 2013, 162, 601–613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albertini, A.A.V.; Baquero, E.; Ferlin, A.; Gaudin, Y. Molecular and Cellular Aspects of Rhabdovirus Entry. Viruses 2012, 4, 117–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasaki, M.; Anindita, P.D.; Ito, N.; Sugiyama, M.; Carr, M.; Fukuhara, H.; Ose, T.; Maenaka, K.; Takada, A.; Hall, W.W.; et al. The Role of Heparan Sulfate Proteoglycans as an Attachment Factor for Rabies Virus Entry and Infection. J. Infect. Dis. 2018, 217, 1740–1749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lentz, T.L.; Burrage, T.G.; Smith, A.L.; Crick, J.; Tignor, G.H. Is the Acetylcholine Receptor a Rabies Virus Receptor? Science 1982, 215, 182–184. [Google Scholar] [CrossRef]
- Thoulouze, M.-I.; Lafage, M.; Schachner, M.; Hartmann, U.; Cremer, H.; Lafon, M. The Neural Cell Adhesion Molecule Is a Receptor for Rabies Virus. J. Virol. 1998, 72, 7181–7190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuffereau, C.; Bénéjean, J.; Blondel, D.; Kieffer, B.; Flamand, A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998, 17, 7250–7259. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Liu, R.; Shuai, L.; Wang, X.; Luo, J.; Wang, C.; Chen, W.; Wang, X.; Ge, J.; et al. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog. 2018, 14, e1007189. [Google Scholar] [CrossRef]
- Etessami, R.; Conzelmann, K.-K.; Fadai-Ghotbi, B.; Natelson, B.; Tsiang, H.; Ceccaldi, P.-E. Spread and pathogenic characteristics of a G-deficient rabies virus recombinant: An in vitro and in vivo study. J. Gen. Virol. 2000, 81, 2147–2153. [Google Scholar] [CrossRef]
- Ito, N.; Takayama, M.; Yamada, K.; Sugiyama, M.; Minamoto, N. Rescue of Rabies Virus from Cloned cDNA and Identification of the Pathogenicity-Related Gene: Glycoprotein Gene Is Associated with Virulence for Adult Mice. J. Virol. 2001, 75, 9121–9128. [Google Scholar] [CrossRef] [Green Version]
- Takayama-Ito, M.; Inoue, K.-I.; Shoji, Y.; Inoue, S.; Iijima, T.; Sakai, T.; Kurane, I.; Morimoto, K. A highly attenuated rabies virus HEP-Flury strain reverts to virulent by single amino acid substitution to arginine at position 333 in glycoprotein. Virus Res. 2006, 119, 208–215. [Google Scholar] [CrossRef]
- Takayama-Ito, M.; Ito, N.; Yamada, K.; Sugiyama, M.; Minamoto, N. Multiple amino acids in the glycoprotein of rabies virus are responsible for pathogenicity in adult mice. Virus Res. 2006, 115, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, B.; Lyu, Z.; Wu, Y.; Guo, X. Single amino acid change at position 255 in rabies virus glycoprotein decreases viral pathogenicity. FASEB J. 2020, 34, 9650–9663. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Liu, H.; Zhang, X.; Baolige, D.; Zhao, S.; Hu, W.; Yang, Y. Change in the Single Amino Acid Site 83 in Rabies Virus Glycoprotein Enhances the BBB Permeability and Reduces Viral Pathogenicity. Front. Cell Dev. Biol. 2021, 8, 632957. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhang, B.; Wu, Y.; Guo, X. Amino Acid Mutation in Position 349 of Glycoprotein Affect the Pathogenicity of Rabies Virus. Front. Microbiol. 2020, 11, 481. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, S.; Asgari, T.; Mirzapour-Delavar, H.; Aliakbari, S.; Pourbadie, H.G.; Prehaud, C.; Lafon, M.; Gholami, A.; Azadmanesh, K.; Naderi, N.; et al. Lentiviral Expression of Rabies Virus Glycoprotein in the Rat Hippocampus Strengthens Synaptic Plasticity. Cell. Mol. Neurobiol. 2021, 27, 1–12. [Google Scholar] [CrossRef]
- Katz, I.S.S.; Dias, M.H.; Lima, I.F.; Chaves, L.B.; Ribeiro, O.G.; Scheffer, K.C.; Iwai, L.K. Large protein as a potential target for use in rabies diagnostics. Acta Virol. 2017, 61, 280–288. [Google Scholar]
- Zhao, W.; Su, J.; Zhao, N.; Liu, J.; Su, S. Development of Monoclonal Antibodies for Detection of Conserved and Variable Epitopes of Large Protein of Rabies Virus. Viruses 2021, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.; Nolden, T.; Nemitz, S.; Perlson, E.; Finke, S. A Dynein Light Chain 1 Binding Motif in Rabies Virus Polymerase L Protein Plays a Role in Microtubule Reorganization and Viral Primary Transcription. J. Virol. 2015, 89, 9591–9600. [Google Scholar] [CrossRef] [Green Version]
- Tian, D.; Luo, Z.; Zhou, M.; Li, M.; Yu, L.; Wang, C.; Yuan, J.; Li, F.; Tian, B.; Sui, B.; et al. Critical Role of K1685 and K1829 in the Large Protein of Rabies Virus in Viral Pathogenicity and Immune Evasion. J. Virol. 2016, 90, 232–244. [Google Scholar] [CrossRef] [Green Version]
- Schnell, M.; Mebatsion, T.; Conzelmann, K.-K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994, 13, 4195–4203. [Google Scholar] [CrossRef] [Green Version]
- Inoue, K.-I.; Shoji, Y.; Kurane, I.; Iijima, T.; Sakai, T.; Morimoto, K. An improved method for recovering rabies virus from cloned cDNA. J. Virol. Methods 2003, 107, 229–236. [Google Scholar] [CrossRef]
- Le Mercier, P.; Jacob, Y.; Tanner, K.; Tordo, N. A Novel Expression Cassette of Lyssavirus Shows that the Distantly Related Mokola Virus Can Rescue a Defective Rabies Virus Genome. J. Virol. 2002, 76, 2024–2027. [Google Scholar] [CrossRef] [Green Version]
- Ming, P.-G.; Huang, Y.; Tang, Q.; Du, J.-L.; Tao, X.-Y.; Yan, J.-X.; Hu, R.-L. Construction and identification of the helper plasmids for reverse genetic system of rabies virus street strain. Virol. Sin. 2009, 24, 559–565. [Google Scholar] [CrossRef]
- Ghanem, A.; Kern, A.; Conzelmann, K.-K. Significantly improved rescue of rabies virus from cDNA plasmids. Eur. J. Cell Biol. 2012, 91, 10–16. [Google Scholar] [CrossRef]
- Morimoto, K.; Shoji, Y.; Inoue, S. Characterization of P gene-deficient rabies virus: Propagation, pathogenicity and antigenicity. Virus Res. 2005, 111, 61–67. [Google Scholar] [CrossRef]
- Mcgettigan, J.P.; David, F.; Figueiredo, M.D.; Minke, J.; Mebatsion, T.; Schnell, M. Safety and serological response to a matrix gene-deleted rabies virus-based vaccine vector in dogs. Vaccine 2014, 32, 1716–1719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holcomb, K.; Weinberg, J.M. A novel vaccine (Zostavax) to prevent herpes zoster and postherpetic neuralgia. J. Drugs Dermatol. JDD 2006, 5, 863–866. [Google Scholar] [PubMed]
- Gomme, E.A.; Wanjalla, C.N.; Wirblich, C.; Schnell, M.J. Rabies Virus as a Research Tool and Viral Vaccine Vector. Adv Virus Res. 2011, 79, 139–164. [Google Scholar] [PubMed]
- Faber, M.; Pulmanausahakul, R.; Hodawadekar, S.S.; Spitsin, S.; McGettigan, J.P.; Schnell, M.J.; Dietzschold, B. Overexpression of the Rabies Virus Glycoprotein Results in Enhancement of Apoptosis and Antiviral Immune Response. J. Virol. 2002, 76, 3374–3381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, L.; Ge, J.; Wang, X.; Wen, Z.; Zhai, H.; Hua, T.; Zhao, B.; Kong, D.; Yang, C.; Bu, Z. Generation of a recombinant rabies Flury LEP virus carrying an additional G gene creates an improved seed virus for inactivated vaccine production. Virol. J. 2011, 8, 454. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-N.; Chen, C.; Deng, C.-L.; Zhang, C.-G.; Li, N.; Wang, Z.; Zhao, L.; Zhang, B. A novel rabies vaccine based on infectious propagating particles derived from hybrid VEEV-Rabies replicon. EBioMedicine 2020, 56, 102819. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Shin, H.-J. Immunogenicity of replication-deficient vesicular stomatitis virus based rabies vaccine in mice. Veter. Q. 2021, 41, 202–209. [Google Scholar] [CrossRef]
- Zhou, M.; Zhang, G.; Ren, G.; Gnanadurai, C.W.; Li, Z.; Chai, Q.; Yang, Y.; Leyson, C.M.; Wu, W.; Cui, M.; et al. Recombinant Rabies Viruses Expressing GM-CSF or Flagellin Are Effective Vaccines for Both Intramuscular and Oral Immunizations. PLoS ONE 2013, 8, e63384. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, L.; Zhou, S.; Wang, Z.; Ruan, J.; Tang, L.; Jia, Z.; Cui, M.; Zhao, L.; Fu, Z.F. Recombinant rabies virus expressing dog GM-CSF is an efficacious oral rabies vaccine for dogs. Oncotarget 2015, 6, 38504–38516. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Liang, Q.; Zhang, Y.; Yang, J.; Li, M.; Wang, K.; Cui, M.; Chen, H.; Fu, Z.F.; Zhao, L.; et al. An optimized HMGB1 expressed by recombinant rabies virus enhances immunogenicity through activation of dendritic cells in mice. Oncotarget 2017, 8, 83539–83554. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Zhou, M.; Li, Y.; Luo, Z.; Chen, H.; Cui, M.; Fu, Z.F.; Zhao, L. Recombinant rabies virus with the glycoprotein fused with a DC-binding peptide is an efficacious rabies vaccine. Oncotarget 2017, 9, 831–841. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, J.; Li, M.; Cui, M.; Fu, Z.F.; Zhao, L.; Zhou, M. A Recombinant Rabies Virus Expressing Fms-like Tyrosine Kinase 3 Ligand (Flt3L) Induces Enhanced Immunogenicity in Mice. Virol. Sin. 2019, 34, 662–672. [Google Scholar] [CrossRef]
- Wang, Z.; Li, M.; Zhou, M.; Zhang, Y.; Yang, J.; Cao, Y.; Wang, K.; Cui, M.; Chen, H.; Fu, Z.F.; et al. A Novel Rabies Vaccine Expressing CXCL13 Enhances Humoral Immunity by Recruiting both T Follicular Helper and Germinal Center B Cells. J. Virol. 2017, 91, e01956-16. [Google Scholar] [CrossRef] [Green Version]
- Luo, J.; Zhang, B.; Wu, Y.; Tian, Q.; Zhao, J.; Lyu, Z.; Zhang, Q.; Mei, M.; Luo, Y.; Guo, X. Expression of interleukin-6 by a recombinant rabies virus enhances its immunogenicity as a potential vaccine. Vaccine 2017, 35, 938–944. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, M.; Luo, Z.; Zhang, Y.; Cui, M.; Chen, H.; Fu, Z.F.; Zhao, L. Overexpression of Interleukin-7 Extends the Humoral Immune Response Induced by Rabies Vaccination. J. Virol. 2017, 91, e02324-16. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Zhang, Y.; Wang, Z.; Yang, J.; Li, M.; Wang, K.; Cui, M.; Fu, Z.F.; Zhao, L.; Zhou, M. Recombinant rabies virus expressing IL-15 enhances immunogenicity through promoting the activation of dendritic cells in mice. Virol. Sin. 2017, 32, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Zheng, W.; Wang, C.; Wong, G.; Song, Y.; Zheng, X. Immunization with recombinant rabies virus expressing Interleukin-18 exhibits enhanced immunogenicity and protection in mice. Oncotarget 2017, 8, 91505–91515. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, M.; Wang, Z.; Yang, J.; Li, M.; Wang, K.; Cui, M.; Chen, H.; Fu, Z.F.; Zhao, L. Recombinant rabies virus expressing IL-21 enhances immunogenicity through activation of T follicular helper cells and germinal centre B cells. J. Gen. Virol. 2016, 97, 3154–3160. [Google Scholar] [CrossRef] [PubMed]
- Pei, J.; Huang, F.; Wu, Q.; Luo, Z.; Zhang, Y.; Ruan, J.; Li, Y.; Zhou, M.; Fu, Z.; Zhao, L. Codon optimization of G protein enhances rabies virus-induced humoral immunity. J. Gen. Virol. 2019, 100, 1222–1233. [Google Scholar] [CrossRef] [PubMed]
- Bankovskiy, D.; Safonov, G.; Kurilchuk, Y. Immunogenicity of the ERA G 333 rabies virus strain in foxes and raccoon dogs. Dev. Boil. 2008, 131, 461–466. [Google Scholar]
- Faber, M.; Faber, M.-L.; Papaneri, A.; Bette, M.; Weihe, E.; Dietzschold, B.; Schnell, M.J. A Single Amino Acid Change in Rabies Virus Glycoprotein Increases Virus Spread and Enhances Virus Pathogenicity. J. Virol. 2005, 79, 14141–14148. [Google Scholar] [CrossRef] [Green Version]
- Freuling, C.M.; Eggerbauer, E.; Finke, S.; Kaiser, C.; Kretzschmar, A.; Nolden, T.; Ortmann, S.; Schröder, C.; Teifke, J.P.; Schuster, P.; et al. Efficacy of the oral rabies virus vaccine strain SPBN GASGAS in foxes and raccoon dogs. Vaccine 2017, 37, 4750–4757. [Google Scholar] [CrossRef]
- Mebatsion, T. Extensive Attenuation of Rabies Virus by Simultaneously Modifying the Dynein Light Chain Binding Site in the P Protein and Replacing Arg333 in the G Protein. J. Virol. 2001, 75, 11496–11502. [Google Scholar] [CrossRef] [Green Version]
- Faber, M.; Dietzschold, B.; Li, J. Immunogenicity and Safety of Recombinant Rabies Viruses Used for Oral Vaccination of Stray Dogs and Wildlife. Zoonoses Public Health 2009, 56, 262–269. [Google Scholar] [CrossRef]
- Freuling, C.M.; Kamp, V.T.; Klein, A.; Gunther, M.; Zaeck, L.; Potratz, M.; Eggerbauer, E.; Bobe, K.; Kaiser, C.; Kretzschmar, A.; et al. Long-Term Immunogenicity and Efficacy of the Oral Rabies Virus Vaccine Strain SPBN GASGAS in Foxes. Viruses 2019, 11, 790. [Google Scholar] [CrossRef] [Green Version]
- Černe, D.; Hostnik, P.; Toplak, I. The Successful Elimination of Sylvatic Rabies Using Oral Vaccination of Foxes in Slovenia. Viruses 2021, 13, 405. [Google Scholar] [CrossRef] [PubMed]
- Foley, H.D.; McGettigan, J.P.; Siler, C.A.; Dietzschold, B.; Schnell, M.J. A recombinant rabies virus expressing vesicular stomatitis virus glycoprotein fails to protect against rabies virus infection. Proc. Natl. Acad. Sci. USA 2000, 97, 14680–14685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKenna, P.M.; Aye, P.P.; Dietzschold, B.; Montefiori, D.C.; Martin, L.N.; Marx, P.A.; Pomerantz, R.J.; Lackner, A.; Schnell, M.J. Immunogenicity study of glycoprotein-deficient rabies virus expressing simian/human immunodeficiency virus SHIV89.6P envelope in a rhesus macaque. J. Virol. 2004, 78, 13455–13459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Yan, L.; Zheng, W.; Lei, X.; Fu, Q.; Xue, X.; Wang, X.; Xia, X.; Zheng, X. A rabies virus vectored severe fever with thrombocytopenia syndrome (SFTS) bivalent candidate vaccine confers protective immune responses in mice. Veter. Microbiol. 2021, 257, 109076. [Google Scholar] [CrossRef] [PubMed]
- Chi, H.; Zheng, X.; Wang, X.; Wang, C.; Wang, H.; Gai, W.; Perlman, S.; Yang, S.; Zhao, J.; Xia, X. DNA vaccine encoding Middle East respiratory syndrome coronavirus S1 protein induces protective immune responses in mice. Vaccine 2017, 35, 2069–2075. [Google Scholar] [CrossRef]
- Papaneri, A.B.; Bernbaum, J.G.; Blaney, J.E.; Jahrling, P.B.; Schnell, M.J.; Johnson, R.F. Controlled viral glycoprotein expression as a safety feature in a bivalent rabies-ebola vaccine. Virus Res. 2014, 197, 54–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.; Jiao, C.; Cao, Z.; Huang, P.; Chi, H.; Bai, Y.; Liu, D.; Wang, J.; Feng, N.; Li, N.; et al. An inactivated recombinant rabies virus displaying the Zika virus prM-E induces protective immunity against both pathogens. PLoS Negl. Trop. Dis. 2021, 15, e0009484. [Google Scholar] [CrossRef]
- Zheng, W.; Zhao, Z.; Tian, L.; Liu, L.; Xu, T.; Wang, X.; He, H.; Xia, X.; Zheng, Y.; Wei, Y.; et al. Genetically modified rabies virus vector-based bovine ephemeral fever virus vaccine induces protective immune responses against BEFV and RABV in mice. Transbound. Emerg. Dis. 2021, 68, 1353–1362. [Google Scholar] [CrossRef]
- McKenna, P.M.; Mcgettigan, J.P.; Pomerantz, R.J.; Dietzschold, B.; Schnell, M.J. Recombinant rhabdoviruses as potential vaccines for HIV-1 and other diseases. Curr. HIV Res. 2003, 1, 229–237. [Google Scholar] [CrossRef]
- McGettigan, J.P.; Koser, M.L.; McKenna, P.M.; Smith, M.E.; Marvin, J.M.; Eisenlohr, L.C.; Dietzschold, B.; Schnell, M.J. Enhanced humoral HIV-1-specific immune responses generated from recombinant rhabdoviral-based vaccine vectors co-expressing HIV-1 proteins and IL-2. Virology 2006, 344, 363–377. [Google Scholar] [CrossRef]
- Papaneri, A.; Wirblich, C.; Cann, J.A.; Cooper, K.; Jahrling, P.B.; Schnell, M.J.; Blaney, J.E. A replication-deficient rabies virus vaccine expressing Ebola virus glycoprotein is highly attenuated for neurovirulence. Virology. 2012, 434, 18–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keshwara, R.; Johnson, R.F.; Schnell, M.J. Toward an Effective Ebola Virus Vaccine. Annu. Rev. Med. 2017, 68, 371–386. [Google Scholar] [CrossRef] [PubMed]


| Structural Protein | Function |
|---|---|
| N (450aa) | (1) assembling viral RNA, forming RNP together with L and P |
| (2) protecting viral genomic RNA from the cleavage of host cell nuclease | |
| (3) co-localization of Bax protein effectively inhibits mitochondrial apoptosis in the early stage of infection, prevents caspase-dependent and non-dependent apoptosis, and provides time for virus replication and transcription | |
| (4) inhibits the activation of RIG-Ⅰ and the expression of interferon regulatory factor 3 (IRF) and downstream antiviral genes | |
| P (297aa) | (1) interacts with L, forms an active RNA polymerase, regulates the replication and packaging of virus RNA |
| (2) interacts with LC8 not only promotes the transcription of viral RNA, but also facilitates the axonal transport of virus in neurons | |
| (3) interaction with L9 protein can inhibit the transcription and replication of virus | |
| (4) combined with BECN1, induces incomplete autophagy to destroy the host immune system and promote the replication of viral genome | |
| (5) interacts with FAK to regulate RABV infection | |
| (6) weakening the phosphorylation of IRF-3 to inhibit IFN production and block IFN mediated JAK/STAT signal transduction, play the role of type I interferon (IFN) antagonist, helping the virus to proliferate in host cells | |
| M (202aa) | (1) tight connection with RNP, helps G protein to complete the budding of RABV virus particles |
| (2) increases the expression of histone deacetylase 6 (HDAC6) | |
| (3) acts with P65 / Rel A to inhibit the response of NF-κB to IFN-β and block the replication of viral RNA | |
| (4) binds to the C-terminal domain of P43 / Rel A subunit in NF-κB reaction, resulting in the inhibition of NF-κB dependent gene regulatory factors | |
| G (505aa) | (1) binds to the specific receptor (e.g., heparan sulfate, acetylcholine receptor (nAChR), nerve cell adhesion molecule (NCAM) or low affinity neurotrophic receptor p75NTR, metabotropic glutamate receptor subtype II (mGluR2)) on the cell |
| (2) mediates the endocytosis of the virus into the cell | |
| induces the virus to produce neutralizing antibody, and determines the neurophagocytic property of the virus | |
| L (2130aa) | (1) together with P protein, is responsible for viral genome replication, transcription and post-transcriptional processing |
| (2) influences microtubule organization of and mediates cytoskeleton reorganization; | |
| contains a conserved catalytic tetramer region of K-D-K-E, which mainly performs the function of N-7 and 2′-O methyltransferase (MTase) during viral mRNA capping | |
| (4) the important factor in pathogenicity | |
| (5) escapes the innate immunity of host |
| Years | Type of Vaccine |
|---|---|
| 1885 | Pasteur seedlings |
| 1911 | Sheep brain vaccine |
| 1955 | Suckling rat brain vaccine |
| 1956 | Duck embryo vaccine |
| 1960 | Primary hamster kidney vaccine |
| 1965 | Human diploid seedlings |
| 1985 | Vero cell vaccine |
| Characteristics | Virus | Reason |
|---|---|---|
| (1) RABV genome contains five genes and with short transcription stop/start sequences flanking the genes | Human immunodeficiency virus-1 (HIV-1) | an intracellular life cycle and ability to stably express foreign antigens [103]. |
| Hepatitis C virus (HCV) | induce both a humoral and cellular response [10]. | |
| (2) The life cycle of RABV is exclusively in cytoplasmic, no recombination, reversion or integration observed | Severe Fever with Thrombocytopenia Syndrome virus (SFTSV) | induce high neutralizing antibodies in mice [104]. |
| Middle East respiratory syndrome coronavirus (MERS-CoV) | stable heredity and high growth titer [105]. | |
| (3) Stable expression of large and multiple foreign genes of up to 6.5 kb | Ebola virus (EBOV) | safe and immunogenic to non-human primates [106]. |
| (4) RABV can induce a protective immune response in a variety of animals | Henipaviruses (HeV) | a killed RABV vaccine would be highly effective against HeV infections [9]. |
| Zika virus (ZIKV) | induced VNA against RABV and ZIKV and induced a specific cellular immune response, with the potential to prevent ZIKV and RABV infection. [107]. | |
| Bovine Ephemeral fever virus (BEFV) | induce high neutralizing antibody against RABV and high specific antibodies against BEFV [108]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yin, J.; Wang, X.; Mao, R.; Zhang, Z.; Gao, X.; Luo, Y.; Sun, Y.; Yin, X. Research Advances on the Interactions between Rabies Virus Structural Proteins and Host Target Cells: Accrued Knowledge from the Application of Reverse Genetics Systems. Viruses 2021, 13, 2288. https://doi.org/10.3390/v13112288
Yin J, Wang X, Mao R, Zhang Z, Gao X, Luo Y, Sun Y, Yin X. Research Advances on the Interactions between Rabies Virus Structural Proteins and Host Target Cells: Accrued Knowledge from the Application of Reverse Genetics Systems. Viruses. 2021; 13(11):2288. https://doi.org/10.3390/v13112288
Chicago/Turabian StyleYin, Juanbin, Xiangwei Wang, Ruoqing Mao, Zhixiong Zhang, Xin Gao, Yingying Luo, Yuefeng Sun, and Xiangping Yin. 2021. "Research Advances on the Interactions between Rabies Virus Structural Proteins and Host Target Cells: Accrued Knowledge from the Application of Reverse Genetics Systems" Viruses 13, no. 11: 2288. https://doi.org/10.3390/v13112288





