Inositol Phosphates and Retroviral Assembly: A Cellular Perspective
Abstract
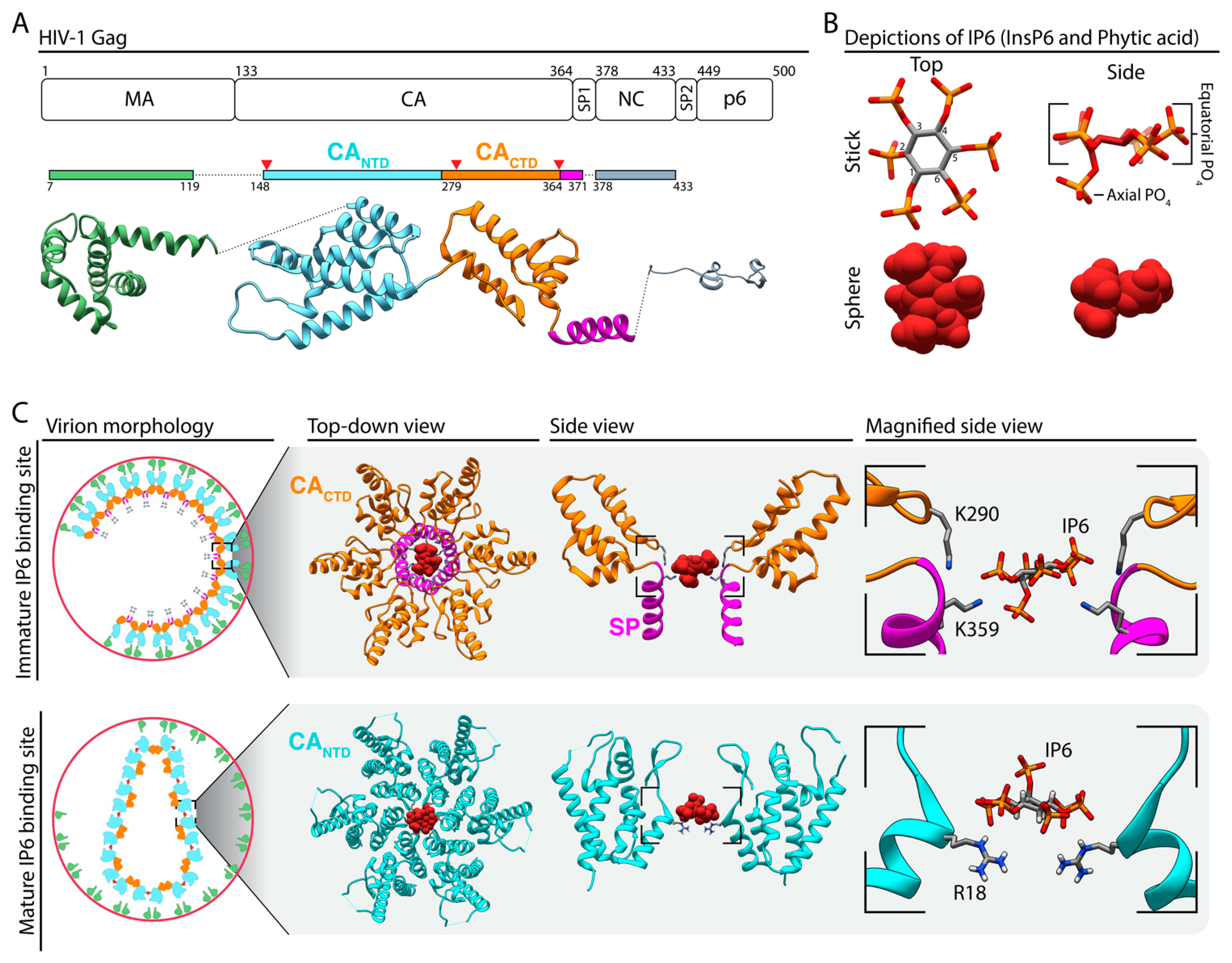
1. Retroviral Assembly and Inositol Phosphates
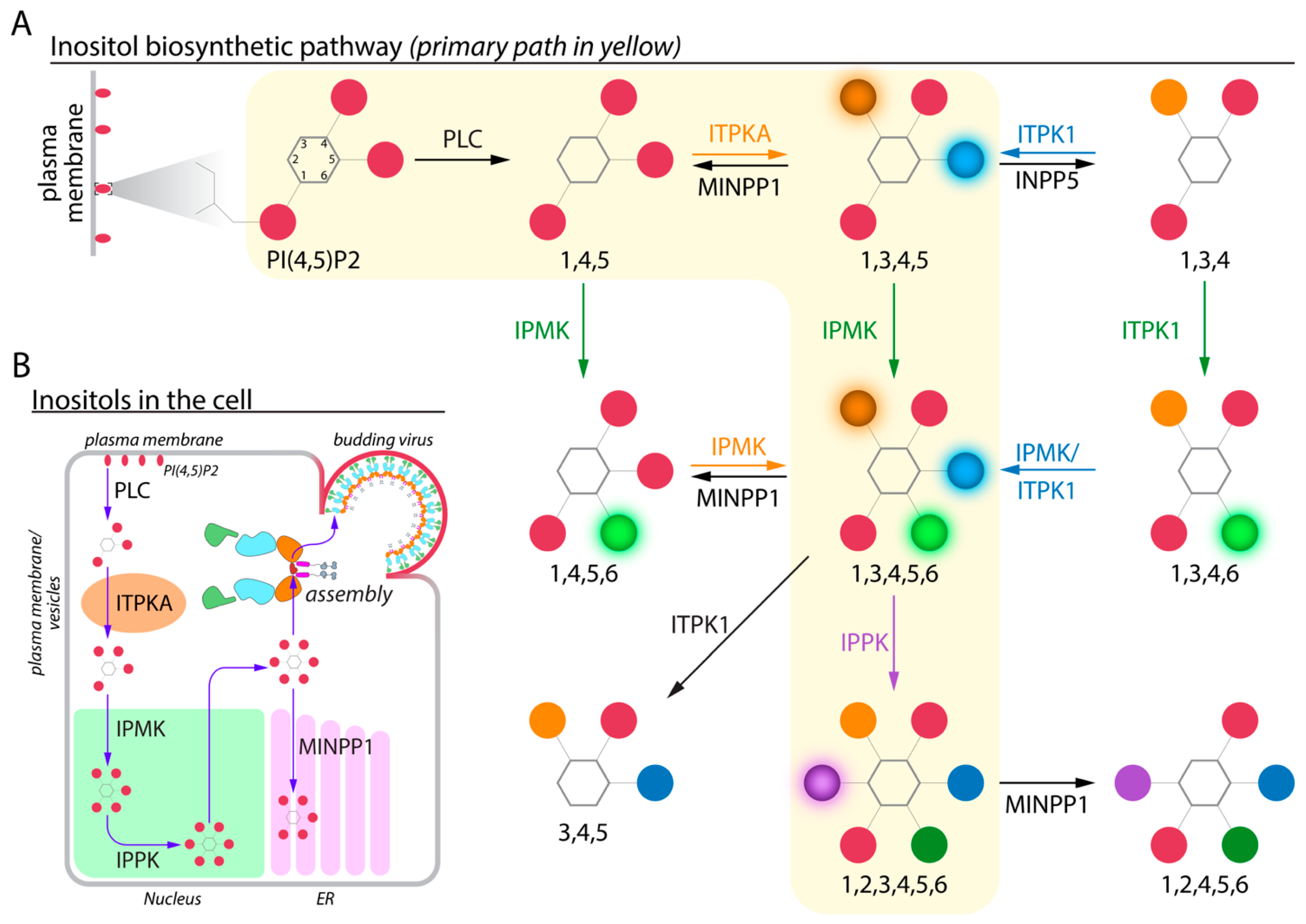
2. Biosynthesis of Higher-Order IPs in Cells
3. Measuring Cellular and Viral IP Levels
4. Current Progress in Identifying the Roles of IP6 and IP5 in HIV Assembly
5. The Role of IPs in Post-Entry Steps
6. The Role of IPs in Retroviruses Other Than HIV
7. Future Directions for Studying IPs in the Context of Retroviral Infection
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briggs, J.A.G.; Simon, M.N.; Gross, I.; Kräusslich, H.-G.; Fuller, S.D.; Vogt, V.M.; Johnson, M.C. The stoichiometry of Gag protein in HIV-1. Nat. Struct. Mol. Biol. 2004, 11, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Wright, E.R.; Schooler, J.B.; Ding, H.J.; Kieffer, C.; Fillmore, C.; Sundquist, W.I.; Jensen, G.J. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 2007, 26, 2218–2226. [Google Scholar] [CrossRef] [PubMed]
- Briggs, J.A.G.; Riches, J.D.; Glass, B.; Bartonova, V.; Zanetti, G.; Krausslich, H.-G. Structure and assembly of immature HIV. Proc. Natl. Acad. Sci. USA 2009, 106, 11090–11095. [Google Scholar] [CrossRef]
- Kleinpeter, A.B.; Freed, E.O. HIV-1 Maturation: Lessons Learned from Inhibitors. Viruses 2020, 12, 940. [Google Scholar] [CrossRef]
- Votteler, J.; Sundquist, W.I. Virus Budding and the ESCRT Pathway. Cell Host Microbe 2013, 14, 232–241. [Google Scholar] [CrossRef]
- Meng, B.; Lever, A.M.L. The Interplay between ESCRT and Viral Factors in the Enveloped Virus Life Cycle. Viruses 2021, 13, 324. [Google Scholar] [CrossRef] [PubMed]
- Rose, K.M.; Spada, S.J.; Hirsch, V.M.; Bouamr, F. When in Need of an ESCRT: The Nature of Virus Assembly Sites Suggests Mechanistic Parallels between Nuclear Virus Egress and Retroviral Budding. Viruses 2021, 13, 1138. [Google Scholar] [CrossRef]
- Hill, C.P.; Worthylake, D.; Bancroft, D.P.; Christensen, A.M.; Sundquist, W.I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: Implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 1996, 93, 3099–3104. [Google Scholar] [CrossRef] [PubMed]
- Schur, F.K.M.; Obr, M.; Hagen, W.J.H.; Wan, W.; Jakobi, A.J.; Kirkpatrick, J.M.; Sachse, C.; Kräusslich, H.-G.; Briggs, J.A.G. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 2016, 353, 506–508. [Google Scholar] [CrossRef]
- Wouters, J.; Oudjama, Y.; Barkley, S.J.; Tricot, C.; Stalon, V.; Droogmans, L.; Poulter, C.D. Catalytic Mechanism of Escherichia coli Isopentenyl Diphosphate Isomerase Involves Cys-67, Glu-116, and Tyr-104 as Suggested by Crystal Structures of Complexes with Transition State Analogues and Irreversible Inhibitors. J. Biol. Chem. 2003, 278, 11903–11908. [Google Scholar] [CrossRef]
- Qu, K.; Ke, Z.; Zila, V.; Anders-Össwein, M.; Glass, B.; Mücksch, F.; Müller, R.; Schultz, C.; Müller, B.; Kräusslich, H.-G.; et al. Maturation of the matrix and viral membrane of HIV-1. Science 2021, 373, 700–704. [Google Scholar] [CrossRef]
- Alfadhli, A.; Barklis, R.L.; Barklis, E. HIV-1 matrix organizes as a hexamer of trimers on membranes containing phosphatidylinositol-(4,5)-bisphosphate. Virology 2009, 387, 466–472. [Google Scholar] [CrossRef]
- De Marco, A.; Muller, B.; Glass, B.; Riches, J.D.; Kräusslich, H.-G.; Briggs, J.A.G. Structural Analysis of HIV-1 Maturation Using Cryo-Electron Tomography. PLoS Pathog. 2010, 6, e1001215. [Google Scholar] [CrossRef] [PubMed]
- Dick, R.A.; Zadrozny, K.K.; Xu, C.; Schur, F.K.M.; Lyddon, T.D.; Ricana, C.L.; Wagner, J.M.; Perilla, J.R.; Ganser-Pornillos, B.K.; Johnson, M.C.; et al. Inositol phosphates are assembly co-factors for HIV-1. Nature 2018, 560, 509–512. [Google Scholar] [CrossRef] [PubMed]
- Obr, M.; Schur, F.K.M.; Dick, R.A. A Structural Perspective of the Role of IP6 in Immature and Mature Retroviral Assembly. Viruses 2021, 13, 1853. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Huckhagel, C.; Kräusslich, H.-G. N-Terminal Extension of Human Immunodeficiency Virus Capsid Protein Converts the In Vitro Assembly Phenotype from Tubular to Spherical Particles. J. Virol. 1998, 72, 4798–4810. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Müller, B.; Hell, S.W.; Kräusslich, H.-G. Maturation-Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef]
- Buttler, C.A.; Pezeshkian, N.; Fernandez, M.V.; Aaron, J.; Norman, S.; Freed, E.O.; Van Engelenburg, S.B. Single molecule fate of HIV-1 envelope reveals late-stage viral lattice incorporation. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Izadmehr, S.; Kamau, E.; Kong, X.-P.; Chen, B.K. Sequential trafficking of Env and Gag to HIV-1 T cell virological synapses revealed by live imaging. Retrovirology 2019, 16, 1–16. [Google Scholar] [CrossRef]
- Mattei, S.; Glass, B.; Hagen, W.J.H.; Kräusslich, H.-G.; Briggs, J.A.G. The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 2016, 354, 1434–1437. [Google Scholar] [CrossRef]
- Bush, D.L.; Vogt, V.M. In Vitro Assembly of Retroviruses. Annu. Rev. Virol. 2014, 1, 561–580. [Google Scholar] [CrossRef]
- Qu, K.; Glass, B.; Doležal, M.; Schur, F.K.M.; Murciano, B.; Rein, A.; Rumlová, M.; Ruml, T.; Kräusslich, H.-G.; Briggs, J.A.G. Structure and architecture of immature and mature murine leukemia virus capsids. Proc. Natl. Acad. Sci. USA 2018, 115, E11751–E11760. [Google Scholar] [CrossRef]
- Bharat, T.A.M.; Menendez, L.R.C.; Hagen, W.J.H.; Lux, V.; Igonet, S.; Schorb, M.; Schur, F.K.M.; Kräusslich, H.-G.; Briggs, J.A.G. Cryo-electron microscopy of tubular arrays of HIV-1 Gag resolves structures essential for immature virus assembly. Proc. Natl. Acad. Sci. USA 2014, 111, 8233–8238. [Google Scholar] [CrossRef] [PubMed]
- Füzik, T.; Píchalová, R.; Schur, F.K.M.; Strohalmová, K.; Křížová, I.; Hadravová, R.; Rumlová, M.; Briggs, J.A.G.; Ulbrich, P.; Ruml, T. Nucleic Acid Binding by Mason-Pfizer Monkey Virus CA Promotes Virus Assembly and Genome Packaging. J. Virol. 2016, 90, 4593–4603. [Google Scholar] [CrossRef][Green Version]
- Dick, R.A.; Xu, C.; Morado, D.R.; Kravchuk, V.; Ricana, C.L.; Lyddon, T.D.; Broad, A.M.; Feathers, J.R.; Johnson, M.C.; Vogt, V.M.; et al. Structures of immature EIAV Gag lattices reveal a conserved role for IP6 in lentivirus assembly. PLoS Pathog. 2020, 16, e1008277. [Google Scholar] [CrossRef] [PubMed]
- Obr, M.; Ricana, C.L.; Nikulin, N.; Feathers, J.-P.R.; Klanschnig, M.; Thader, A.; Johnson, M.C.; Vogt, V.M.; Schur, F.K.M.; Dick, R.A. Structure of the mature Rous sarcoma virus lattice reveals a role for IP6 in the formation of the capsid hexamer. Nat. Commun. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Vogt, V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [CrossRef] [PubMed]
- Klikova, M.; Rhee, S.S.; Hunter, E.; Ruml, T. Efficient in vivo and in vitro assembly of retroviral capsids from Gag precursor proteins expressed in bacteria. J. Virol. 1995, 69, 1093–1098. [Google Scholar] [CrossRef]
- Bharat, T.A.M.; Davey, N.E.; Ulbrich, P.; Riches, J.D.; de Marco, A.; Rumlova, M.; Sachse, C.; Ruml, T.; Briggs, J.A.G. Structure of the immature retroviral capsid at 8 Å resolution by cryo-electron microscopy. Nature 2012, 487, 385–389. [Google Scholar] [CrossRef]
- Campbell, S.; Rein, A. In Vitro Assembly Properties of Human Immunodeficiency Virus Type 1 Gag Protein Lacking the p6 Domain. J. Virol. 1999, 73, 2270–2279. [Google Scholar] [CrossRef]
- Purdy, J.G.; Flanagan, J.M.; Ropson, I.J.; Rennoll-Bankert, K.E.; Craven, R.C. Critical Role of Conserved Hydrophobic Residues within the Major Homology Region in Mature Retroviral Capsid Assembly. J. Virol. 2008, 82, 5951–5961. [Google Scholar] [CrossRef]
- Ehrlich, L.S.; Agresta, B.E.; Carter, C.A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J. Virol. 1992, 66, 4874–4883. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Krausslich, H.-G. In Vitro Assembly Properties of Purified Bacterially Expressed Capsid Proteins of Human Immunodeficiency Virus. Eur. J. Biochem. 1997, 249, 592–600. [Google Scholar] [CrossRef]
- Campbell, S.; Fisher, R.J.; Towler, E.M.; Fox, S.; Issaq, H.J.; Wolfe, T.; Phillips, L.R.; Rein, A. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 2001, 98, 10875–10879. [Google Scholar] [CrossRef]
- French, P.J.; Bunce, C.M.; Stephens, L.R.; Lord, J.M.; McConnell, F.M.; Brown, G.; Creba, J.A.; Michell, R.H. Changes in the levels of inositol lipids and phosphates during the differentiation of HL60 promyelocytic cells towards neutrophils or monocytes. Proc. Biol. Sci. 1991, 245, 193–201. [Google Scholar] [CrossRef]
- Mountford, J.; Bunce, C.; French, P.J.; Michell, R.H.; Brown, G. Intracellular concentrations of inositol, glycerophosphoinositol and inositol pentakisphosphate increase during haemopoietic cell differentiation. Biochim. Biophys. Acta 1994, 1222, 101–108. [Google Scholar] [CrossRef]
- Shamsuddin, A.M. Metabolism and cellular functions of IP6: A review. Anticancer Res. 1999, 19, 3733–3736. [Google Scholar] [PubMed]
- Letcher, A.J.; Schell, M.J.; Irvine, R.F. Do mammals make all their own inositol hexakisphosphate? Biochem. J. 2008, 416, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.S.C.; Bulley, S.J.; Pisani, F.; Irvine, R.F.; Saiardi, A. A novel method for the purification of inositol phosphates from biological samples reveals that no phytate is present in human plasma or urine. Open Biol. 2015, 5, 150014. [Google Scholar] [CrossRef] [PubMed]
- Fäcke, M.; Janetzko, A.; Shoeman, R.L.; Kräusslich, H.G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J. Virol. 1993, 67, 4972–4980. [Google Scholar] [CrossRef]
- Gross, I.; Hohenberg, H.; Wilk, T.; Wiegers, K.; Grättinger, M.; Müller, B.; Fuller, S.; Kräusslich, H.G. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 2000, 19, 103–113. [Google Scholar] [CrossRef]
- Qualley, D.F.; Lackey, C.M.; Paterson, J.P. Inositol phosphates compete with nucleic acids for binding to bovine leukemia virus matrix protein: Implications for deltaretroviral assembly. Proteins 2013, 81, 1377–1385. [Google Scholar] [CrossRef]
- Ealfadhli, A.; Ebarklis, E. The roles of lipids and nucleic acids in HIV-1 assembly. Front. Microbiol. 2014, 5, 253. [Google Scholar] [CrossRef] [PubMed]
- Tateishi, H.; Anraku, K.; Koga, R.; Okamoto, Y.; Fujita, M.; Otsuka, M. Design and synthesis of lipid-coupled inositol 1,2,3,4,5,6-hexakisphosphate derivatives exhibiting high-affinity binding for the HIV-1 MA domain. Org. Biomol. Chem. 2014, 12, 5006–5022. [Google Scholar] [CrossRef]
- Datta, S.A.K.; Zhao, Z.; Clark, P.K.; Tarasov, S.; Alexandratos, J.N.; Campbell, S.J.; Kvaratskhelia, M.; Lebowitz, J.; Rein, A. Interactions between HIV-1 Gag Molecules in Solution: An Inositol Phosphate-mediated Switch. J. Mol. Biol. 2007, 365, 799–811. [Google Scholar] [CrossRef]
- Mallery, D.L.; Márquez, C.L.; McEwan, W.A.; Dickson, C.F.; Jacques, D.A.; Anandapadamanaban, M.; Bichel, K.; Towers, G.; Saiardi, A.; Böcking, T.; et al. IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis. eLife 2018, 7, e35335. [Google Scholar] [CrossRef]
- Springer. Inositol Phosphates: Methods and Protocols; Miller, G.J., Ed.; Springer: New York, NY, USA, 2020; Volume 2091, ISBN 978-1-07-160166-2. [Google Scholar]
- Rapoport, S. Phytic acid in avian erythrocytes. J. Biol. Chem. 1940, 135, 403–406. [Google Scholar] [CrossRef]
- Streb, H.; Irvine, R.F.; Berridge, M.J.; Schulz, I. Release of Ca2+ from a nonmitochondrial intracellular store in pancreatic acinar cells by inositol-1,4,5-trisphosphate. Nature 1983, 306, 67–69. [Google Scholar] [CrossRef]
- Shears, S.B. The versatility of inositol phosphates as cellular signals. Biochim. Biophys. Acta 1998, 1436, 49–67. [Google Scholar] [CrossRef]
- Shears, S.B. Assessing the omnipotence of inositol hexakisphosphate. Cell. Signal. 2001, 13, 151–158. [Google Scholar] [CrossRef]
- Irvine, R.F.; Schell, M.J. Back in the water: The return of the inositol phosphates. Nat. Rev. Mol. Cell Biol. 2001, 2, 327–338. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Ahn, H.; Kim, M.G.; Lee, H.; Kim, S. The Expanding Significance of Inositol Polyphosphate Multikinase as a Signaling Hub. Mol. Cells 2017, 40, 315–321. [Google Scholar] [CrossRef]
- Chatree, S.; Thongmaen, N.; Tantivejkul, K.; Sitticharoon, C.; Vucenik, I. Role of Inositols and Inositol Phosphates in Energy Metabolism. Molecules 2020, 25, 5079. [Google Scholar] [CrossRef]
- Marolt, G.; Kolar, M. Analytical Methods for Determination of Phytic Acid and Other Inositol Phosphates: A Review. Molecules 2020, 26, 174. [Google Scholar] [CrossRef]
- Maffucci, T.; Falasca, M. Signalling Properties of Inositol Polyphosphates. Molecules 2020, 25, 5281. [Google Scholar] [CrossRef]
- Wilson, M.P.; Majerus, P.W. Isolation of Inositol 1,3,4-Trisphosphate 5/6-Kinase, cDNA Cloning, and Expression of the Recombinant Enzyme. J. Biol. Chem. 1996, 271, 11904–11910. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Reece, J.; Gabriel, S.E.; Shears, S.B. Apical localization of ITPK1 enhances its ability to be a modifier gene product in a murine tracheal cell model of cystic fibrosis. J. Cell Sci. 2006, 119, 1320–1328. [Google Scholar] [CrossRef]
- Saiardi, A.; Cockcroft, S. Human ITPK1: A Reversible Inositol Phosphate Kinase/Phosphatase That Links Receptor-Dependent Phospholipase C to Ca 2+ -Activated Chloride Channels. Sci. Signal. 2008, 1, pe5. [Google Scholar] [CrossRef]
- Majerus, P.W.; Wilson, D.B.; Zhang, C.; Nicholas, P.J.; Wilson, M.P. Expression of inositol 1,3,4-trisphosphate 5/6-kinase (ITPK1) and its role in neural tube defects. Adv. Enzym. Regul. 2010, 50, 365–372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ali, N.; Craxton, A.; Shears, S.B. Hepatic Ins(1,3,4,5)P4 3-phosphatase is compartmentalized inside endoplasmic reticulum. J. Biol. Chem. 1993, 268, 6161–6167. [Google Scholar] [CrossRef]
- Caffrey, J.J.; Hidaka, K.; Matsuda, M.; Hirata, M.; Shears, S.B. The human and rat forms of multiple inositol polyphosphate phosphatase: Functional homology with a histidine acid phosphatase up-regulated during endochondral ossification. FEBS Lett. 1999, 442, 99–104. [Google Scholar] [CrossRef]
- Chi, H.; Tiller, G.E.; Dasouki, M.J.; Romano, P.R.; Wang, J.; O’Keefe, R.J.; Puzas, J.E.; Rosier, R.N.; Reynolds, P.R. Multiple Inositol Polyphosphate Phosphatase: Evolution as a Distinct Group within the Histidine Phosphatase Family and Chromosomal Localization of the Human and Mouse Genes to Chromosomes 10q23 and 19. Genomics 1999, 56, 324–336. [Google Scholar] [CrossRef]
- Chi, H.; Yang, X.; Kingsley, P.D.; O’Keefe, R.J.; Puzas, J.E.; Rosier, R.N.; Shears, S.B.; Reynolds, P.R. Targeted Deletion of Minpp1 Provides New Insight into the Activity of Multiple Inositol Polyphosphate Phosphatase In Vivo. Mol. Cell. Biol. 2000, 20, 6496–6507. [Google Scholar] [CrossRef] [PubMed]
- Brehm, M.A.; Schenk, T.M.H.; Zhou, X.; Fanick, W.; Lin, H.; Windhorst, S.; Nalaskowski, M.M.; Kobras, M.; Shears, S.B.; Mayr, G.W. Intracellular localization of human Ins(1,3,4,5,6)P5 2-kinase. Biochem. J. 2007, 408, 335–345. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Thul, P.J.; Åkesson, L.; Wiking, M.; Mahdessian, D.; Geladaki, A.; Blal, H.A.; Alm, T.; Asplund, A.; Björk, L.; Breckels, L.M.; et al. A subcellular map of the human proteome. Science 2017, 356, eaal3321. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 2507. [Google Scholar] [CrossRef] [PubMed]
- Cell Atlas-IPPK-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000127080-IPPK/cell (accessed on 4 June 2021).
- Blood Atlas-IPPK-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000127080-IPPK/blood (accessed on 4 June 2021).
- Schmiedel, B.J.; Singh, D.; Madrigal, A.; Valdovino-Gonzalez, A.G.; White, B.M.; Zapardiel-Gonzalo, J.; Ha, B.; Altay, G.; Greenbaum, J.A.; McVicker, G.; et al. Impact of Genetic Polymorphisms on Human Immune Cell Gene Expression. Cell 2018, 175, 1701–1715.e16. [Google Scholar] [CrossRef]
- Monaco, G.; Lee, B.; Xu, W.; Mustafah, S.; Hwang, Y.Y.; Carré, C.; Burdin, N.; Visan, L.; Ceccarelli, M.; Poidinger, M.; et al. RNA-Seq Signatures Normalized by mRNA Abundance Allow Absolute Deconvolution of Human Immune Cell Types. Cell Rep. 2019, 26, 1627–1640.e7. [Google Scholar] [CrossRef]
- Stuart, J.A.; Anderson, K.L.; French, P.J.; Kirk, C.J.; Michell, R.H. The intracellular distribution of inositol polyphosphates in HL60 promyeloid cells. Biochem. J. 1994, 303, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Poyner, D.R.; Cooke, F.; Hanley, M.R.; Reynolds, D.J.; Hawkins, P.T. Characterization of metal ion-induced [3H]inositol hexakisphosphate binding to rat cerebellar membranes. J. Biol. Chem. 1993, 268, 1032–1038. [Google Scholar] [CrossRef]
- Nalaskowski, M.M.; Deschermeier, C.; Fanick, W.; Mayr, G.W. The human homologue of yeast ArgRIII protein is an inositol phosphate multikinase with predominantly nuclear localization. Biochem. J. 2002, 366, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Cell Atlas-IPMK-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000151151-IPMK/cell (accessed on 4 June 2021).
- Cell Atlas-ITPK1-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000100605-ITPK1/cell (accessed on 6 June 2021).
- Blood Atlas-IPMK-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000151151-IPMK/blood (accessed on 6 June 2021).
- Blood Atlas-ITPK1-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000100605-ITPK1/blood (accessed on 6 June 2021).
- Windhorst, S.; Lin, H.; Blechner, C.; Fanick, W.; Brandt, L.; Brehm, M.A.; Mayr, G.W. Tumour cells can employ extracellular Ins(1,2,3,4,5,6)P6 and multiple inositol-polyphosphate phosphatase 1 (MINPP1) dephosphorylation to improve their proliferation. Biochem. J. 2013, 450, 115–125. [Google Scholar] [CrossRef]
- Wundenberg, T.; Grabinski, N.; Lin, H.; Mayr, G.W. Discovery of InsP6-kinases as InsP6-dephosphorylating enzymes provides a new mechanism of cytosolic InsP6 degradation driven by the cellular ATP/ADP ratio. Biochem. J. 2014, 462, 173–184. [Google Scholar] [CrossRef] [PubMed]
- Blood Atlas-MINPP1-The Human Protein Atlas. Available online: https://www.proteinatlas.org/ENSG00000107789-MINPP1/blood (accessed on 7 June 2021).
- Gosein, V.; Leung, T.-F.; Krajden, O.; Miller, G.J. Inositol phosphate-induced stabilization of inositol 1,3,4,5,6-pentakisphosphate 2-kinase and its role in substrate specificity. Protein Sci. 2012, 21, 737–742. [Google Scholar] [CrossRef]
- Gosein, V.; Miller, G.J. Conformational Stability of Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase (IPK1) Dictates Its Substrate Selectivity. J. Biol. Chem. 2013, 288, 36788–36795. [Google Scholar] [CrossRef] [PubMed]
- Franco-Echevarría, E.; Sanz-Aparicio, J.; Troffer-Charlier, N.; Poterszman, A.; Perez, B.G. Crystallization and Preliminary X-Ray Diffraction Analysis of a Mammal Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase. Protein J. 2017, 36, 240–248. [Google Scholar] [CrossRef]
- Franco-Echevarría, E.; Sanz-Aparicio, J.; Brearley, C.; Rubio, J.M.G.; González, B. The crystal structure of mammalian inositol 1,3,4,5,6-pentakisphosphate 2-kinase reveals a new zinc-binding site and key features for protein function. J. Biol. Chem. 2017, 292, 10534–10548. [Google Scholar] [CrossRef]
- Wang, H.; Shears, S.B. Structural features of human inositol phosphate multikinase rationalize its inositol phosphate kinase and phosphoinositide 3-kinase activities. J. Biol. Chem. 2017, 292, 18192–18202. [Google Scholar] [CrossRef]
- Endo-Streeter, S.; Tsui, M.-K.M.; Odom, A.R.; Block, J.; York, J.D. Structural Studies and Protein Engineering of Inositol Phosphate Multikinase. J. Biol. Chem. 2012, 287, 35360–35369. [Google Scholar] [CrossRef] [PubMed]
- Seacrist, C.D.; Blind, R.D. Crystallographic and kinetic analyses of human IPMK reveal disordered domains modulate ATP binding and kinase activity. Sci. Rep. 2018, 8, 16672. [Google Scholar] [CrossRef] [PubMed]
- Miller, G.J.; Wilson, M.P.; Majerus, P.W.; Hurley, J.H. Specificity Determinants in Inositol Polyphosphate Synthesis: Crystal Structure of Inositol 1,3,4-Trisphosphate 5/6-Kinase. Mol. Cell 2005, 18, 201–212. [Google Scholar] [CrossRef] [PubMed]
- Chamberlain, P.P.; Qian, X.; Stiles, A.R.; Cho, J.; Jones, D.H.; Lesley, S.A.; Grabau, E.A.; Shears, S.B.; Spraggon, G. Integration of Inositol Phosphate Signaling Pathways via Human ITPK1. J. Biol. Chem. 2007, 282, 28117–28125. [Google Scholar] [CrossRef]
- Stentz, R.; Osborne, S.; Horn, N.; Li, A.W.H.; Hautefort, I.; Bongaerts, R.; Rouyer, M.; Bailey, P.; Shears, S.B.; Hemmings, A.M.; et al. A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut. Cell Rep. 2014, 6, 646–656. [Google Scholar] [CrossRef]
- Acquistapace, I.M.; Zietek, M.A.; Li, A.W.H.; Salmon, M.; Kühn, I.; Bedford, M.R.; Brearley, C.A.; Hemmings, A.M. Snapshots during the catalytic cycle of a histidine acid phytase reveal an induced-fit structural mechanism. J. Biol. Chem. 2020, 295, 17724–17737. [Google Scholar] [CrossRef] [PubMed]
- Blind, R.D. Structural analyses of inositol phosphate second messengers bound to signaling effector proteins. Adv. Biol. Regul. 2020, 75, 100667. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Park, S.J.; Hong, S.; Kim, K.; Kim, S. Inositol Polyphosphate Multikinase Signaling: Multifaceted Functions in Health and Disease. Mol. Cells 2021, 44, 187–194. [Google Scholar] [CrossRef]
- Kim, E.; Beon, J.; Lee, S.; Park, J.; Kim, S. IPMK: A versatile regulator of nuclear signaling events. Adv. Biol. Regul. 2016, 61, 25–32. [Google Scholar] [CrossRef]
- Lee, S.; Kim, M.-G.; Ahn, H.; Kim, S. Inositol Pyrophosphates: Signaling Molecules with Pleiotropic Actions in Mammals. Molecules 2020, 25, 2208. [Google Scholar] [CrossRef]
- Losito, O.; Szijgyarto, Z.; Resnick, A.C.; Saiardi, A. Inositol Pyrophosphates and Their Unique Metabolic Complexity: Analysis by Gel Electrophoresis. PLoS ONE 2009, 4, e5580. [Google Scholar] [CrossRef]
- Azevedo, C.; Saiardi, A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat. Protoc. 2006, 1, 2416–2422. [Google Scholar] [CrossRef]
- Tang, C.; Loeliger, E.; Luncsford, P.; Kinde, I.; Beckett, D.; Summers, M.F. Entropic switch regulates myristate exposure in the HIV-1 matrix protein. Proc. Natl. Acad. Sci. USA 2004, 101, 517–522. [Google Scholar] [CrossRef]
- Saad, J.S.; Miller, J.; Tai, J.; Kim, A.; Ghanam, R.H.; Summers, M.F. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11364–11369. [Google Scholar] [CrossRef] [PubMed]
- Tedbury, P.R.; Novikova, M.; Ablan, S.D.; Freed, E.O. Biochemical evidence of a role for matrix trimerization in HIV-1 envelope glycoprotein incorporation. Proc. Natl. Acad. Sci. USA 2016, 113, E182–E190. [Google Scholar] [CrossRef]
- Alfadhli, A.; Staubus, A.O.; Tedbury, P.R.; Novikova, M.; Freed, E.O.; Barklis, E. Analysis of HIV-1 Matrix-Envelope Cytoplasmic Tail Interactions. J. Virol. 2019, 93, e01079-19. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.I.; Sierra, R.G.; Yoon, C.H.; Su, Z.; Tateishi, H.; Koga, R.; Kotaro, K.; Yumoto, F.; Senda, T.; Liang, M.; et al. Serial Femtosecond X-Ray Diffraction of HIV-1 Gag MA-IP6 Microcrystals at Ambient Temperature. Int. J. Mol. Sci. 2019, 20, 1675. [Google Scholar] [CrossRef] [PubMed]
- Ciftci, H.; Tateishi, H.; Koiwai, K.; Koga, R.; Anraku, K.; Monde, K.; Dağ, Ç.; Destan, E.; Yuksel, B.; Ayan, E.; et al. Structural insight into host plasma membrane association and assembly of HIV-1 matrix protein. Sci. Rep. 2021, 11, 1–14. [Google Scholar] [CrossRef]
- Mallery, D.L.; Faysal, K.M.R.; Kleinpeter, A.; Wilson, M.S.C.; Vaysburd, M.; Fletcher, A.J.; Novikova, M.; Böcking, T.; Freed, E.O.; Saiardi, A.; et al. Cellular IP6 Levels Limit HIV Production while Viruses that Cannot Efficiently Package IP6 Are Attenuated for Infection and Replication. Cell Rep. 2019, 29, 3983–3996.e4. [Google Scholar] [CrossRef]
- Ricana, C.L.; Lyddon, T.D.; Dick, R.A.; Johnson, M.C. Primate lentiviruses require Inositol hexakisphosphate (IP6) or inositol pentakisphosphate (IP5) for the production of viral particles. PLoS Pathog. 2020, 16, e1008646. [Google Scholar] [CrossRef]
- Dostálková, A.; Kaufman, F.; Křížová, I.; Vokatá, B.; Ruml, T.; Rumlová, M. In Vitro Quantification of the Effects of IP6 and Other Small Polyanions on Immature HIV-1 Particle Assembly and Core Stability. J. Virol. 2020, 94, e00991-20. [Google Scholar] [CrossRef]
- Yu, A.; Lee, E.M.Y.; Jin, J.; Voth, G.A. Atomic-scale characterization of mature HIV-1 capsid stabilization by inositol hexakisphosphate (IP 6 ). Sci. Adv. 2020, 6, eabc6465. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Fischer, D.K.; Rankovic, S.; Li, W.; Dick, R.A.; Runge, B.; Zadorozhnyi, R.; Ahn, J.; Aiken, C.; Polenova, T.; et al. Permeability of the HIV-1 capsid to metabolites modulates viral DNA synthesis. PLoS Biol. 2020, 18, e3001015. [Google Scholar] [CrossRef]
- Kucharska, I.; Ding, P.; Zadrozny, K.K.; Dick, R.A.; Summers, M.F.; Ganser-Pornillos, B.K.; Pornillos, O. Biochemical Reconstitution of HIV-1 Assembly and Maturation. J. Virol. 2020, 94, e01844-19. [Google Scholar] [CrossRef]
- Mallery, D.L.; Kleinpeter, A.B.; Renner, N.; Faysal, K.M.R.; Novikova, M.; Kiss, L.; Wilson, M.S.C.; Ahsan, B.; Ke, Z.; Briggs, J.A.G.; et al. A stable immature lattice packages IP 6 for HIV capsid maturation. Sci. Adv. 2021, 7, eabe4716. [Google Scholar] [CrossRef]
- Poston, D.; Zang, T.; Bieniasz, P. Derivation and characterization of an HIV-1 mutant that rescues IP6 binding deficiency. Retrovirology 2021, 18, 1–14. [Google Scholar] [CrossRef]
- Sowd, G.A.; Aiken, C. Inositol phosphates promote HIV-1 assembly and maturation to facilitate viral spread in human CD4+ T cells. PLoS Pathog. 2021, 17, e1009190. [Google Scholar] [CrossRef]
- Huang, P.-T.; Summers, B.J.; Xu, C.; Perilla, J.R.; Malikov, V.; Naghavi, M.H.; Xiong, Y. FEZ1 Is Recruited to a Conserved Cofactor Site on Capsid to Promote HIV-1 Trafficking. Cell Rep. 2019, 28, 2373–2385.e7. [Google Scholar] [CrossRef]
- Christensen, D.E.; Ganser-Pornillos, B.K.; Johnson, J.S.; Pornillos, O.; Sundquist, W.I. Reconstitution and visualization of HIV-1 capsid-dependent replication and integration in vitro. Science 2020, 370, 8420. [Google Scholar] [CrossRef]
- Ramalho, R.; Rankovic, S.; Zhou, J.; Aiken, C.; Rousso, I. Analysis of the mechanical properties of wild type and hyperstable mutants of the HIV-1 capsid. Retrovirology 2016, 13, 17. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, S.; Varadarajan, J.; Ramalho, R.; Aiken, C.; Rousso, I. Reverse Transcription Mechanically Initiates HIV-1 Capsid Disassembly. J. Virol. 2017, 91, e00289-17. [Google Scholar] [CrossRef]
- Rankovic, S.; Ramalho, R.; Aiken, C.; Rousso, I. PF74 Reinforces the HIV-1 Capsid To Impair Reverse Transcription-Induced Uncoating. J. Virol. 2018, 92, e00845-18. [Google Scholar] [CrossRef]
- Márquez, C.L.; Lau, D.; Walsh, J.; Shah, V.; McGuinness, C.; Wong, A.; Aggarwal, A.; Parker, M.W.; Jacques, D.A.; Turville, S.; et al. Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife 2018, 7, e34772. [Google Scholar] [CrossRef]
- Yant, S.R.; Mulato, A.; Hansen, D.; Tse, W.C.; Niedziela-Majka, A.; Zhang, J.R.; Stepan, G.J.; Jin, D.; Wong, M.H.; Perreira, J.M.; et al. A highly potent long-acting small-molecule HIV-1 capsid inhibitor with efficacy in a humanized mouse model. Nat. Med. 2019, 25, 1377–1384. [Google Scholar] [CrossRef] [PubMed]
- Link, J.O.; Rhee, M.S.; Tse, W.C.; Zheng, J.; Somoza, J.R.; Rowe, W.; Begley, R.; Chiu, A.; Mulato, A.; Hansen, D.; et al. Clinical targeting of HIV capsid protein with a long-acting small molecule. Nature 2020, 584, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Jennings, J.; Shi, J.; Varadarajan, J.; Jamieson, P.J.; Aiken, C. The Host Cell Metabolite Inositol Hexakisphosphate Promotes Efficient Endogenous HIV-1 Reverse Transcription by Stabilizing the Viral Capsid. mBio 2020, 11, e02820-20. [Google Scholar] [CrossRef] [PubMed]
- Rankovic, S.; Deshpande, A.; Harel, S.; Aiken, C.; Rousso, I. HIV-1 Uncoating Occurs via a Series of Rapid Biomechanical Changes in the Core Related to Individual Stages of Reverse Transcription. J. Virol. 2021, 95, e00166-21. [Google Scholar] [CrossRef]
- Dostálková, A.; Vokatá, B.; Kaufman, F.; Ulbrich, P.; Ruml, T.; Rumlová, M. Effect of Small Polyanions on In Vitro Assembly of Selected Members of Alpha-, Beta- and Gammaretroviruses. Viruses 2021, 13, 129. [Google Scholar] [CrossRef]
- Riley, A.M.; Windhorst, S.; Lin, H.-Y.; Potter, B.V.L. Cellular Internalisation of an Inositol Phosphate Visualised by Using Fluorescent InsP5. ChemBioChem 2014, 15, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, R.; Lindwasser, O.W.; Smith, W.J.; Hurley, J.H.; Bonifacino, J.S. Downregulation of CD4 by Human Immunodeficiency Virus Type 1 Nef Is Dependent on Clathrin and Involves Direct Interaction of Nef with the AP2 Clathrin Adaptor. J. Virol. 2007, 81, 3877–3890. [Google Scholar] [CrossRef]
- Zhang, F.; Zang, T.; Wilson, S.J.; Johnson, M.C.; Bieniasz, P.D. Clathrin Facilitates the Morphogenesis of Retrovirus Particles. PLoS Pathog. 2011, 7, e1002119. [Google Scholar] [CrossRef]
- Stoneham, C.A.; Singh, R.; Jia, X.; Xiong, Y.; Guatelli, J. Endocytic activity of HIV-1 Vpu: Phosphoserine-dependent interactions with clathrin adaptors. Traffic 2017, 18, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Park, S.Y.; Bonifacino, J.S.; Hurley, J.H. How HIV-1 Nef hijacks the AP-2 clathrin adaptor to downregulate CD4. eLife 2014, 3, e01754. [Google Scholar] [CrossRef] [PubMed]
- Tourdot, R.; Radhakrishnan, R. Clathrin Mediated Endocytosis and its Role in Viral Entry. Atlas Genet. Cytogenet. Oncol. Haematol. 2013. [Google Scholar] [CrossRef]
- Pulloor, N.K.; Nair, S.; Kostic, A.; Bist, P.; Weaver, J.D.; Riley, A.; Tyagi, R.; Uchil, P.; York, J.D.; Snyder, S.H.; et al. Human Genome-Wide RNAi Screen Identifies an Essential Role for Inositol Pyrophosphates in Type-I Interferon Response. PLoS Pathog. 2014, 10, e1003981. [Google Scholar] [CrossRef]
- Kim, E.; Beon, J.; Lee, S.; Park, S.J.; Ahn, H.; Kim, M.G.; Park, J.E.; Kim, W.; Yuk, J.-M.; Kang, S.-J.; et al. Inositol polyphosphate multikinase promotes Toll-like receptor–induced inflammation by stabilizing TRAF6. Sci. Adv. 2017, 3, e1602296. [Google Scholar] [CrossRef]
- Wang, Q.; Vogan, E.; Nocka, L.M.; Rosen, C.E.; Zorn, J.A.; Harrison, S.C.; Kuriyan, J. Autoinhibition of Bruton’s tyrosine kinase (Btk) and activation by soluble inositol hexakisphosphate. eLife 2015, 4, e06074. [Google Scholar] [CrossRef]
- Kim, W.; Kim, E.; Min, H.; Kim, M.G.; Eisenbeis, V.B.; Dutta, A.K.; Pavlovic, I.; Jessen, H.J.; Kim, S.; Seong, R.H. Inositol polyphosphates promote T cell-independent humoral immunity via the regulation of Bruton’s tyrosine kinase. Proc. Natl. Acad. Sci. USA 2019, 116, 12952–12957. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricaña, C.L.; Dick, R.A. Inositol Phosphates and Retroviral Assembly: A Cellular Perspective. Viruses 2021, 13, 2516. https://doi.org/10.3390/v13122516
Ricaña CL, Dick RA. Inositol Phosphates and Retroviral Assembly: A Cellular Perspective. Viruses. 2021; 13(12):2516. https://doi.org/10.3390/v13122516
Chicago/Turabian StyleRicaña, Clifton L., and Robert A. Dick. 2021. "Inositol Phosphates and Retroviral Assembly: A Cellular Perspective" Viruses 13, no. 12: 2516. https://doi.org/10.3390/v13122516
APA StyleRicaña, C. L., & Dick, R. A. (2021). Inositol Phosphates and Retroviral Assembly: A Cellular Perspective. Viruses, 13(12), 2516. https://doi.org/10.3390/v13122516





