Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Processing
2.2. Genome Assembly and Annotation
2.3. Infectivity Assays
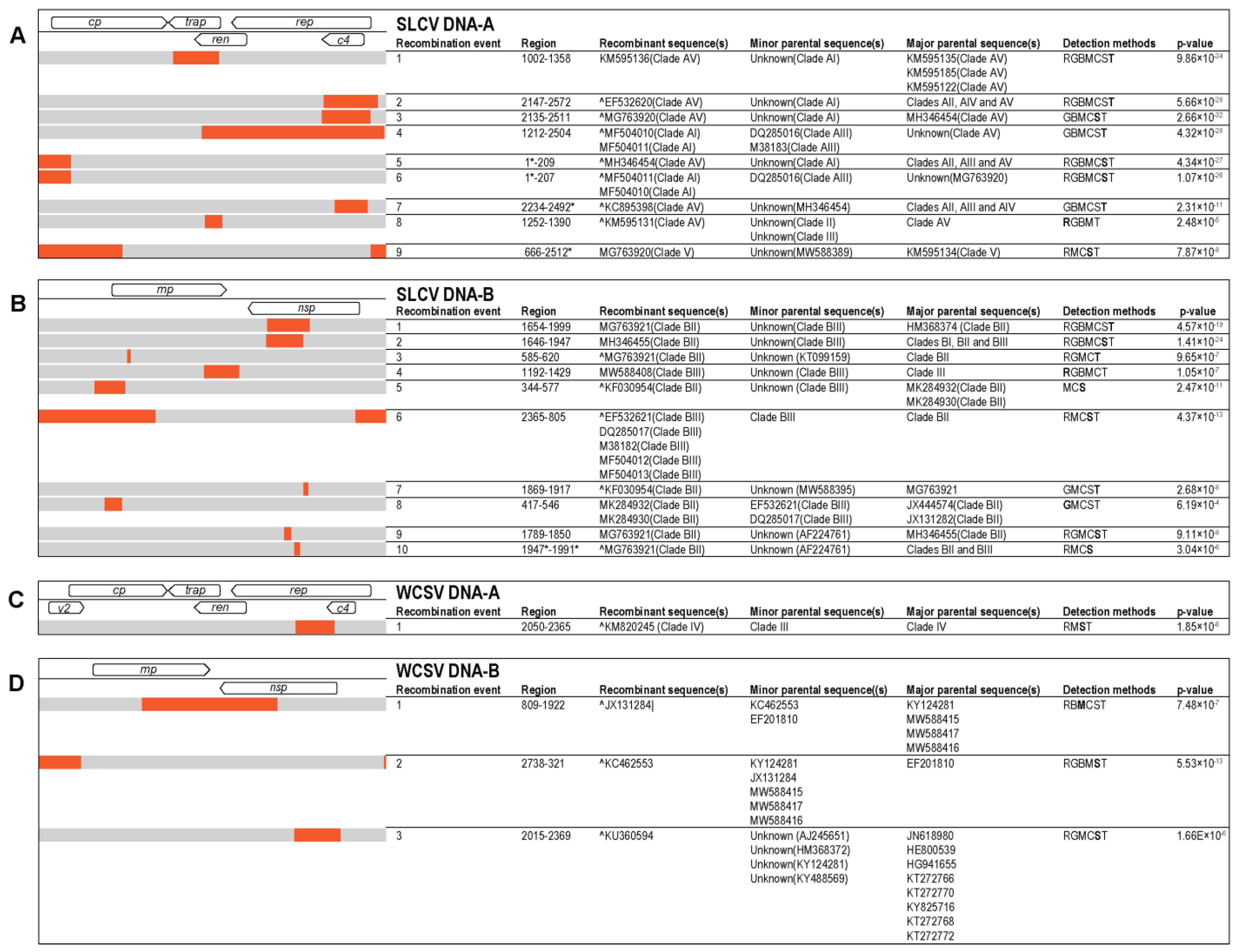
2.4. Sequence Analyses
3. Results and Discussion
3.1. Squash Leaf Curl Virus
3.2. Watermelon Chlorotic Stunt Virus
3.3. Infectivity Assays for Pseudo-Recombination
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Olson, N.H.; Baker, T.S.; Faulkner, L.; Agbandje-McKenna, M.; Boulton, M.I.; Davies, J.W.; McKenna, R. Structure of the Maize streak virus geminate particle. Virology 2001, 279, 471–477. [Google Scholar] [CrossRef] [Green Version]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Rojas, M.R.; Macedo, M.A.; Maliano, M.R.; Soto-Aguilar, M.; Souza, J.O.; Briddon, R.W.; Kenyon, L.; Rivera Bustamante, R.F.; Zerbini, F.M.; Adkins, S.; et al. World Management of Geminiviruses. Annu. Rev. Phytopathol. 2018, 56, 637–677. [Google Scholar] [CrossRef] [PubMed]
- Lazarowitz, S.G. Molecular characterization of two bipartite geminiviruses causing squash leaf curl disease: Role of viral replication and movement functions in determining host range. Virology 1991, 180, 70–80. [Google Scholar] [CrossRef]
- Flock, R.A.; Mayhew, D.E. Squash leaf curl, a new disease of cucurbits in California. Plant Dis. 1981, 65, 75–76. [Google Scholar] [CrossRef]
- Brown, J.K.; Idris, A.M.; Alteri, C.; Stenger, D.C. Emergence of a New Cucurbit-Infecting Begomovirus Species Capable of Forming Viable Reassortants with Related Viruses in the Squash leaf curl virus Cluster. Phytopathology 2002, 92, 734–742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, Y.W.; Rojas, M.R.; Gilbertson, R.L.; Wintermantel, W.M. First Report of Cucurbit yellow stunting disorder virus in California and Arizona, in Association with Cucurbit leaf crumple virus and Squash leaf curl virus. Plant Dis. 2007, 91, 330. [Google Scholar] [CrossRef] [PubMed]
- Medina-Hernández, D.; Caamal-Chan, M.G.; Vargas-Salinas, M.; Loera-Muro, A.; Barraza, A.; Holguín-Peña, R.J. Molecular characterization and phylogenetic analysis of a Squash leaf curl virus isolate from Baja California Sur, Mexico. PeerJ 2019, 7, e6774. [Google Scholar] [CrossRef] [Green Version]
- Antignus, Y.; Lachman, O.; Pearlsman, M.; Omer, S.; Yunis, H.; Messika, Y.; Uko, O.; Koren, A. Squash leaf curl geminivirus—A new illegal immigrant from the Western Hemisphere and a threat to cucurbit crops in Israel. Phytoparasitica 2003, 31, 415. [Google Scholar]
- Al-Musa, A.; Anfoka, G.; Misbeh, S.; Abhary, M.; Ahmad, F.H. Detection and Molecular Characterization of Squash leaf curl virus (SLCV) in Jordan: Molecular Characterization of Squash leaf curl virus. J. Phytopathol. 2008, 156, 311–316. [Google Scholar] [CrossRef]
- Idris, A.M.; Abdel-Salam, A.; Brown, J.K. Introduction of the New World Squash leaf curl virus to Squash (Cucurbita pepo) in Egypt: A Potential Threat to Important Food Crops. Plant Dis. 2006, 90, 1262. [Google Scholar] [CrossRef] [PubMed]
- Shahid, M.S.; Al-Sulaimani, H.; Al-Sadi, A.M. Squash Leaf Curl Virus: A New World Bipartite Begomovirus Threatening Squash Production in Oman. Plant Dis. 2020, 104, 2533. [Google Scholar] [CrossRef] [Green Version]
- Sobh, H.; Samsatly, J.; Jawhari, M.; Najjar, C.; Haidar, A.; Abou-Jawdah, Y. First Report of Squash leaf curl virus in Cucurbits in Lebanon. Plant Dis. 2012, 96, 1231. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.S.; Jamous, R.M.; Husein, E.Y.; Alkhader, M.Y. First Report of Squash leaf curl virus in Squash (Cucurbita pepo), Melon (Cucumis melo), and Cucumber (Cucumis sativa) in the Northern West Bank of the Palestinian Authority. Plant Disease 2010, 94, 640. [Google Scholar] [CrossRef]
- Abudy, A.; Sufrin-Ringwald, T.; Dayan-Glick, C.; Guenoune-Gelbart, D.; Livneh, O.; Zaccai, M.; Lapidot, M. Watermelon chlorotic stunt and Squash leaf curl begomoviruses—New threats to cucurbit crops in the Middle East. Isr. J. Plant Sci. 2010, 58, 33–42. [Google Scholar] [CrossRef]
- Ahmad, F.H.; Odeh, W.; Anfoka, G. First Report on the Association of Squash leaf curl virus and Watermelon chlorotic stunt virus with Tomato Yellow Leaf Curl Disease. Plant Dis. 2013, 97, 428. [Google Scholar] [CrossRef]
- Sufrin-Ringwald, T.; Lapidot, M. Characterization of a Synergistic Interaction Between Two Cucurbit-Infecting Begomoviruses: Squash leaf curl virus and Watermelon chlorotic stunt virus. Phytopathology 2011, 101, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Bedford, I.D.; Briddon, R.W.; Jones, P.; Alkaff, N.; Markham, P.G. Differentiation of three whitefly-transmitted geminiviruses from the Republic of Yemen. Eur. J. Plant Pathol. 1994, 100, 243–257. [Google Scholar] [CrossRef]
- Al-Musa, A.; Anfoka, G.; Al-Abdulat, A.; Misbeh, S.; Haj Ahmed, F.; Otri, I. Watermelon chlorotic stunt virus (WmCSV): A serious disease threatening watermelon production in Jordan. Virus Genes 2011, 43, 79–89. [Google Scholar] [CrossRef]
- Ali-Shtayeh, M.S.; Jamous, R.M.; Mallah, O.B.; Abu-Zeitoun, S.Y. Molecular characterization of watermelon chlorotic stunt virus (WmCSV) from Palestine. Viruses 2014, 6, 2444–2462. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.J.; Akhtar, S.; Briddon, R.W.; Ammara, U.; Al-Matrooshi, A.M.; Mansoor, S. Complete nucleotide sequence of watermelon chlorotic stunt virus originating from Oman. Viruses 2012, 4, 1169–1181. [Google Scholar] [CrossRef]
- Kheyr-Pour, A.; Bananej, K.; Dafalla, G.A.; Caciagli, P.; Noris, E.; Ahoonmanesh, A.; Lecoq, H.; Gronenborn, B. Watermelon chlorotic stunt virus from the Sudan and Iran: Sequence Comparisons and Identification of a Whitefly-Transmission Determinant. Phytopathology 2000, 90, 629–635. [Google Scholar] [CrossRef] [Green Version]
- Rezk, A.A.; Sattar, M.N.; Alhudaib, K.A.; Soliman, A.M. Identification of watermelon chlorotic stunt virus from watermelon and zucchini in Saudi Arabia. Can. J. Plant Pathol. 2019, 41, 285–290. [Google Scholar] [CrossRef]
- Bananej, K.; Ahoonmanesh, A.; Kheyr-Pour, A. Host Range of an Iranian Isolate of Watermelon Chlorotic Stunt Virus as Determined by Whitefly-mediated Inoculation and Agroinfection, and its Geographical Distribution. J. Phytopathol. 2002, 150, 423–430. [Google Scholar] [CrossRef]
- Abou-Jawdah, Y.; Sobh, H.; Haidar, A.; Samsatly, J. First report in Lebanon on detection of two whitefly transmitted cucurbit viruses and their molecular characterization. In Petria, Giornale di Patologia delle Piante 20, Proceedings of the 13th Congress of the Mediterranean Phytopathological Union, MPU, Plant Pathology Research Centre in Rome, Rome, Italy, 20–25 June 2010; Barba, M., Motta, E., Tomassoli, L., Riccioni, L., Eds.; CRA-Centro di Ricerca per la Patologia Vegetale: Rome, Italy, 2010; p. 268. [Google Scholar]
- Domínguez-Durán, G.; Rodríguez-Negrete, E.A.; Morales-Aguilar, J.J.; Camacho-Beltrán, E.; Romero-Romero, J.L.; Rivera-Acosta, M.A.; Leyva-López, N.E.; Arroyo-Becerra, A.; Méndez-Lozano, J. Molecular and biological characterization of Watermelon chlorotic stunt virus (WmCSV): An Eastern Hemisphere begomovirus introduced in the Western Hemisphere. Crop Prot. 2018, 103, 51–55. [Google Scholar] [CrossRef]
- Fontenele, R.S.; Salywon, A.M.; Majure, L.C.; Cobb, I.N.; Bhaskara, A.; Avalos-Calleros, J.A.; Argüello-Astorga, G.R.; Schmidlin, K.; Khalifeh, A.; Smith, K.; et al. A Novel Divergent Geminivirus Identified in Asymptomatic New World Cactaceae Plants. Viruses 2020, 12, 398. [Google Scholar] [CrossRef] [Green Version]
- Rojas, M.R. Use of degenerate primers in the polymerase chain reaction to detect Whitefly-transmitted geminiviruses. Plant Dis. 1993, 77, 340. [Google Scholar] [CrossRef]
- Lindbo, J.A. TRBO: A high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007, 145, 1232–1240. [Google Scholar] [CrossRef] [Green Version]
- Ferro, M.M.M.; Ramos-Sobrinho, R.; Xavier, C.A.D.; Zerbini, F.M.; Lima, G.S.A.; Nagata, T.; Assunção, I.P. New approach for the construction of infectious clones of a circular DNA plant virus using Gibson Assembly. J. Virol. Methods 2019, 263, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T.; Kelz, Z.; Kumar, V.; Murrell, B. RDP5: A computer program for analysing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 2020. [Google Scholar] [CrossRef]
- Martin, D.; Rybicki, E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics 2000, 16, 562–563. [Google Scholar] [CrossRef] [PubMed]
- Padidam, M.; Sawyer, S.; Fauquet, C.M. Possible emergence of new geminiviruses by frequent recombination. Virology 1999, 265, 218–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, D.P.; Posada, D.; Crandall, K.A.; Williamson, C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res. Hum. Retrovir. 2005, 21, 98–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, J.M. Analyzing the mosaic structure of genes. J. Mol. Evol. 1992, 34, 126–129. [Google Scholar] [CrossRef]
- Posada, D.; Crandall, K.A. Evaluation of methods for detecting recombination from DNA sequences: Computer simulations. Proc. Natl. Acad. Sci. USA 2001, 98, 13757–13762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibbs, M.J.; Armstrong, J.S.; Gibbs, A.J. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics 2000, 16, 573–582. [Google Scholar] [CrossRef]
- Boni, M.F.; Posada, D.; Feldman, M.W. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics 2007, 176, 1035–1047. [Google Scholar] [CrossRef] [Green Version]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Stöver, B.C.; Müller, K.F. TreeGraph 2: Combining and visualizing evidence from different phylogenetic analyses. BMC Bioinform. 2010, 11, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [Green Version]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Lapidot, M.; Gelbart, D.; Gal-On, A.; Sela, N.; Anfoka, G.; Haj Ahmed, F.; Abou-Jawada, Y.; Sobh, H.; Mazyad, H.; Aboul-Ata, A.-A.E.; et al. Frequent migration of introduced cucurbit-infecting begomoviruses among Middle Eastern countries. Virol. J. 2014, 11, 181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.; Baumann, K.; Idris, A. Characterization of squash leaf curl and squash mild leaf curl viruses: Host range and reassortment for four SLCV clade viruses. Phytopathology 2005, 95, S14. [Google Scholar]
- Briddon, R.W.; Patil, B.L.; Bagewadi, B.; Nawaz-ul-Rehman, M.S.; Fauquet, C.M. Distinct evolutionary histories of the DNA-A and DNA-B components of bipartite begomoviruses. BMC Evol. Biol. 2010, 10, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Bruyn, A.; Villemot, J.; Lefeuvre, P.; Villar, E.; Hoareau, M.; Harimalala, M.; Abdoul-Karime, A.L.; Abdou-Chakour, C.; Reynaud, B.; Harkins, G.W.; et al. East African cassava mosaic-like viruses from Africa to Indian ocean islands: Molecular diversity, evolutionary history and geographical dissemination of a bipartite begomovirus. BMC Evol. Biol. 2012, 12, 228. [Google Scholar] [CrossRef] [Green Version]
- Mohammed, H.S.; El Siddig, M.A.; El Hussein, A.A.; Navas-Castillo, J.; Fiallo-Olivé, E. First Report of Datura innoxia as a Natural Host of Watermelon chlorotic stunt virus in Sudan. Plant Dis. 2017, 101, 1334. [Google Scholar] [CrossRef]
- Esmaeili, M.; Heydarnejad, J.; Massumi, H.; Varsani, A. Analysis of watermelon chlorotic stunt virus and tomato leaf curl Palampur virus mixed and pseudo-recombination infections. Virus Genes 2015, 51, 408–416. [Google Scholar] [CrossRef]
- Argüello-Astorga, G.R.; Guevara-González, R.G.; Herrera-Estrella, L.R.; Rivera-Bustamante, R.F. Geminivirus replication origins have a group-specific organization of iterative elements: A model for replication. Virology 1994, 203, 90–100. [Google Scholar] [CrossRef]
- Al-Saleh, M.A.; Ahmad, M.H.; Al-Shahwan, I.M.; Brown, J.K.; Idris, A.M. First Report of Watermelon chlorotic stunt virus Infecting Watermelon in Saudi Arabia. Plant Dis. 2014, 98, 1451. [Google Scholar] [CrossRef]
- Galvão, R.M.; Mariano, A.C.; Luz, D.F.; Alfenas, P.F.; Andrade, E.C.; Zerbini, F.M.; Almeida, M.R.; Fontes, E.P.B. A naturally occurring recombinant DNA-A of a typical bipartite begomovirus does not require the cognate DNA-B to infect Nicotiana benthamiana systemically. J. Gen. Virol. 2003, 84, 715–726. [Google Scholar] [CrossRef] [PubMed]
- Duffy, S.; Holmes, E.C. Multiple introductions of the Old World begomovirus Tomato yellow leaf curl virus into the New World. Appl. Environ. Microbiol. 2007, 73, 7114–7117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mabvakure, B.; Martin, D.P.; Kraberger, S.; Cloete, L.; van Brunschot, S.; Geering, A.D.W.; Thomas, J.E.; Bananej, K.; Lett, J.-M.; Lefeuvre, P.; et al. Ongoing geographical spread of Tomato yellow leaf curl virus. Virology 2016, 498, 257–264. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Harkins, G.; Lemey, P.; Gray, A.J.A.; Meredith, S.; Lakay, F.; Monjane, A.; Lett, J.-M.; Varsani, A.; et al. The spread of tomato yellow leaf curl virus from the Middle East to the world. PLoS Pathog. 2010, 6, e1001164. [Google Scholar] [CrossRef] [Green Version]
- Fontenele, R.S.; Salywon, A.M.; Majure, L.C.; Cobb, I.N.; Bhaskara, A.; Avalos-Calleros, J.A.; Arguello-Astorga, G.R.; Schmidlin, K.; Khalifeh, A.; Smith, K.; et al. New World Cactaceae plants harbor diverse geminiviruses. Viruses 2021, 3, 694. [Google Scholar] [CrossRef]




| Begomovirus | Accession # | Host | Isolate | Region of Collection | Collection Year | Component |
|---|---|---|---|---|---|---|
| Squash leaf curl virus KPB1 SB | MW588396 | Capsicum annuum Jalapeño | KPB1_SB | USA | 2019 | B |
| Squash leaf curl virus SF6 | MW588388 | Capsicum sp. | SF6_SA | USA | 2019 | A |
| MW588413 | SF6_SB | USA | 2019 | B | ||
| Squash leaf curl virus SF3 SA | MW588385 | Solanum melongena | SF3_SA | USA | 2019 | A |
| Squash leaf curl virus SF4 | MW588386 | Cucurbita pepo | SF4_SA | USA | 2019 | A |
| MW588411 | SF4_SB | USA | 2019 | B | ||
| Squash leaf curl virus LCM95 SP41 | MW588383 | Cylindropuntia whipplei | LCM95_SP41 | USA | 2015 | A |
| Squash leaf curl virus LCM93 | MW588409 | Ferocactus cylindraceus | LCM93_SP58 | USA | 2016 | B |
| MW588410 | LCM93_SP2634 | USA | 2016 | B | ||
| Squash leaf curl virus LCM90 SP344 | MW588382 | Leptocereus quadricostatus | LCM90_SP344 | USA | 2015 | A |
| Squash leaf curl virus LCM67 SP209 | MW588400 | Opuntia andersoni | LCM67_SP209 | Mexico | 2009 | B |
| Squash leaf curl virus LCM65 | MW588372 | Opuntia arenaria | LCM65_SP41 | USA | 2009 | A |
| MW588373 | LCM65_SP344_1 | USA | 2009 | A | ||
| MW588374 | LCM65_SP344_2 | USA | 2009 | A | ||
| MW588398 | LCM65_SP209 | USA | 2009 | B | ||
| Squash leaf curl virus LCM69 | MW588375 | Opuntia atrispina | LCM69_SP41 | USA | 2009 | A |
| MW588376 | LCM69_SP341 | USA | 2009 | A | ||
| MW588377 | LCM69_SP344 | USA | 2009 | A | ||
| Squash leaf curl virus LCM66 SP209 | MW588399 | Opuntia aureispina | LCM66_SP209 | USA | 2006 | B |
| Squash leaf curl virus LCM89 | MW588381 | Opuntia basilaris var. longiareolata | LCM89_SP41 | USA | 2015 | A |
| MW588407 | LCM89_SP58 | USA | 2015 | B | ||
| MW588408 | LCM89_SP2634 | USA | 2015 | B | ||
| Squash leaf curl virus LCM68 SP209 | MW588401 | Opuntia boldinghii | LCM68_SP209 | USA | 2009 | B |
| Squash leaf curl virus LCM74 SP1332 | MW588402 | Opuntia bravoana | LCM74_SP1332 | Mexico | 2009 | B |
| Squash leaf curl virus LCM76 | MW588380 | Opuntia caracassana | LCM76_SA | USA | 2009 | A |
| MW588404 | LCM76_SP1332 | USA | 2009 | B | ||
| Squash leaf curl virus LCM75 | MW588378 | Opuntia carstenii | LCM75_SP41 | Mexico | 2009 | A |
| MW588379 | LCM75_SP344 | Mexico | 2009 | A | ||
| MW588403 | LCM75_SP1332 | Mexico | 2009 | B | ||
| Squash leaf curl virus LCM78 SP1332 | MW588405 | Opuntia chaffeyi | LCM78_SP1332 | Mexico | 2006 | B |
| Squash leaf curl virus LCM79 SP1332 | MW588406 | Opuntia chaffeyi | LCM79_SP1332 | Mexico | 2006 | B |
| Squash leaf curl virus LCM56 SP341 | MW588369 | Opuntia guatemalensis | LCM56_SP341 | USA | 2009 | A |
| Squash leaf curl virus LCM58 SP41 | MW588370 | Opuntia hondurensis | LCM58_SP41 | USA | 2009 | A |
| Squash leaf curl virus LCM60 SP341 | MW588371 | Opuntia inaperta | LCM60_SP341 | USA | 2009 | A |
| Squash leaf curl virus LCM62 SP8848 | MW588397 | Opuntia karwinskiana | LCM62_SP8848 | USA | 2009 | B |
| Squash leaf curl virus LCM29 SP41 | MW588368 | Opuntia puberula | LCM29_SP41 | Mexico | 2009 | A |
| Squash leaf curl virus DBG34 SP15973 | MW588395 | Opuntia robusta | DBG34_SP15973 | USA | 2018 | B |
| Squash leaf curl virus LCM96 SP41 | MW588384 | Pereskiopsis kellermannii | LCM96_SP41 | USA | 2016 | A |
| Squash leaf curl virus SF5 | MW588387 | Raphanus sativus | SF5_SA | USA | 2019 | A |
| MW588412 | SF5_SB | USA | 2019 | B | ||
| Squash leaf curl virus CG5 | MW588366 | Solanum lycopersicum | CG5_SA | USA | 2018 | A |
| MW588393 | CG5_SB | USA | 2018 | B | ||
| Squash leaf curl virus CG6 | MW588367 | Solanum lycopersicum | CG6_SA | USA | 2018 | A |
| MW588394 | CG6_SB | USA | 2018 | B | ||
| Squash leaf curl virus SWAT | MW588389 | Citrullus lanatus | SWAT_SA | USA | 2019 | A |
| MW588414 | SWAT_SB | USA | 2019 | B | ||
| Watermelon chlorotic stunt virus HERB15 | MW588391 | Solanum sp. | HERB15 | USA | 2019 | A |
| Watermelon chlorotic stunt virus HERB28 | MW588392 | Stachys byzantina | HERB28 | USA | 2019 | A |
| Watermelon chlorotic stunt virus LCM53 | MW588417 | Opuntia cochenillifera | LCM53 | USA | 2006 | B |
| Watermelon chlorotic stunt virus LCM 52 | MW588390 | Opuntia auberi | LCM_52 | Mexico | 2006 | A |
| Watermelon chlorotic stunt virus LCM51 | MW588416 | Opuntia auberi | LCM51 | Mexico | 2006 | B |
| Watermelon chlorotic stunt virus LCM50 | MW588415 | Consolea spinosissima | LCM50 | USA | 2006 | B |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fontenele, R.S.; Bhaskara, A.; Cobb, I.N.; Majure, L.C.; Salywon, A.M.; Avalos-Calleros, J.A.; Argüello-Astorga, G.R.; Schmidlin, K.; Roumagnac, P.; Ribeiro, S.G.; et al. Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America. Viruses 2021, 13, 810. https://doi.org/10.3390/v13050810
Fontenele RS, Bhaskara A, Cobb IN, Majure LC, Salywon AM, Avalos-Calleros JA, Argüello-Astorga GR, Schmidlin K, Roumagnac P, Ribeiro SG, et al. Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America. Viruses. 2021; 13(5):810. https://doi.org/10.3390/v13050810
Chicago/Turabian StyleFontenele, Rafaela S., Amulya Bhaskara, Ilaria N. Cobb, Lucas C. Majure, Andrew M. Salywon, Jesús A. Avalos-Calleros, Gerardo R. Argüello-Astorga, Kara Schmidlin, Philippe Roumagnac, Simone G. Ribeiro, and et al. 2021. "Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America" Viruses 13, no. 5: 810. https://doi.org/10.3390/v13050810
APA StyleFontenele, R. S., Bhaskara, A., Cobb, I. N., Majure, L. C., Salywon, A. M., Avalos-Calleros, J. A., Argüello-Astorga, G. R., Schmidlin, K., Roumagnac, P., Ribeiro, S. G., Kraberger, S., Martin, D. P., Lefeuvre, P., & Varsani, A. (2021). Identification of the Begomoviruses Squash Leaf Curl Virus and Watermelon Chlorotic Stunt Virus in Various Plant Samples in North America. Viruses, 13(5), 810. https://doi.org/10.3390/v13050810








