Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Viruses
2.2. Plasmids
2.3. Generation of Reassortant Virus
2.4. Passaging of Reassortant Virus
2.5. qRT-PCR, RT-PCR, and Restriction Digest Analyses
2.6. Replication Kinetics
2.7. Endpoint Dilution Assays
2.8. Sequence Analyses and Protein Structure Visualization
2.9. Statistics
3. Results
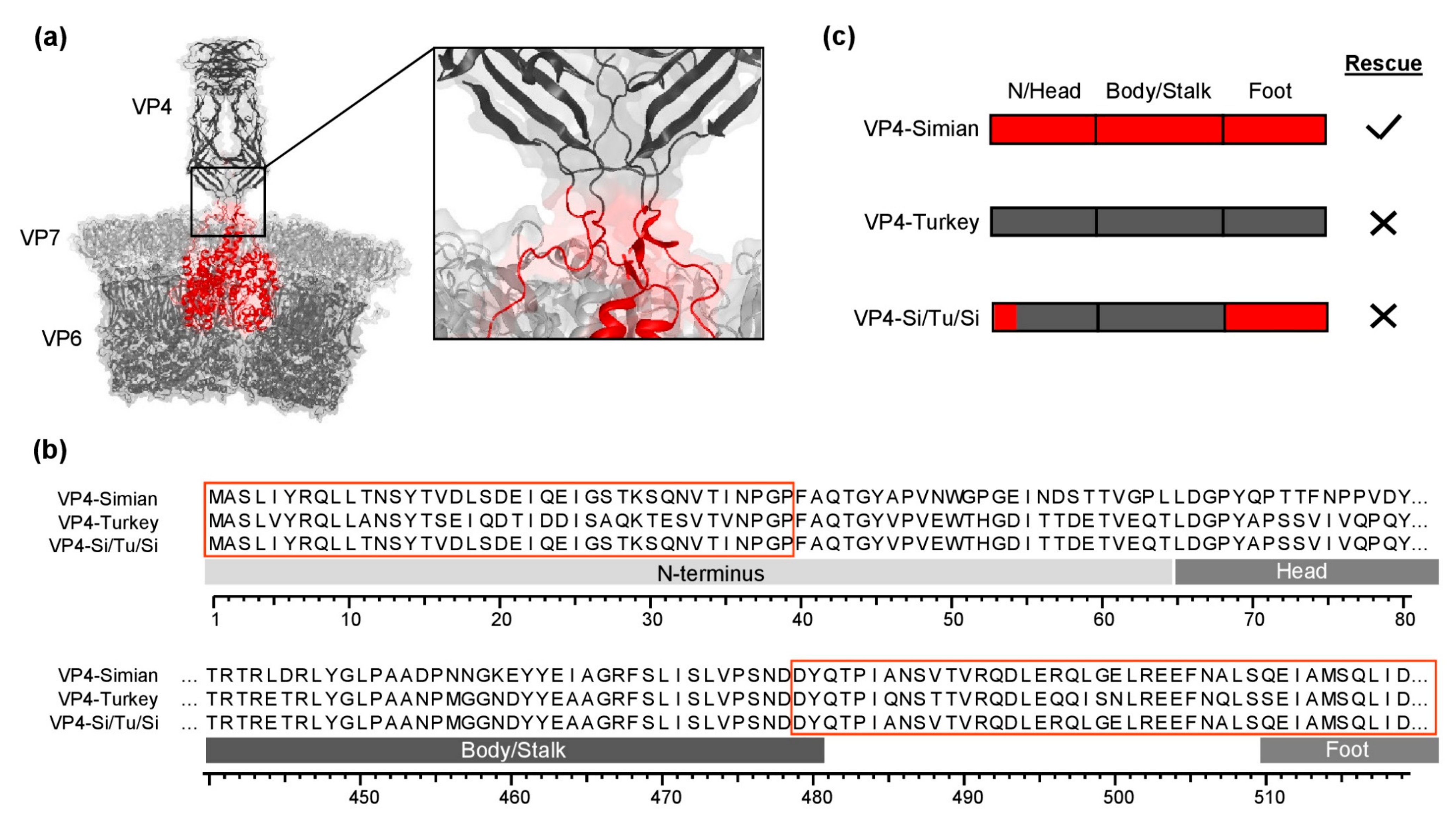
3.1. Chimeric Simian/Turkey VP4
3.2. Chimeric Chicken/Turkey VP4
3.3. Chimeric Bat/Human VP4
3.4. Chimeric Simian/Human VP4
3.5. Growth Kinetics of Cell-Culture Adapted Parent Viruses and their Rescued Reassortant Viruses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Troeger, C.; Khalil, I.A.; Rao, P.C.; Cao, S.; Blacker, B.F.; Ahmed, T.; Armah, G.; Bines, J.E.; Brewer, T.G.; Colombara, D.V.; et al. Rotavirus Vaccination and the Global Burden of Rotavirus Diarrhea Among Children Younger Than 5 Years. JAMA Pediatr. 2018, 172, 958–965. [Google Scholar] [CrossRef] [Green Version]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]
- Dhama, K.; Chauhan, R.S.; Mahendran, M.; Malik, S.V. Rotavirus diarrhea in bovines and other domestic animals. Vet. Res. Commun. 2009, 33, 1–23. [Google Scholar] [CrossRef]
- Dhama, K.; Saminathan, M.; Karthik, K.; Tiwari, R.; Shabbir, M.Z.; Kumar, N.; Malik, Y.S.; Singh, R.K. Avian rotavirus enteritis—An updated review. Vet. Q. 2015, 35, 142–158. [Google Scholar] [CrossRef] [Green Version]
- Martella, V.; Banyai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef] [Green Version]
- McDonald, S.M.; Nelson, M.I.; Turner, P.E.; Patton, J.T. Reassortment in segmented RNA viruses: Mechanisms and outcomes. Nat. Rev. Microbiol. 2016, 14, 448–460. [Google Scholar] [CrossRef] [Green Version]
- Gentsch, J.R.; Laird, A.R.; Bielfelt, B.; Griffin, D.D.; Banyai, K.; Ramachandran, M.; Jain, V.; Cunliffe, N.A.; Nakagomi, O.; Kirkwood, C.D.; et al. Serotype diversity and reassortment between human and animal rotavirus strains: Implications for rotavirus vaccine programs. J. Infect. Dis. 2005, 192 (Suppl. 1), S146–S159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, N.; Sen, A.; Wolf, M.; Vo, P.; Hoshino, Y.; Greenberg, H.B. Roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J. Virol. 2011, 85, 2686–2694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Settembre, E.C.; Chen, J.Z.; Dormitzer, P.R.; Grigorieff, N.; Harrison, S.C. Atomic model of an infectious rotavirus particle. EMBO J. 2011, 30, 408–416. [Google Scholar] [CrossRef]
- Estes, M.K.; Graham, D.Y.; Mason, B.B. Proteolytic enhancement of rotavirus infectivity: Molecular mechanisms. J. Virol. 1981, 39, 879–888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crawford, S.E.; Ramani, S.; Tate, J.E.; Parashar, U.D.; Svensson, L.; Hagbom, M.; Franco, M.A.; Greenberg, H.B.; O’Ryan, M.; Kang, G.; et al. Rotavirus infection. Nat. Rev. Dis. Primers 2017, 3, 17083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, M.; Goto, K.; Uemukai, H.; Mori, Y.; Ito, N.; Minamoto, N. Attachment and infection to MA104 cells of avian rotaviruses require the presence of sialic acid on the cell surface. J. Vet. Med. Sci. 2004, 66, 461–463. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ahmed, L.U.; Stuckert, M.R.; McGinnis, K.R.; Liu, Y.; Tan, M.; Huang, P.; Zhong, W.; Zhao, D.; Jiang, X.; et al. Molecular basis of P[II] major human rotavirus VP8* domain recognition of histo-blood group antigens. PLoS Pathog. 2020, 16, e1008386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Sankaran, B.; Laucirica, D.R.; Patil, K.; Salmen, W.; Ferreon, A.C.M.; Tsoi, P.S.; Lasanajak, Y.; Smith, D.F.; Ramani, S.; et al. Glycan recognition in globally dominant human rotaviruses. Nat. Commun. 2018, 9, 2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, L.; Crawford, S.E.; Czako, R.; Cortes-Penfield, N.W.; Smith, D.F.; Le Pendu, J.; Estes, M.K.; Prasad, B.V. Cell attachment protein VP8* of a human rotavirus specifically interacts with A-type histo-blood group antigen. Nature 2012, 485, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Bohm, R.; Fleming, F.E.; Maggioni, A.; Dang, V.T.; Holloway, G.; Coulson, B.S.; von Itzstein, M.; Haselhorst, T. Revisiting the role of histo-blood group antigens in rotavirus host-cell invasion. Nat. Commun. 2015, 6, 5907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, N.; Hu, L.; Ding, S.; Sanyal, M.; Zhao, B.; Sankaran, B.; Ramani, S.; McNeal, M.; Yasukawa, L.L.; Song, Y.; et al. Human VP8* mAbs neutralize rotavirus selectively in human intestinal epithelial cells. J. Clin. Investig. 2019, 130, 3839–3851. [Google Scholar] [CrossRef]
- Barbe, L.; Le Moullac-Vaidye, B.; Echasserieau, K.; Bernardeau, K.; Carton, T.; Bovin, N.; Nordgren, J.; Svensson, L.; Ruvoen-Clouet, N.; Le Pendu, J. Histo-blood group antigen-binding specificities of human rotaviruses are associated with gastroenteritis but not with in vitro infection. Sci. Rep. 2018, 8, 12961. [Google Scholar] [CrossRef]
- Johne, R.; Reetz, J.; Kaufer, B.B.; Trojnar, E. Generation of an Avian-Mammalian Rotavirus Reassortant by Using a Helper Virus-Dependent Reverse Genetics System. J. Virol. 2016, 90, 1439–1443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komoto, S.; Sasaki, J.; Taniguchi, K. Reverse genetics system for introduction of site-specific mutations into the double-stranded RNA genome of infectious rotavirus. Proc. Natl. Acad. Sci. USA 2006, 103, 4646–4651. [Google Scholar] [CrossRef] [Green Version]
- Trask, S.D.; Taraporewala, Z.F.; Boehme, K.W.; Dermody, T.S.; Patton, J.T. Dual selection mechanisms drive efficient single-gene reverse genetics for rotavirus. Proc. Natl. Acad. Sci. USA 2010, 107, 18652–18657. [Google Scholar] [CrossRef] [Green Version]
- Kanai, Y.; Komoto, S.; Kawagishi, T.; Nouda, R.; Nagasawa, N.; Onishi, M.; Matsuura, Y.; Taniguchi, K.; Kobayashi, T. Entirely plasmid-based reverse genetics system for rotaviruses. Proc. Natl. Acad. Sci. USA 2017, 114, 2349–2354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komoto, S.; Kanai, Y.; Fukuda, S.; Kugita, M.; Kawagishi, T.; Ito, N.; Sugiyama, M.; Matsuura, Y.; Kobayashi, T.; Taniguchi, K. Reverse Genetics System Demonstrates that Rotavirus Nonstructural Protein NSP6 Is Not Essential for Viral Replication in Cell Culture. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komoto, S.; Fukuda, S.; Ide, T.; Ito, N.; Sugiyama, M.; Yoshikawa, T.; Murata, T.; Taniguchi, K. Generation of Recombinant Rotaviruses Expressing Fluorescent Proteins by Using an Optimized Reverse Genetics System. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Kanai, Y.; Kawagishi, T.; Nouda, R.; Onishi, M.; Pannacha, P.; Nurdin, J.A.; Nomura, K.; Matsuura, Y.; Kobayashi, T. Development of Stable Rotavirus Reporter Expression Systems. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Komoto, S.; Fukuda, S.; Kugita, M.; Hatazawa, R.; Koyama, C.; Katayama, K.; Murata, T.; Taniguchi, K. Generation of Infectious Recombinant Human Rotaviruses from Just 11 Cloned cDNAs Encoding the Rotavirus Genome. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [Green Version]
- Philip, A.A.; Perry, J.L.; Eaton, H.E.; Shmulevitz, M.; Hyser, J.M.; Patton, J.T. Generation of Recombinant Rotavirus Expressing NSP3-UnaG Fusion Protein by a Simplified Reverse Genetics System. J. Virol. 2019, 93. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Hatazawa, R.; Kawamura, Y.; Yoshikawa, T.; Murata, T.; Taniguchi, K.; Komoto, S. Rapid generation of rotavirus single-gene reassortants by means of eleven plasmid-only based reverse genetics. J. Gen. Virol. 2020, 101, 806–815. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Onishi, M.; Kawagishi, T.; Pannacha, P.; Nurdin, J.; Nouda, R.; Yamasaki, M.; Lusiany, T.; Khamrin, P.; Okitsu, S.; et al. Reverse genetics approach for developing rotavirus vaccine candidates carrying VP4 and VP7 genes cloned from clinical isolates of human rotavirus. J. Virol. 2020, 95. [Google Scholar] [CrossRef]
- Kawagishi, T.; Nurdin, J.A.; Onishi, M.; Nouda, R.; Kanai, Y.; Tajima, T.; Ushijima, H.; Kobayashi, T. Reverse Genetics System for a Human Group A Rotavirus. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tacuba, L.; Feng, N.; Meade, N.J.; Mellits, K.H.; Jais, P.H.; Yasukawa, L.L.; Resch, T.K.; Jiang, B.; Lopez, S.; Ding, S.; et al. An Optimized Reverse Genetics System Suitable for Efficient Recovery of Simian, Human, and Murine-Like Rotaviruses. J. Virol. 2020, 94. [Google Scholar] [CrossRef]
- Papa, G.; Venditti, L.; Arnoldi, F.; Schraner, E.M.; Potgieter, C.; Borodavka, A.; Eichwald, C.; Burrone, O.R. Recombinant Rotaviruses Rescued by Reverse Genetics Reveal the Role of NSP5 Hyperphosphorylation in the Assembly of Viral Factories. J. Virol. 2019, 94. [Google Scholar] [CrossRef] [Green Version]
- Patzina-Mehling, C.; Falkenhagen, A.; Trojnar, E.; Gadicherla, A.K.; Johne, R. Potential of avian and mammalian species A rotaviruses to reassort as explored by plasmid only-based reverse genetics. Virus Res. 2020, 286, 198027. [Google Scholar] [CrossRef] [PubMed]
- Criglar, J.M.; Crawford, S.E.; Zhao, B.; Smith, H.G.; Stossi, F.; Estes, M.K. A Genetically Engineered Rotavirus NSP2 Phosphorylation Mutant Impaired in Viroplasm Formation and Replication Shows an Early Interaction between vNSP2 and Cellular Lipid Droplets. J. Virol. 2020, 94. [Google Scholar] [CrossRef] [PubMed]
- Falkenhagen, A.; Patzina-Mehling, C.; Ruckner, A.; Vahlenkamp, T.W.; Johne, R. Generation of simian rotavirus reassortants with diverse VP4 genes using reverse genetics. J. Gen. Virol. 2019, 100, 1595–1604. [Google Scholar] [CrossRef]
- Falkenhagen, A.; Patzina-Mehling, C.; Gadicherla, A.K.; Strydom, A.; O’Neill, H.G.; Johne, R. Generation of Simian Rotavirus Reassortants with VP4- and VP7-Encoding Genome Segments from Human Strains Circulating in Africa Using Reverse Genetics. Viruses 2020, 12, 201. [Google Scholar] [CrossRef] [Green Version]
- Otto, P.H.; Reetz, J.; Eichhorn, W.; Herbst, W.; Elschner, M.C. Isolation and propagation of the animal rotaviruses in MA-104 cells—30 years of practical experience. J. Virol. Methods 2015, 223, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Dormitzer, P.R.; Sun, Z.Y.; Wagner, G.; Harrison, S.C. The rhesus rotavirus VP4 sialic acid binding domain has a galectin fold with a novel carbohydrate binding site. EMBO J. 2002, 21, 885–897. [Google Scholar] [CrossRef]
- Ciarlet, M.; Schodel, F. Development of a rotavirus vaccine: Clinical safety, immunogenicity, and efficacy of the pentavalent rotavirus vaccine, RotaTeq. Vaccine 2009, 27 (Suppl. 6), G72–G81. [Google Scholar] [CrossRef]
- Ward, R.L.; Bernstein, D.I. Rotarix: A rotavirus vaccine for the world. Clin. Infect. Dis. 2009, 48, 222–228. [Google Scholar] [CrossRef] [PubMed]
- Zeller, M.; Patton, J.T.; Heylen, E.; de Coster, S.; Ciarlet, M.; van Ranst, M.; Matthijnssens, J. Genetic analyses reveal differences in the VP7 and VP4 antigenic epitopes between human rotaviruses circulating in Belgium and rotaviruses in Rotarix and RotaTeq. J. Clin. Microbiol. 2012, 50, 966–976. [Google Scholar] [CrossRef] [Green Version]
- Taniguchi, K.; Maloy, W.L.; Nishikawa, K.; Green, K.Y.; Hoshino, Y.; Urasawa, S.; Kapikian, A.Z.; Chanock, R.M.; Gorziglia, M. Identification of cross-reactive and serotype 2-specific neutralization epitopes on VP3 of human rotavirus. J. Virol. 1988, 62, 2421–2426. [Google Scholar] [CrossRef] [Green Version]
- Nair, N.; Feng, N.; Blum, L.K.; Sanyal, M.; Ding, S.; Jiang, B.; Sen, A.; Morton, J.M.; He, X.S.; Robinson, W.H.; et al. VP4- and VP7-specific antibodies mediate heterotypic immunity to rotavirus in humans. Sci. Transl. Med. 2017, 9. [Google Scholar] [CrossRef] [Green Version]
- Padilla-Noriega, L.; Dunn, S.J.; Lopez, S.; Greenberg, H.B.; Arias, C.F. Identification of two independent neutralization domains on the VP4 trypsin cleavage products VP5* and VP8* of human rotavirus ST3. Virology 1995, 206, 148–154. [Google Scholar] [CrossRef] [Green Version]
- Komoto, S.; Kugita, M.; Sasaki, J.; Taniguchi, K. Generation of recombinant rotavirus with an antigenic mosaic of cross-reactive neutralization epitopes on VP4. J. Virol. 2008, 82, 6753–6757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pando, V.; Isa, P.; Arias, C.F.; Lopez, S. Influence of calcium on the early steps of rotavirus infection. Virology 2002, 295, 190–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pesavento, J.B.; Billingsley, A.M.; Roberts, E.J.; Ramig, R.F.; Prasad, B.V. Structures of rotavirus reassortants demonstrate correlation of altered conformation of the VP4 spike and expression of unexpected VP4-associated phenotypes. J. Virol. 2003, 77, 3291–3296. [Google Scholar] [CrossRef] [Green Version]
- McDonald, S.M.; Matthijnssens, J.; McAllen, J.K.; Hine, E.; Overton, L.; Wang, S.; Lemey, P.; Zeller, M.; Van Ranst, M.; Spiro, D.J.; et al. Evolutionary dynamics of human rotaviruses: Balancing reassortment with preferred genome constellations. PLoS Pathog. 2009, 5, e1000634. [Google Scholar] [CrossRef] [PubMed]
- McDonald, S.M.; Patton, J.T. Assortment and packaging of the segmented rotavirus genome. Trends Microbiol. 2011, 19, 136–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mingo, R.; Zhang, S.; Long, C.P.; LaConte, L.E.W.; McDonald, S.M. Genetic determinants restricting the reassortment of heterologous NSP2 genes into the simian rotavirus SA11 genome. Sci. Rep. 2017, 7, 9301. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heiman, E.M.; McDonald, S.M.; Barro, M.; Taraporewala, Z.F.; Bar-Magen, T.; Patton, J.T. Group A human rotavirus genomics: Evidence that gene constellations are influenced by viral protein interactions. J. Virol. 2008, 82, 11106–11116. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Falkenhagen, A.; Huyzers, M.; van Dijk, A.A.; Johne, R. Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4. Viruses 2021, 13, 363. https://doi.org/10.3390/v13030363
Falkenhagen A, Huyzers M, van Dijk AA, Johne R. Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4. Viruses. 2021; 13(3):363. https://doi.org/10.3390/v13030363
Chicago/Turabian StyleFalkenhagen, Alexander, Marno Huyzers, Alberdina A. van Dijk, and Reimar Johne. 2021. "Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4" Viruses 13, no. 3: 363. https://doi.org/10.3390/v13030363
APA StyleFalkenhagen, A., Huyzers, M., van Dijk, A. A., & Johne, R. (2021). Rescue of Infectious Rotavirus Reassortants by a Reverse Genetics System Is Restricted by the Receptor-Binding Region of VP4. Viruses, 13(3), 363. https://doi.org/10.3390/v13030363





