Identification of a New Badnavirus in the Chinaberry (Melia azedarach) Tree and Establishment of a LAMP-LFD Assay for Its Rapid and Visual Detection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and High Throughput Sequencing (HTS)
2.2. Virus Identification
2.3. Genome and Phylogenetic Analysis
2.4. Design of LAMP Primers
2.5. Optimization of LAMP Conditions
2.6. LAMP-LFD and Analytical Sensitivity Assay
2.7. LAMP-LFD Detection in Chinaberry Tree Field Samples
3. Results
3.1. Identification of a Novel Badnavirus Infecting Chinaberry Trees
3.2. Complete Genome Sequence of ChTBV1
3.3. Genome Organization and Phylogenetic Analyses
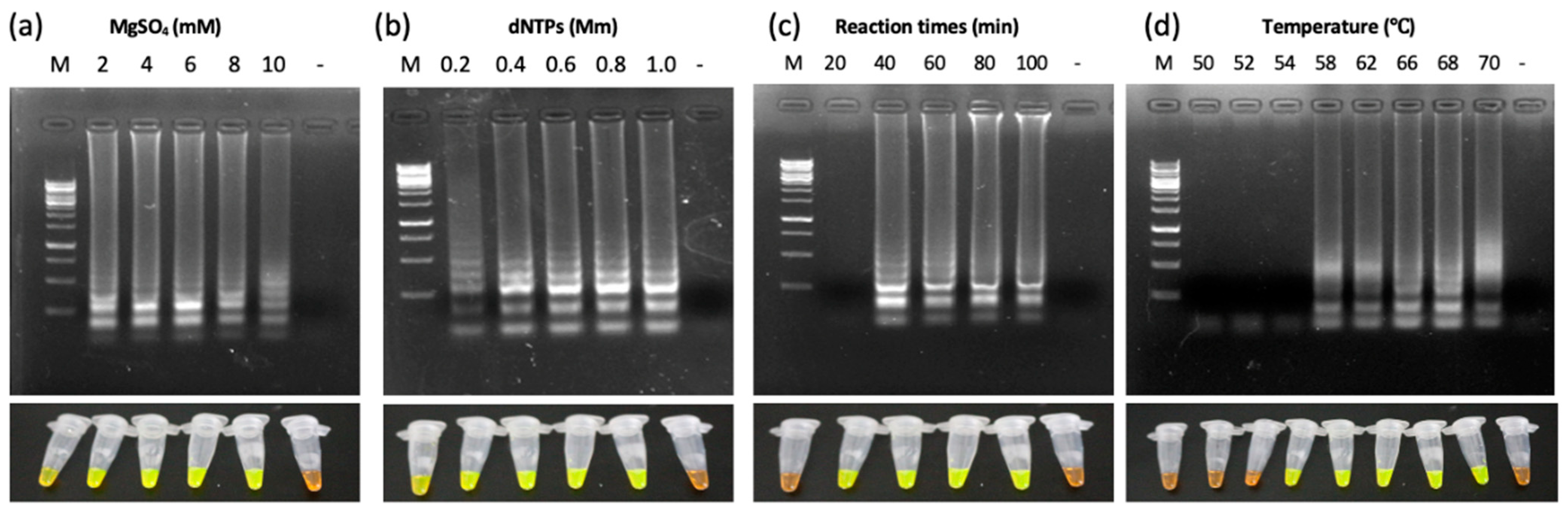
3.4. Standardization and Optimization of LAMP Reaction Conditions
3.5. Analytical Specificity and Sensitivity of the LAMP-LFD Method
3.6. Application of the LAMP-LFD Method for Detection in Field Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Qiu, L.; Heng, L.; Xu, R.; Luo, J.; Li, Y. Two new nimbolinin- and trichilin-class limonoids isolated from the fruits of Melia azedarach. Chin. J. Nat. Med. 2019, 17, 227–230. [Google Scholar] [CrossRef]
- Zhang, S.N.; Huang, L.; Ma, R.J.; Yang, M.F.; Wei, B.F.; Song, H.Z.; Wang, H.S.; Tan, Q.G. Chemical constituents from the barks of Melia azedarach and their PTP1B inhibitory activity. Nat. Prod. Res. 2020, 35, 4442–4447. [Google Scholar] [CrossRef] [PubMed]
- Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The current global scenario. Viruses 2016, 8, 177. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology; University of Florida: Gainesville, FL, USA, 2005. [Google Scholar]
- Jeevalatha, A.; Kaundal, P.; Kumar, R. Optimized loop-mediated isothermal amplification assay for Tomato leaf curl New Delhi virus-[potato] detection in potato leaves and tubers. Eur. J. Plant Pathol. 2018, 150, 565–573. [Google Scholar] [CrossRef]
- Lu, Y.; Yao, B.; Wang, G.; Hong, N. The detection of ACLSV and ASPV in pear plants by RT-LAMP assays. J. Virol. Methods 2018, 252, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.L.; Deng, Q.Q.; Chen, J.W.; Shen, W.K. Development of a reverse transcription loop-mediated isothermal amplification assay for rapid and visual detection of Sugarcane streak mosaic virus in sugarcane. Crop Prot. 2019, 119, 38–45. [Google Scholar] [CrossRef]
- Lalle, M.; Possenti, A.; Dubey, J.P.; Pozio, E. Loop-Mediated Isothermal Amplification-Lateral-Flow Dipstick (LAMP-LFD) to detect Toxoplasma gondii oocyst in ready-to-eat salad. Food Microbiol. 2018, 70, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.N.; Jia, C.X.; Qian, Y.; Yang, T.; Wang, Y.R.; Zheng, T.R.; Zhuang, Q.G.; Xi, D.H. Development and application of a reverse transcription loop-mediated isothermal amplification combined with lateral flow dipstick for rapid and visual detection of Citrus leaf blotch virus in kiwifruit. Crop Prot. 2021, 143, 105555. [Google Scholar] [CrossRef]
- Allgöwer, S.M.; Hartmann, C.A.; Lipinski, C.; Mahler, V.; Randow, S.; Völker, E.; Holzhauser, T. LAMP-LFD Based on Isothermal Amplification of Multicopy Gene ORF160b: Applicability for Highly Sensitive Low-Tech Screening of Allergenic Soybean (Glycine max) in Food. Foods 2020, 9, 1741. [Google Scholar] [CrossRef] [PubMed]
- Lindgreen, S. AdapterRemoval: Easy cleaning of next-generation sequencing reads. BMC Res. Notes 2012, 5, 337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Gao, S.; Padmanabhan, C.; Li, R.; Galvez, M.; Gutierrez, D.; Fuentes, S.; Ling, K.S.; Kreuze, J.; Fei, Z. VirusDetect: An automated pipeline for efficient virus discovery using deep sequencing of small RNAs. Virology 2017, 500, 130–138. [Google Scholar] [CrossRef] [PubMed]
- Sun, K.; Liang, Y.; Zhong, X.; Hu, X.; Zhang, P.; Yu, X. Nightshade Curly Top Virus: A Possible New Virus of the Genus Topocuvirus Infecting Solanum nigrum in China. Plant Dis. 2021, 105, 1006–1012. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 3, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, E.; Ravel, S.; Agret, C.; Abrokwah, F.; Dzahini-Obiatey, H.; Galyuon, I.; Kouakou, K.; Jeyaseelan, E.C.; Allainguillaume, J.; Wetten, A. Next generation sequencing elucidates cacao badnavirus diversity and reveals the existence of more than ten viral species. Virus Res. 2018, 244, 235–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, J.H.; Guo, Q.X.; Jian, H.; Chen, C.L.; Dan, Y.; Liu, Q.; Guo, Y.D. Rapid detection of Meloidogyne spp. by LAMP assay in soil and roots. Crop Prot. 2011, 30, 1063–1069. [Google Scholar] [CrossRef]
- King, A.M.Q.; Adams, M.J.; Lefkowitz, E.J.; Carstens, E.B. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses; Academic Press: San Diego, CA, USA, 2012; p. 1463. [Google Scholar]
- Zheng, H.; Wu, Y.; Ding, J.; Binion, D.; Fu, W.; Reardon, R. Invasive Plants Established in the United States that are Found in Asia and Their Associated Natural Enemies; USDA Forest Service: Morgantown, WV, USA, 2006; Volume 2, p. 175. [Google Scholar]
- Lin, Z.; Lei, Y. Bionomics of Erythroneura melia Kuoh and Its Control. Entomol. Knowl. 2001, 38, 47–49. [Google Scholar]
- Zhao, Z. Economic Insect Fauna of China, Volume 42, Lepidoptera. Lymantriidae (II); Science Press: Beijing, China, 1994; p. 165. [Google Scholar]




| Primer Type | Primer Name | Sequence (5′–3′) | Length (bp) | Usage |
|---|---|---|---|---|
| PCR | ChTBV1-F | CTGCAGCTTCTGCAAGAG | 18 | PCR Detection of ChTBV1 |
| ChTBV1-R | GAAAATGTAGGGCTCATTGT | 20 | ||
| LAMP | F3 | GAGAAATGGAGAAACAAACGT | 21 | LAMP Detection of ChTBV1 |
| B3 | CAACCTTTCCAAATCTCTGT | 20 | ||
| LB | TCGCTAGAGGATCCTGCTAC | 21 | ||
| FIP (F1c + F2) a | GTCCCTTCTCTTCCCTCAGCGAGAATGATGTATCCCACGG | 40 | ||
| BIP (B1c + B2) | CTGTCCCAGATAAGAAGAGTGTTCCCTGATCCTGAATATGTGTTGT | 46 | ||
| Probe b | GGACAGAATATTCTGTGTC | 19 |
| ORF | Genome Position (5′-3′) | Protein | Function | Top Two Viruses with the Highest Amino Acid Sequence Identity | |
|---|---|---|---|---|---|
| Amino Acid | kDa | ||||
| 1 | 252–683 | 144 | 16.5 | Unknown | 67.1%, ORF1 of Cacao swollen shoot Ghana L virus at a query coverage of 99%; 61.5%, hypothetical protein of Cacao swollen shoot virus at a query coverage of 99%. |
| 2 | 680–1108 | 143 | 16.1 | Unknown | 51.7%, ORF2 of Cacao swollen shoot Ghana L virus at a query coverage of 99%; 42.9%, ORF2 of Cacao swollen shoot Ghana K virus at a query coverage of 98%. |
| 3 | 1071–6647 | 1858 | 213 | Polyprotein | 57.6%, ORF1 of Cacao swollen shoot Ghana L virus at a query coverage of 99%; 56.1%, ORF1 of Cacao swollen shoot virus at a query coverage of 97%. |
| Y | 6278–6670 | 130 | 14.3 | Unknown | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, H.; Tang, J.; Sun, K.; Yu, X. Identification of a New Badnavirus in the Chinaberry (Melia azedarach) Tree and Establishment of a LAMP-LFD Assay for Its Rapid and Visual Detection. Viruses 2021, 13, 2408. https://doi.org/10.3390/v13122408
Lu H, Tang J, Sun K, Yu X. Identification of a New Badnavirus in the Chinaberry (Melia azedarach) Tree and Establishment of a LAMP-LFD Assay for Its Rapid and Visual Detection. Viruses. 2021; 13(12):2408. https://doi.org/10.3390/v13122408
Chicago/Turabian StyleLu, Huixin, Jintian Tang, Kai Sun, and Xiaoping Yu. 2021. "Identification of a New Badnavirus in the Chinaberry (Melia azedarach) Tree and Establishment of a LAMP-LFD Assay for Its Rapid and Visual Detection" Viruses 13, no. 12: 2408. https://doi.org/10.3390/v13122408
APA StyleLu, H., Tang, J., Sun, K., & Yu, X. (2021). Identification of a New Badnavirus in the Chinaberry (Melia azedarach) Tree and Establishment of a LAMP-LFD Assay for Its Rapid and Visual Detection. Viruses, 13(12), 2408. https://doi.org/10.3390/v13122408





