Functions of Coronavirus Accessory Proteins: Overview of the State of the Art
Abstract
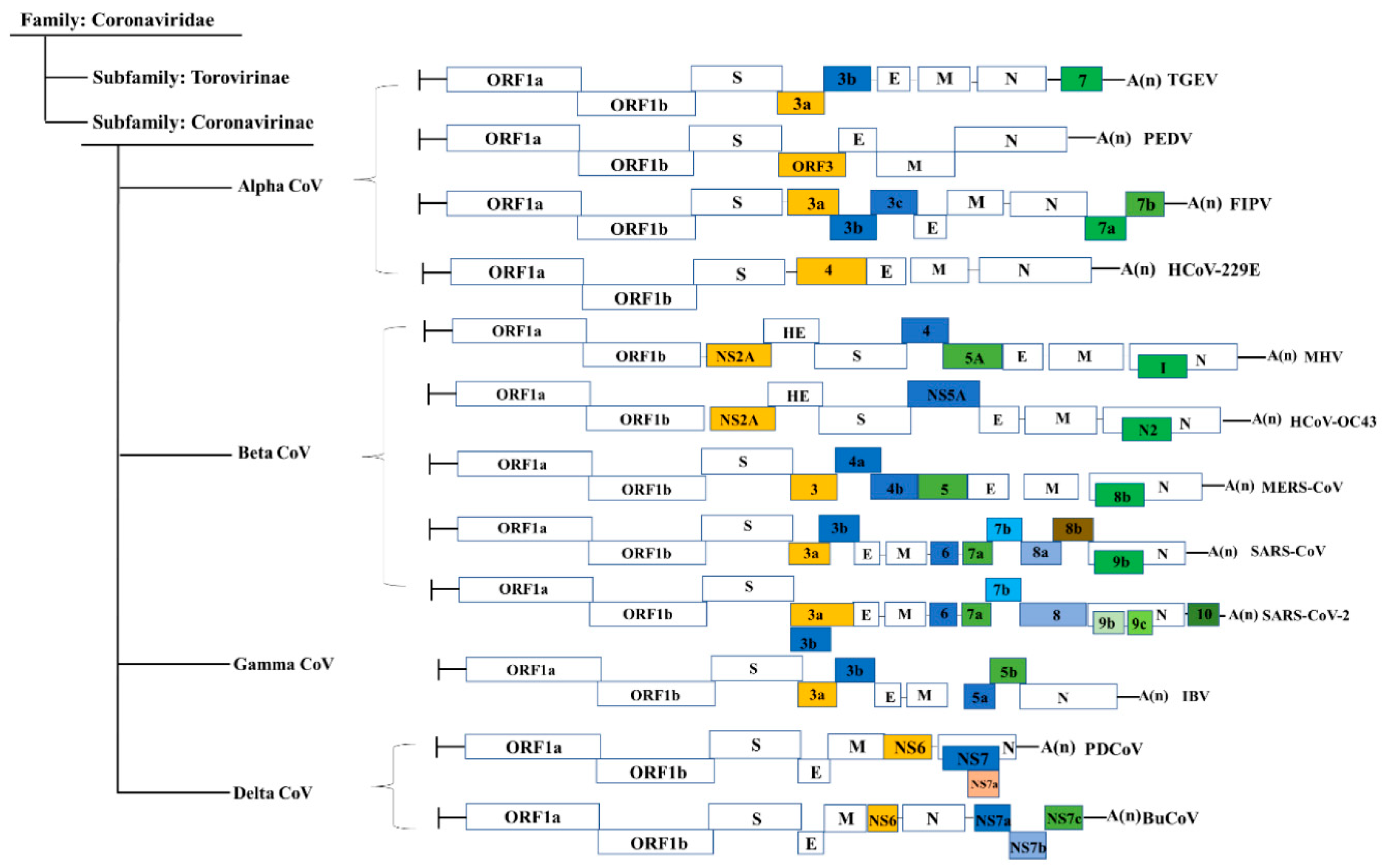
1. Introduction
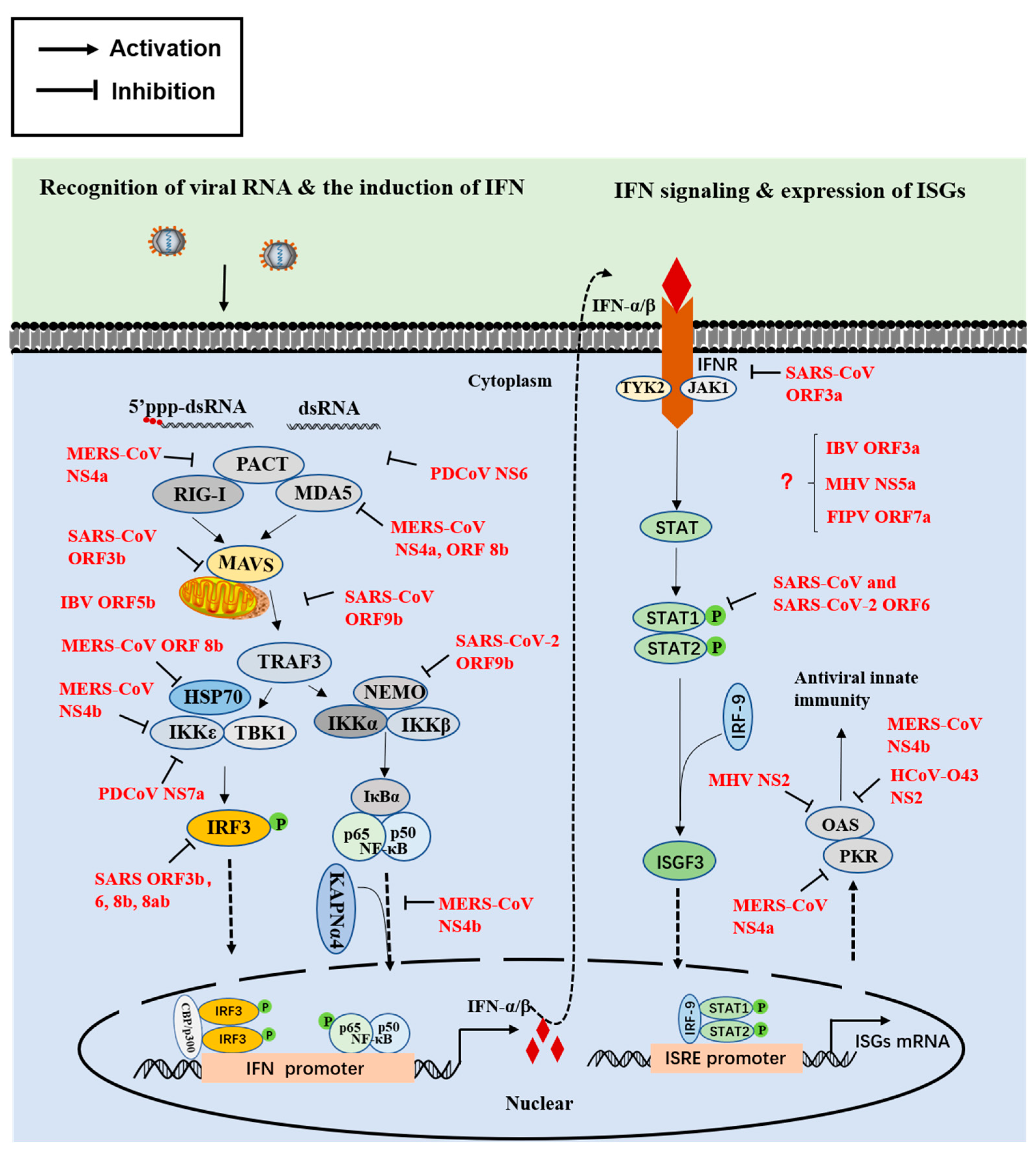
2. Innate Immune Interferon (IFN) Responses
2.1. The First Step Involves IFN Induction
2.2. The Second Step Involves Signal Transduction
2.3. The Final Step Involves the Activity of Antiviral Proteins
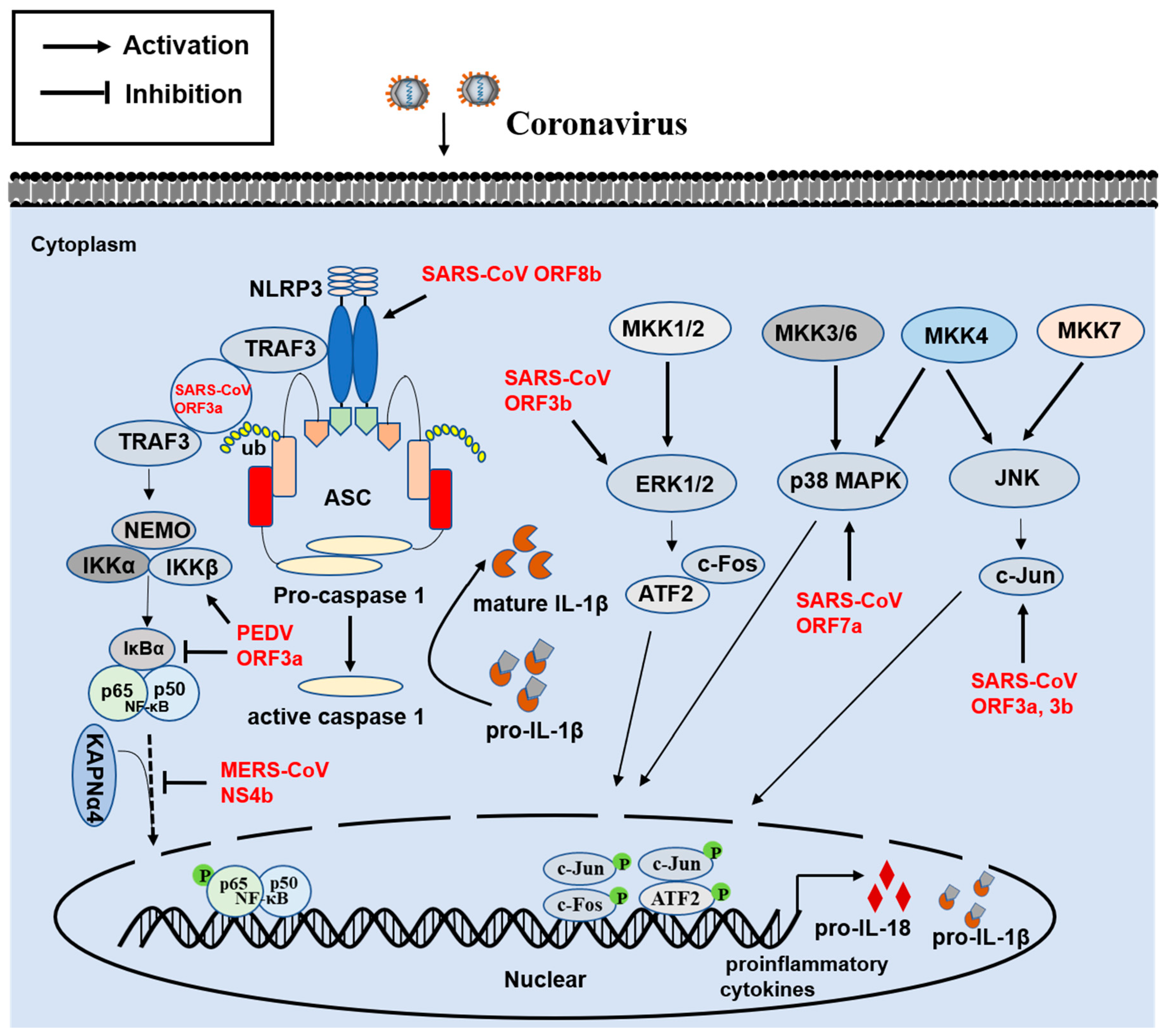
3. Innate Proinflammatory Immune Response
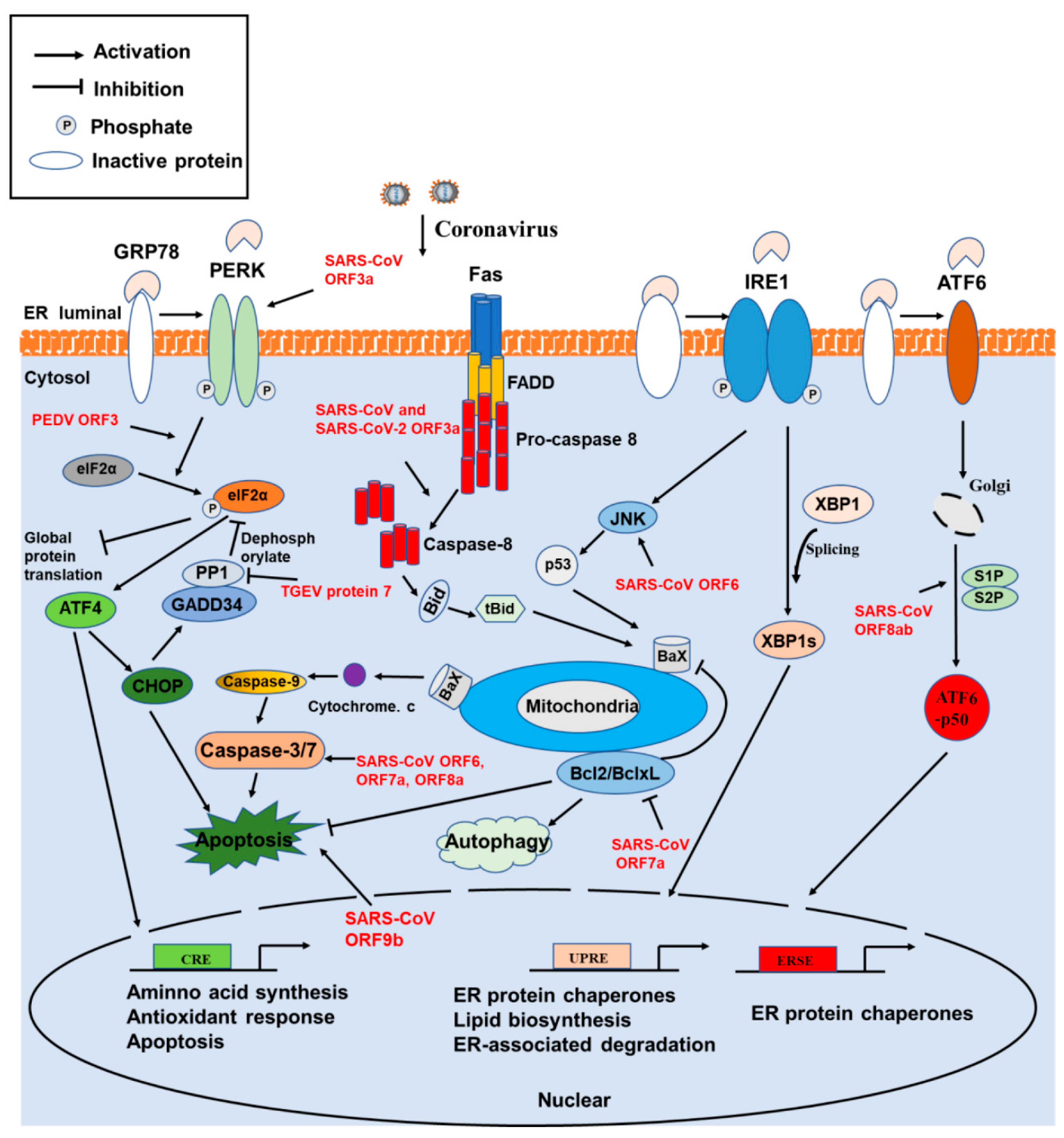
4. Involvement of ER Stress, Apoptosis, and Autophagy
5. The Action Mediating the Ion Channel Activity of Viroporins
6. Involvement of Virulence
7. Concluding Remarks and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Forni, D.; Cagliani, R.; Clerici, M.; Sironi, M. Molecular Evolution of Human Coronavirus Genomes. Trends Microbiol. 2017, 25, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Coleman, C.; Frieman, M.B.; Pauck, A.; Lener, B.; Hoell, M.; Kaiser, A.; Kaufmann, A.M.; Zwerschke, W.; Jansen-Dürr, P.; Imperiale, M.J. Coronaviruses: Important Emerging Human Pathogens. J. Virol. 2014, 88, 5209–5212. [Google Scholar] [CrossRef]
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045. [Google Scholar] [CrossRef]
- Bi, J.; Zeng, S.; Xiao, S.; Chen, H.; Fang, L. Complete Genome Sequence of Porcine Epidemic Diarrhea Virus Strain AJ1102 Isolated from a Suckling Piglet with Acute Diarrhea in China. J. Virol. 2012, 86, 10910–10911. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Pei, S.; Chen, B.; Song, Y.; Zhang, T.; Yang, W.; Shaman, J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020, 368, 489–493. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Wen, Z.; Zhong, G.; Yang, H.; Wang, C.; Huang, B.; Liu, R.; He, X.; Shuai, L.; Sun, Z.; et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science 2020, 368, 1016–1020. [Google Scholar] [CrossRef]
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008. [Google Scholar] [CrossRef]
- Lim, Y.X.; Ng, Y.L.; Tam, J.P.; Liu, D.X. Human Coronaviruses: A Review of Virus–Host Interactions. Diseases 2016, 4, 26. [Google Scholar] [CrossRef]
- Perlman, S.; Netland, J. Coronaviruses post-SARS: Update on replication and pathogenesis. Nat. Rev. Genet. 2009, 7, 439–450. [Google Scholar] [CrossRef]
- Hu, B.; Ge, X.; Wang, L.-F.; Shi, Z. Bat origin of human coronaviruses. Virol. J. 2015, 12, 1–10. [Google Scholar] [CrossRef]
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Genet. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-L.; Yu, J.-Q.; Huang, Y.-W. Swine enteric alphacoronavirus (swine acute diarrhea syndrome coronavirus): An update three years after its discovery. Virus Res. 2020, 285, 198024. [Google Scholar] [CrossRef]
- Legnardi, M.; Tucciarone, C.M.; Franzo, G.; Cecchinato, M. Infectious Bronchitis Virus Evolution, Diagnosis and Control. Vet. Sci. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J. Porcine deltacoronavirus: Overview of infection dynamics, diagnostic methods, prevalence and genetic evolution. Virus Res. 2016, 226, 71–84. [Google Scholar] [CrossRef]
- Wille, M.; Holmes, E.C. Wild birds as reservoirs for diverse and abundant gamma- and deltacoronaviruses. FEMS Microbiol. Rev. 2020, 44, 631–644. [Google Scholar] [CrossRef]
- Jung, K.; Hu, H.; Saif, L.J. Calves are susceptible to infection with the newly emerged porcine deltacoronavirus, but not with the swine enteric alphacoronavirus, porcine epidemic diarrhea virus. Arch. Virol. 2017, 162, 2357–2362. [Google Scholar] [CrossRef]
- Liang, Q.; Zhang, H.; Li, B.; Ding, Q.; Wang, Y.; Gao, W.; Guo, D.; Wei, Z.; Hu, H. Susceptibility of Chickens to Porcine Deltacoronavirus Infection. Viruses 2019, 11, 573. [Google Scholar] [CrossRef]
- Boley, P.A.; Alhamo, M.A.; Lossie, G.; Yadav, K.K.; Vasquez-Lee, M.; Saif, L.J.; Kenney, S.P.; Boley, P. Porcine Deltacoronavirus Infection and Transmission in Poultry, United States1. Emerg. Infect. Dis. 2020, 26, 255–265. [Google Scholar] [CrossRef]
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. medRxiv 2021. [Google Scholar] [CrossRef]
- Li, W.; Hulswit, R.J.G.; Kenney, S.P.; Widjaja, I.; Jung, K.; Alhamo, M.A.; van Dieren, B.; van Kuppeveld, F.J.M.; Saif, L.J.; Bosch, B.-J. Broad receptor engagement of an emerging global coronavirus may potentiate its diverse cross-species transmissibility. Proc. Natl. Acad. Sci. USA 2018, 115, E5135–E5143. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. 2014, 109, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 1–13. [Google Scholar] [CrossRef]
- Fang, P.; Fang, L.; Hong, Y.; Liu, X.; Dong, N.; Ma, P.; Bi, J.; Wang, D.; Xiao, S. Discovery of a novel accessory protein NS7a encoded by porcine deltacoronavirus. J. Gen. Virol. 2017, 98, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Fang, P.; Fang, L.; Liu, X.; Hong, Y.; Wang, Y.; Dong, N.; Ma, P.; Bi, J.; Wang, D.; Xiao, S. Identification and subcellular localization of porcine deltacoronavirus accessory protein NS6. Virology 2016, 499, 170–177. [Google Scholar] [CrossRef]
- McBride, R.; Fielding, B.C. The Role of Severe Acute Respiratory Syndrome (SARS)-Coronavirus Accessory Proteins in Virus Pathogenesis. Viruses 2012, 4, 2902–2923. [Google Scholar] [CrossRef]
- Tan, Y.-J.; Lim, S.G.; Hong, W. Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus. Antivir. Res. 2006, 72, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef] [PubMed]
- Streicher, F.; Jouvenet, N. Stimulation of Innate Immunity by Host and Viral RNAs. Trends Immunol. 2019, 40, 1134–1148. [Google Scholar] [CrossRef]
- Kawai, T.; Takahashi, K.; Sato, S.; Coban, C.; Kumar, H.; Kato, H.; Ishii, K.J.; Takeuchi, O.; Akira, S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 2005, 6, 981–988. [Google Scholar] [CrossRef]
- Takaoka, A.; Yanai, H. Interferon signalling network in innate defence. Cell. Microbiol. 2006, 8, 907–922. [Google Scholar] [CrossRef]
- Chan, R.W.; Chan, M.C.; Agnihothram, S.; Chan, L.L.; Kuok, D.I.; Fong, J.H.; Guan, Y.; Poon, L.L.; Baric, R.S.; Nicholls, J.M.; et al. Tropism of and innate immune responses to the novel human betacoronavirus lineage C virus in human ex vivo respiratory organ cultures. J. Virol. 2013, 87, 6604–6614. [Google Scholar] [CrossRef]
- Luo, J.; Fang, L.; Dong, N.; Fang, P.; . Ding, Z.; Wang, D.; Chen, H.; Xiao, S. Porcine deltacoronavirus (PDCoV) infection suppresses RIG-I-mediated interferon-beta production. Virology 2016, 495, 10–17. [Google Scholar] [CrossRef] [PubMed]
- Roth-Cross, J.K.; Martínez-Sobrido, L.; Scott, E.P.; García-Sastre, A.; Weiss, S.R. Inhibition of the Alpha/Beta Interferon Response by Mouse Hepatitis Virus at Multiple Levels. J. Virol. 2007, 81, 7189–7199. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef]
- Sen, G.C. Viruses and Interferons. Annu. Rev. Microbiol. 2001, 55, 255–281. [Google Scholar] [CrossRef] [PubMed]
- Zinzula, L.; Tramontano, E. Strategies of highly pathogenic RNA viruses to block dsRNA detection by RIG-I-like receptors: Hide, mask, hit. Antivir. Res. 2013, 100, 615–635. [Google Scholar] [CrossRef] [PubMed]
- Niemeyer, D.; Zillinger, T.; Muth, D.; Zielecki, F.; Horvath, G.; Suliman, T.; Barchet, W.; Weber, F.; Drosten, C.; Müller, M.A. Middle East Respiratory Syndrome Coronavirus Accessory Protein 4a Is a Type I Interferon Antagonist. J. Virol. 2013, 87, 12489–12495. [Google Scholar] [CrossRef]
- Siu, K.-L.; Yeung, M.L.; Kok, K.-H.; Yuen, K.-Y.; Kew, C.; Lui, P.-Y.; Chan, C.-P.; Tse, H.; Woo, P.C.Y.; Jin, D.-Y. Middle East Respiratory Syndrome Coronavirus 4a Protein Is a Double-Stranded RNA-Binding Protein That Suppresses PACT-Induced Activation of RIG-I and MDA5 in the Innate Antiviral Response. J. Virol. 2014, 88, 4866–4876. [Google Scholar] [CrossRef]
- Fang, P.; Fang, L.; Ren, J.; Hong, Y.; Liu, X.; Zhao, Y.; Wang, D.; Peng, G.; Xiao, S. Porcine Deltacoronavirus Accessory Protein NS6 Antagonizes Interferon Beta Production by Interfering with the Binding of RIG-I/MDA5 to Double-Stranded RNA. J. Virol. 2018, 92, e00712-18. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, S.-J.; Myoung, J. Middle East Respiratory Syndrome Coronavirus-Encoded ORF8b Inhibits RIG-I-Like Receptors in a Differential Mechanism. J. Microbiol. Biotechnol. 2019, 29, 2014–2021. [Google Scholar] [CrossRef] [PubMed]
- Mibayashi, M.; Martínez-Sobrido, L.; Loo, Y.-M.; Caárdenas, W.B.; Gale, M.; García-Sastre, A. Inhibition of Retinoic Acid-Inducible Gene I-Mediated Induction of Beta Interferon by the NS1 Protein of Influenza A Virus. J. Virol. 2007, 81, 514–524. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Mao, A.; Li, C.; Li, Y.; Tien, P. Hepatitis B virus X protein suppresses virus-triggered IRF3 activation and IFN-beta induction by disrupting the VISA-associated complex. Cell Mol. Immunol. 2010, 7, 341–348. [Google Scholar] [CrossRef] [PubMed]
- Kopecky-Bromberg, S.A.; Martínez-Sobrido, L.; Frieman, M.; Baric, R.A.; Palese, P. Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame (ORF) 3b, ORF 6, and Nucleocapsid Proteins Function as Interferon Antagonists. J. Virol. 2007, 81, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Freundt, E.C.; Yu, L.; Park, E.; Lenardo, M.J.; Xu, X.-N. Molecular Determinants for Subcellular Localization of the Severe Acute Respiratory Syndrome Coronavirus Open Reading Frame 3b Protein. J. Virol. 2009, 83, 6631–6640. [Google Scholar] [CrossRef]
- Shi, C.-S.; Qi, H.-Y.; Boularan, C.; Huang, N.-N.; Abu-Asab, M.; Shelhamer, J.H.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-9b Suppresses Innate Immunity by Targeting Mitochondria and the MAVS/TRAF3/TRAF6 Signalosome. J. Immunol. 2014, 193, 3080–3089. [Google Scholar] [CrossRef] [PubMed]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Wu, J.; Shi, Y.; Pan, X.; Wu, S.; Hou, R.; Zhang, Y.; Zhong, T.; Tang, H.; Du, W.; Wang, L.; et al. SARS-CoV-2 ORF9b inhibits RIG-I-MAVS antiviral signaling by interrupting K63-linked ubiquitination of NEMO. Cell Rep. 2021, 34, 108761. [Google Scholar] [CrossRef]
- Yang, Y.; Ye, F.; Zhu, N.; Wang, W.; Deng, Y.; Zhao, Z.; Tan, W. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci. Rep. 2015, 5, 17554. [Google Scholar] [CrossRef]
- Wong, L.R.; Ye, Z.W.; Lui, P.Y.; Zheng, X.; Yuan, S.; Zhu, L.; Fung, S.Y.; Yuen, K.S.; Siu, K.L.; Yeung, M.L.; et al. Middle East Respiratory Syndrome Coronavirus ORF8b Accessory Protein Suppresses Type I IFN Expression by Impeding HSP70-Dependent Activation of IRF3 Kinase IKKepsilon. J. Immunol. 2020, 205, 1564–1579. [Google Scholar] [CrossRef]
- Fang, P.; Fang, L.; Xia, S.; Ren, J.; Zhang, J.; Bai, D.; Zhou, Y.; Peng, G.; Zhao, S.; Xiao, S. Porcine Deltacoronavirus Accessory Protein NS7a Antagonizes IFN-beta Production by Competing With TRAF3 and IRF3 for Binding to IKKepsilon. Front. Cell. Infect. Microbiol. 2020, 10, 257. [Google Scholar] [CrossRef]
- Wong, H.H.; Fung, T.S.; Fang, S.; Huang, M.; Le, M.T.; Liu, D.X. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 2018, 515, 165–175. [Google Scholar] [CrossRef]
- Canton, J.; Fehr, A.R.; Fernandez-Delgado, R.; Gutierrez-Alvarez, F.J.; Sanchez-Aparicio, M.T.; Garcia-Sastre, A.; Perlman, S.; Enjuanes, L.; Sola, I. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog. 2018, 14, e1006838. [Google Scholar] [CrossRef]
- Beidas, M.; Chehadeh, W. Effect of Human Coronavirus OC43 Structural and Accessory Proteins on the Transcriptional Activation of Antiviral Response Elements. Intervirology 2018, 61, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Zhang, L.; Geng, H.; Deng, Y.; Huang, B.; Guo, Y.; Zhao, Z.; Tan, W. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 2013, 4, 951–961. [Google Scholar] [CrossRef]
- Li, J.Y.; Liao, C.H.; Wang, Q.; Tan, Y.J.; Luo, R.; Qiu, Y.; Ge, X.Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Konno, Y.; Kimura, I.; Uriu, K.; Fukushi, M.; Irie, T.; Koyanagi, Y.; Sauter, D.; Gifford, R.J.; Consortium, U.-C.; Nakagawa, S.; et al. SARS-CoV-2 ORF3b Is a Potent Interferon Antagonist Whose Activity Is Increased by a Naturally Occurring Elongation Variant. Cell Rep. 2020, 32, 108–185. [Google Scholar] [CrossRef]
- Flower, T.G.; Buffalo, C.Z.; Hooy, R.M.; Allaire, M.; Ren, X.; Hurley, J.H. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Minakshi, R.; Padhan, K.; Rani, M.; Khan, N.; Ahmad, F.; Jameel, S. The SARS Coronavirus 3a Protein Causes Endoplasmic Reticulum Stress and Induces Ligand-Independent Downregulation of the Type 1 Interferon Receptor. PLoS ONE 2009, 4, e8342. [Google Scholar] [CrossRef] [PubMed]
- Frieman, M.; Yount, B.; Heise, M.; Kopecky-Bromberg, S.A.; Palese, P.; Baric, R.S. Severe Acute Respiratory Syndrome Coronavirus ORF6 Antagonizes STAT1 Function by Sequestering Nuclear Import Factors on the Rough Endoplasmic Reticulum/Golgi Membrane. J. Virol. 2007, 81, 9812–9824. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Chen, S.; Li, R.; Chen, Y.; Wang, M.; Guo, D. Severe acute respiratory syndrome coronavirus protein 6 mediates ubiquitin-dependent proteosomal degradation of N-Myc (and STAT) interactor. Virol. Sin. 2015, 30, 153–161. [Google Scholar] [CrossRef][Green Version]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Nat. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef] [PubMed]
- Dedeurwaerder, A.; Olyslaegers, D.A.J.; Desmarets, L.M.B.; Roukaerts, I.D.M.; Theuns, S.; Nauwynck, H.J. ORF7-encoded accessory protein 7a of feline infectious peritonitis virus as a counteragent against IFN-α-induced antiviral response. J. Gen. Virol. 2014, 95, 393–402. [Google Scholar] [CrossRef]
- Koetzner, C.A.; Kuo, L.; Goebel, S.J.; Dean, A.B.; Parker, M.M.; Masters, P.S. Accessory Protein 5a Is a Major Antagonist of the Antiviral Action of Interferon against Murine Coronavirus. J. Virol. 2010, 84, 8262–8274. [Google Scholar] [CrossRef] [PubMed]
- Kint, J.; Dickhout, A.; Kutter, J.; Maier, H.J.; Britton, P.; Koumans, J.; Pijlman, G.P.; Fros, J.; Wiegertjes, G.; Forlenza, M. Infectious Bronchitis Coronavirus Inhibits STAT1 Signaling and Requires Accessory Proteins for Resistance to Type I Interferon Activity. J. Virol. 2015, 89, 12047–12057. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Han, N.; Chen, Z.; Peng, Y.; Li, L.; Zhou, H.; Ji, C.; Meng, J.; Jiang, T.; Wu, A. Compositional diversity and evolutionary pattern of coronavirus accessory proteins. Briefings Bioinform. 2021, 22, 1267–1278. [Google Scholar] [CrossRef]
- Cruz, J.L.G.; Sola, I.; Becares, M.; Alberca, B.; Plana, J.; Enjuanes, L.; Zuñiga, S. Coronavirus Gene 7 Counteracts Host Defenses and Modulates Virus Virulence. PLoS Pathog. 2011, 7, e1002090. [Google Scholar] [CrossRef]
- Rabouw, H.H.; Langereis, M.A.; Knaap, R.C.M.; Dalebout, T.J.; Canton, J.; Sola, I.; Enjuanes, L.; Bredenbeek, P.J.; Kikkert, M.; De Groot, R.J.; et al. Middle East Respiratory Coronavirus Accessory Protein 4a Inhibits PKR-Mediated Antiviral Stress Responses. PLoS Pathog. 2016, 12, e1005982. [Google Scholar] [CrossRef]
- Nakagawa, K.; Narayanan, K.; Wada, M.; Makino, S. Inhibition of Stress Granule Formation by Middle East Respiratory Syndrome Coronavirus 4a Accessory Protein Facilitates Viral Translation, Leading to Efficient Virus Replication. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Jha, B.K.; Wu, A.; Elliott, R.; Ziebuhr, J.; Gorbalenya, A.; Silverman, R.H.; Weiss, S.R. Antagonism of the Interferon-Induced OAS-RNase L Pathway by Murine Coronavirus ns2 Protein Is Required for Virus Replication and Liver Pathology. Cell Host Microbe 2012, 11, 607–616. [Google Scholar] [CrossRef]
- Goldstein, S.A.; Thornbrough, J.M.; Zhang, R.; Jha, B.K.; Li, Y.; Elliott, R.; Quiroz-Figueroa, K.; Chen, A.I.; Silverman, R.H.; Weiss, S.R. Lineage A Betacoronavirus NS2 Proteins and the Homologous Torovirus Berne pp1a Carboxy-Terminal Domain Are Phosphodiesterases That Antagonize Activation of RNase L. J. Virol. 2017, 91, e02201-16. [Google Scholar] [CrossRef] [PubMed]
- Thornbrough, J.M.; Jha, B.K.; Yount, B.; Goldstein, S.A.; Li, Y.; Elliott, R.; Sims, A.C.; Baric, R.S.; Silverman, R.H.; Weiss, S.R. Middle East Respiratory Syndrome Coronavirus NS4b Protein Inhibits Host RNase L Activation. mBio 2016, 7, e00258-16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Freeman, T.L.; Swartz, T.H. Targeting the NLRP3 Inflammasome in Severe COVID-19. Front. Immunol. 2020, 11, 1518. [Google Scholar] [CrossRef] [PubMed]
- Sendler, M.; Brandt, C.V.D.; Glaubitz, J.; Wilden, A.; Golchert, J.; Weiss, F.U.; Homuth, G.; Chama, L.L.D.F.; Mishra, N.; Mahajan, U.M.; et al. NLRP3 Inflammasome Regulates Development of Systemic Inflammatory Response and Compensatory Anti-Inflammatory Response Syndromes in Mice With Acute Pancreatitis. Gastroenterology 2020, 158, 253–269.e14. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426. [Google Scholar] [CrossRef]
- Siu, K.; Yuen, K.; Castano-Rodriguez, C.; Ye, Z.; Yeung, M.; Fung, S.; Yuan, S.; Chan, C.; Yuen, K.; Enjuanes, L.; et al. Severe acute respiratory syndrome Coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019, 33, 8865–8877. [Google Scholar] [CrossRef]
- Shi, C.-S.; Nabar, N.R.; Huang, N.-N.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019, 5, 1–12. [Google Scholar] [CrossRef]
- Paniri, A.; Akhavan-Niaki, H. Emerging role of IL-6 and NLRP3 inflammasome as potential therapeutic targets to combat COVID-19: Role of lncRNAs in cytokine storm modulation. Life Sci. 2020, 257, 118114. [Google Scholar] [CrossRef]
- Kanzawa, N.; Nishigaki, K.; Hayashi, T.; Ishii, Y.; Furukawa, S.; Niiro, A.; Yasui, F.; Kohara, M.; Morita, K.; Matsushima, K.; et al. Augmentation of chemokine production by severe acute respiratory syndrome coronavirus 3a/X1 and 7a/X4 proteins through NF-kappaB activation. FEBS Lett. 2006, 580, 6807–6812. [Google Scholar] [CrossRef] [PubMed]
- Obitsu, S.; Ahmed, N.; Nishitsuji, H.; Hasegawa, A.; Nakahama, K.-I.; Morita, I.; Nishigaki, K.; Hayashi, T.; Masuda, T.; Kannagi, M. Potential enhancement of osteoclastogenesis by severe acute respiratory syndrome coronavirus 3a/X1 protein. Arch. Virol. 2009, 154, 1457–1464. [Google Scholar] [CrossRef]
- Kopecky-Bromberg, S.A.; Martinez-Sobrido, L.; Palese, P. 7a Protein of Severe Acute Respiratory Syndrome Coronavirus Inhibits Cellular Protein Synthesis and Activates p38 Mitogen-Activated Protein Kinase. J. Virol. 2006, 80, 785–793. [Google Scholar] [CrossRef]
- Varshney, B.; Agnihothram, S.; Tan, Y.J.; Baric, R.; Lal, S.K. SARS coronavirus 3b accessory protein modulates transcriptional activity of RUNX1b. PLoS ONE 2012, 7, e29542. [Google Scholar] [CrossRef]
- Varshney, B.; Lal, S.K. SARS-CoV accessory protein 3b induces AP-1 transcriptional activity through activation of JNK and ERK pathways. Biochemistry 2011, 50, 5419–5425. [Google Scholar] [CrossRef]
- Ding, Z.; An, K.; Xie, L.; Wu, W.; Zhang, R.; Wang, D.; Fang, Y.; Chen, H.; Xiao, S.; Fang, L. Transmissible gastroenteritis virus infection induces NF-kappaB activation through RLR-mediated signaling. Virology 2017, 507, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Miyazaki, A.; Hu, H.; Saif, L.J. Susceptibility of porcine IPEC-J2 intestinal epithelial cells to infection with porcine deltacoronavirus (PDCoV) and serum cytokine responses of gnotobiotic pigs to acute infection with IPEC-J2 cell culture-passaged PDCoV. Vet. Microbiol. 2018, 221, 49–58. [Google Scholar] [CrossRef]
- Kaewborisuth, C.; Koonpaew, S.; Srisutthisamphan, K.; Viriyakitkosol, R.; Jaru-Ampornpan, P.; Jongkaewwattana, A. PEDV ORF3 Independently Regulates IkappaB Kinase beta-Mediated NF-kappaB and IFN-beta Promoter Activities. Pathogens 2020, 9, 376. [Google Scholar] [CrossRef]
- Wu, Z.; Cheng, L.; Xu, J.; Li, P.; Li, X.; Zou, D.; Zhang, Y.; Wang, X.; Wu, X.; Shen, Y.; et al. The accessory protein ORF3 of porcine epidemic diarrhea virus inhibits cellular interleukin-6 and interleukin-8 productions by blocking the nuclear factor-kappaB p65 activation. Vet. Microbiol. 2020, 251, 108892. [Google Scholar] [CrossRef]
- Cruz, J.L.; Becares, M.; Sola, I.; Oliveros, J.C.; Enjuanes, L.; Zuniga, S. Alphacoronavirus protein 7 modulates host innate immune response. J. Virol. 2013, 87, 9754–9767. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. Coronavirus infection, ER stress, apoptosis and innate immunity. Front. Microbiol. 2014, 5, 296. [Google Scholar] [CrossRef] [PubMed]
- Oakes, S.A.; Papa, F.R. The Role of Endoplasmic Reticulum Stress in Human Pathology. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 173–194. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nieto-Torres, J.L.; Guardeno, J.M.J.; Regla-Nava, J.A.; Álvarez, E.; Oliveros, J.C.; Zhao, J.; Fett, C.; Perlman, S.; Enjuanes, L. Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis. PLoS Pathog. 2011, 7, e1002315. [Google Scholar] [CrossRef]
- Bechill, J.; Chen, Z.; Brewer, J.W.; Baker, S.C. Coronavirus Infection Modulates the Unfolded Protein Response and Mediates Sustained Translational Repression. J. Virol. 2008, 82, 4492–4501. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Fu, F.; Ma, Y.; Zhang, X.; Li, L.; Feng, L.; Liu, P. The PERK Arm of the Unfolded Protein Response Negatively Regulates Transmissible Gastroenteritis Virus Replication by Suppressing Protein Translation and Promoting Type I Interferon Production. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Liao, Y.; Fung, T.S.; Huang, M.; Fang, S.G.; Zhong, Y.; Liu, D.X. Upregulation of CHOP/GADD153 during Coronavirus Infectious Bronchitis Virus Infection Modulates Apoptosis by Restricting Activation of the Extracellular Signal-Regulated Kinase Pathway. J. Virol. 2013, 87, 8124–8134. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Czinn, S.J.; Reiter, R.J.; Blanchard, T.G. Crosstalk between endoplasmic reticulum stress and anti-viral activities: A novel therapeutic target for COVID-19. Life Sci. 2020, 255, 117842. [Google Scholar] [CrossRef] [PubMed]
- Köseler, A.; Sabirli, R.; Gören, T.; Türkçüer, I.; Kurt, Ö. Endoplasmic Reticulum Stress Markers in SARS-COV-2 Infection and Pneumonia: Case-Control Study. In Vivo 2020, 34, 1645–1650. [Google Scholar] [CrossRef]
- Ding, L.; Li, J.; Li, W.; Fang, Z.; Li, N.; Wu, S.; Li, J.; Hong, M. p53- and ROS-mediated AIF pathway involved in TGEV-induced apoptosis. J. Vet. Med. Sci. 2018, 80, 1775–1781. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, C. Porcine epidemic diarrhea virus induces caspase-independent apoptosis through activation of mitochondrial apoptosis-inducing factor. Virology 2014, 460-461, 180–193. [Google Scholar] [CrossRef]
- Xu, X.; Xu, Y.; Zhang, Q.; Yang, F.; Yin, Z.; Wang, L.; Li, Q. Porcine epidemic diarrhea virus infections induce apoptosis in Vero cells via a reactive oxygen species (ROS)/p53, but not p38 MAPK and SAPK/JNK signalling pathways. Vet. Microbiol. 2019, 232, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Shuid, A.N.; Safi, N.; Haghani, A.; Mehrbod, P.; Haron, M.S.R.; Tan, S.W.; Omar, A.R. Apoptosis transcriptional mechanism of feline infectious peritonitis virus infected cells. Apoptosis 2015, 20, 1457–1470. [Google Scholar] [CrossRef] [PubMed]
- Meessen-Pinard, M.; Le Coupanec, A.; Desforges, M.; Talbot, P.J. Pivotal Role of Receptor-Interacting Protein Kinase 1 and Mixed Lineage Kinase Domain-Like in Neuronal Cell Death Induced by the Human Neuroinvasive Coronavirus OC43. J. Virol. 2017, 91. [Google Scholar] [CrossRef] [PubMed]
- Collins, A.R. In Vitro Detection of Apoptosis in Monocytes/Macrophages Infected with Human Coronavirus. Clin. Vaccine Immunol. 2002, 9, 1392–1395. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Xiao, G.; Zhang, J.; Hu, Y.; Yuan, F.; Cole, D.; Zheng, C.; Gao, G.F. SARS coronavirus induces apoptosis in Vero E6 Cells. J. Med. Virol. 2004, 73, 323–331. [Google Scholar] [CrossRef]
- Yeung, M.-L.; Yao, Y.; Jia, L.; Chan, J.F.W.; Chan, K.-H.; Cheung, K.-F.; Chen, H.; Poon, V.K.M.; Tsang, A.K.L.; To, K.K.; et al. MERS coronavirus induces apoptosis in kidney and lung by upregulating Smad7 and FGF2. Nat. Microbiol. 2016, 1, 16004. [Google Scholar] [CrossRef]
- Chu, H.; Zhou, J.; Wong, B.H.-Y.; Li, C.; Chan, J.F.-W.; Cheng, Z.-S.; Yang, D.; Wang, D.; Lee, A.C.Y.; Li, C.; et al. Middle East Respiratory Syndrome Coronavirus Efficiently Infects Human Primary T Lymphocytes and Activates the Extrinsic and Intrinsic Apoptosis Pathways. J. Infect. Dis. 2016, 213, 904–914. [Google Scholar] [CrossRef]
- Chen, C.-J.; Makino, S. Murine Coronavirus-Induced Apoptosis in 17Cl-1 Cells Involves a Mitochondria-Mediated Pathway and Its Downstream Caspase-8 Activation and Bid Cleavage. Virology 2002, 302, 321–332. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X. Murine coronavirus-induced oligodendrocyte apoptosis is mediated through the activation of the Fas signaling pathway. Virology 2007, 360, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Tian, Y.; Guan, R.; Gao, W.; Yang, X.; Zhou, L.; Wang, H. Infectious Bronchitis Virus Infection Induces Apoptosis during Replication in Chicken Macrophage HD11 Cells. Viruses 2017, 9, 198. [Google Scholar] [CrossRef]
- Lee, Y.J.; Lee, C. Porcine deltacoronavirus induces caspase-dependent apoptosis through activation of the cytochrome c -mediated intrinsic mitochondrial pathway. Virus Res. 2018, 253, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Hu, H.; Saif, L.J. Porcine deltacoronavirus induces apoptosis in swine testicular and LLC porcine kidney cell lines in vitro but not in infected intestinal enterocytes in vivo. Vet. Microbiol. 2016, 182, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Cottam, E.M.; Maier, H.J.; Manifava, M.; Vaux, L.C.; Chandra-Schoenfelder, P.; Gerner, W.; Britton, P.; Ktistakis, N.T.; Wileman, T. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy 2011, 7, 1335–1347. [Google Scholar] [CrossRef] [PubMed]
- Qin, P.; Du, E.-Z.; Luo, W.-T.; Yang, Y.-L.; Zhang, Y.-Q.; Wang, B.; Huang, Y.-W. Characteristics of the Life Cycle of Porcine Deltacoronavirus (PDCoV) In Vitro: Replication Kinetics, Cellular Ultrastructure and Virion Morphology, and Evidence of Inducing Autophagy. Viruses 2019, 11, 455. [Google Scholar] [CrossRef] [PubMed]
- García-Pérez, B.E.; González-Rojas, J.A.; Salazar, M.I.; Torres-Torres, C.; Castrejón-Jiménez, N.S. Taming the Autophagy as a Strategy for Treating COVID-19. Cells 2020, 9, 2679. [Google Scholar] [CrossRef]
- Domdom, M.-A.; Brest, P.; Grosjean, I.; Roméo, B.; Landi, M.T.; Gal, J.; Klionsky, D.J.; Hofman, P.; Mograbi, B. A multifactorial score including autophagy for prognosis and care of COVID-19 patients. Autophagy 2020, 16, 2276–2281. [Google Scholar] [CrossRef]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Xu, M.; Pradhan, M.; Tran, B.N.; Zhu, W.; Shamim, K.; Huang, W.; et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv 2020. [Google Scholar] [CrossRef]
- Sung, S.C.; Chao, C.Y.; Jeng, K.S.; Yang, J.Y.; Lai, M.M. The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology 2009, 387, 402–413. [Google Scholar] [CrossRef]
- Ye, Z.; Wong, C.K.; Li, P.; Xie, Y. A SARS-CoV protein, ORF-6, induces caspase-3 mediated, ER stress and JNK-dependent apoptosis. Biochim. Biophys. Acta (BBA) Gen. Subj. 2008, 1780, 1383–1387. [Google Scholar] [CrossRef]
- Padhan, K.; Minakshi, R.; Bin Towheed, M.A.; Jameel, S. Severe acute respiratory syndrome coronavirus 3a protein activates the mitochondrial death pathway through p38 MAP kinase activation. J. Gen. Virol. 2008, 89, 1960–1969. [Google Scholar] [CrossRef]
- Yue, Y.; Nabar, N.R.; Shi, C.-S.; Kamenyeva, O.; Xiao, X.; Hwang, I.-Y.; Wang, M.; Kehrl, J.H. SARS-Coronavirus Open Reading Frame-3a drives multimodal necrotic cell death. Cell Death Dis. 2018, 9, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.X.; Tan, T.H.; Lee, M.J.; Tham, P.Y.; Gunalan, V.; Druce, J.; Birch, C.; Catton, M.; Fu, N.Y.; Yu, V.C.; et al. Induction of apoptosis by the severe acute respiratory syndrome coronavirus 7a protein is dependent on its interaction with the Bcl-XL protein. J. Virol. 2007, 81, 6346–6355. [Google Scholar] [CrossRef]
- Tan, Y.-J.; Fielding, B.C.; Goh, P.-Y.; Shen, S.; Tan, T.H.P.; Lim, S.G.; Hong, W. Overexpression of 7a, a Protein Specifically Encoded by the Severe Acute Respiratory Syndrome Coronavirus, Induces Apoptosis via a Caspase-Dependent Pathway. J. Virol. 2004, 78, 14043–14047. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-Y.; Ping, Y.-H.; Lee, H.-C.; Chen, K.-H.; Lee, Y.-M.; Chan, Y.-J.; Lien, T.-C.; Jap, T.-S.; Lin, C.-H.; Kao, L.-S.; et al. Open Reading Frame 8a of the Human Severe Acute Respiratory Syndrome Coronavirus Not Only Promotes Viral Replication but Also Induces Apoptosis. J. Infect. Dis. 2007, 196, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.; Åkerström, S.; Sharma, A.K.; Chow, V.; Teow, S.; Abrenica, B.; Booth, S.A.; Booth, T.F.; Mirazimi, A.; Lal, S.K. SARS-CoV 9b Protein Diffuses into Nucleus, Undergoes Active Crm1 Mediated Nucleocytoplasmic Export and Triggers Apoptosis When Retained in the Nucleus. PLoS ONE 2011, 6, e19436. [Google Scholar] [CrossRef]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Si, F.; Hu, X.; Wang, C.; Chen, B.; Wang, R.; Dong, S.; Yu, R.; Li, Z. Porcine Epidemic Diarrhea Virus (PEDV) ORF3 Enhances Viral Proliferation by Inhibiting Apoptosis of Infected Cells. Viruses 2020, 12, 214. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, D.X. The ER stress sensor IRE1 and MAP kinase ERK modulate autophagy induction in cells infected with coronavirus infectious bronchitis virus. Virology 2019, 533, 34–44. [Google Scholar] [CrossRef]
- Fung, T.S.; Liao, Y.; Liu, D.X. The Endoplasmic Reticulum Stress Sensor IRE1 Protects Cells from Apoptosis Induced by the Coronavirus Infectious Bronchitis Virus. J. Virol. 2014, 88, 12752–12764. [Google Scholar] [CrossRef]
- Zou, D.; Xu, J.; Duan, X.; Xu, X.; Li, P.; Cheng, L.; Zheng, L.; Li, X.; Zhang, Y.; Wang, X.; et al. Porcine epidemic diarrhea virus ORF3 protein causes endoplasmic reticulum stress to facilitate autophagy. Vet. Microbiol. 2019, 235, 209–219. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and biological functions. Nat. Rev. Genet. 2012, 10, 563–574. [Google Scholar] [CrossRef]
- De Jong, A.S.; Visch, H.J.; de Mattia, F.; van Dommelen, M.M.; Swarts, H.G.; Luyten, T.; Callewaert, G.; Melchers, W.J.; Willems, P.H.; van Kuppeveld, F.J. The coxsackievirus 2B protein increases efflux of ions from the endoplasmic reticulum and Golgi, thereby inhibiting protein trafficking through the Golgi. J Biol. Chem. 2006, 281, 14144–14150. [Google Scholar] [CrossRef]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar] [CrossRef]
- Pavlovic, D.; Neville, D.C.A.; Argaud, O.; Blumberg, B.; Dwek, R.A.; Fischer, W.B.; Zitzmann, N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. USA 2003, 100, 6104–6108. [Google Scholar] [CrossRef]
- Wilson, L.; Mckinlay, C.; Gage, P.; Ewart, G. SARS coronavirus E protein forms cation-selective ion channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef] [PubMed]
- Surya, W.; Li, Y.; Verdià-Bàguena, C.; Aguilella, V.M.; Torres, J. MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res. 2015, 201, 61–66. [Google Scholar] [CrossRef] [PubMed]
- Singh Tomar, P.P.; Arkin, I.T. SARS-CoV-2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem. Biophys. Res. Commun. 2020, 530, 10–14. [Google Scholar] [CrossRef]
- Ye, Y.; Hogue, B.G. Role of the Coronavirus E Viroporin Protein Transmembrane Domain in Virus Assembly. J. Virol. 2007, 81, 3597–3607. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Xie, S.; Sun, B. Viral proteins function as ion channels. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 510–515. [Google Scholar] [CrossRef]
- Chien, T.H.; Chiang, Y.L.; Chen, C.P.; Henklein, P.; Hanel, K.; Hwang, I.S.; Willbold, D.; Fischer, W.B. Assembling an ion channel: ORF 3a from SARS-CoV. Biopolymers 2013, 99, 628–635. [Google Scholar] [CrossRef]
- Chen, C.C.; Kruger, J.; Sramala, I.; Hsu, H.J.; Henklein, P.; Chen, Y.M.; Fischer, W.B. ORF8a of SARS-CoV forms an ion channel: Experiments and molecular dynamics simulations. Biochim. Biophys. Acta 2011, 1808, 572–579. [Google Scholar] [CrossRef]
- Wang, K.; Lu, W.; Chen, J.; Xie, S.; Shi, H.; Hsu, H.; Yu, W.; Xu, K.; Bian, C.; Fischer, W.B.; et al. PEDV ORF3 encodes an ion channel protein and regulates virus production. FEBS Lett. 2012, 586, 384–391. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, K.; Lv, W.; Yu, W.; Xie, S.; Xu, K.; Schwarz, W.; Xiong, S.; Sun, B. The ORF4a protein of human coronavirus 229E functions as a viroporin that regulates viral production. Biochim. Biophys. Acta (BBA) Biomembr. 2014, 1838, 1088–1095. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, K.; Ping, X.; Yu, W.; Qian, Z.; Xiong, S.; Sun, B. The ns12.9 Accessory Protein of Human Coronavirus OC43 Is a Viroporin Involved in Virion Morphogenesis and Pathogenesis. J. Virol. 2015, 89, 11383–11395. [Google Scholar] [CrossRef]
- Almazán, F.; DeDiego, M.L.; Sola, I.; Zuñiga, S.; Nieto-Torres, J.L.; Marquez-Jurado, S.; Andrés, G.; Enjuanes, L. Engineering a Replication-Competent, Propagation-Defective Middle East Respiratory Syndrome Coronavirus as a Vaccine Candidate. mBio 2013, 4, e00650-13. [Google Scholar] [CrossRef]
- DeDiego, M.L.; Álvare, E.; Almazán, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.-J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A Severe Acute Respiratory Syndrome Coronavirus That Lacks the E Gene Is Attenuated In Vitro and In Vivo. J. Virol. 2006, 81, 1701–1713. [Google Scholar] [CrossRef] [PubMed]
- Kuo, L.; Masters, P.S. The Small Envelope Protein E Is Not Essential for Murine Coronavirus Replication. J. Virol. 2003, 77, 4597–4608. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Zheng, B.-J.; Xu, K.; Schwarz, W.; Du, L.; Wong, C.K.L.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 2006, 103, 12540–12545. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.-M.; Tsoi, H.; Chan, W.-M.; Zhai, S.; Wong, C.O.; Yao, X.; Chan, W.Y.; Tsui, S.K.-W.; Chan, H.Y.E. The ion channel activity of the SARS-coronavirus 3a protein is linked to its pro-apoptotic function. Int. J. Biochem. Cell Biol. 2009, 41, 2232–2239. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Rodriguez, C.; Honrubia, J.M.; Gutiérrez-Álvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Báguena, C.; Queralt-Martín, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Bai, D.; Fang, L.; Xia, S.; Ke, W.; Wang, J.; Wu, X.; Fang, P.; Xiao, S. Porcine deltacoronavirus (PDCoV) modulates calcium influx to favor viral replication. Virology 2020, 539, 38–48. [Google Scholar] [CrossRef]
- Hyser, J.M.; Utama, B.; Crawford, S.E.; Broughman, J.R.; Estes, M.K. Activation of the Endoplasmic Reticulum Calcium Sensor STIM1 and Store-Operated Calcium Entry by Rotavirus Requires NSP4 Viroporin Activity. J. Virol. 2013, 87, 13579–13588. [Google Scholar] [CrossRef] [PubMed]
- Stouffer, A.L.; Acharya, R.; Salom, D.; Levine, A.S.; Di Costanzo, L.; Soto, C.S.; Tereshko, V.; Nanda, V.; Stayrook, S.; DeGrado, W.F. Structural basis for the function and inhibition of an influenza virus proton channel. Nat. Cell Biol. 2008, 451, 596–599. [Google Scholar] [CrossRef]
- Luik, P.; Chew, C.; Aittoniemi, J.; Chang, J.; Wentworth, P.; Dwek, R.A.; Biggin, P.C.; Venien-Bryan, C.; Zitzmann, N. The 3-dimensional structure of a hepatitis C virus p7 ion channel by electron microscopy. Proc. Natl. Acad. Sci. USA 2009, 106, 12712–12716. [Google Scholar] [CrossRef]
- Xie, S.; Wang, K.; Yu, W.; Lu, W.; Xu, K.; Wang, J.; Ye, B.; Schwarz, W.; Jin, Q.; Sun, B. DIDS blocks a chloride-dependent current that is mediated by the 2B protein of enterovirus 71. Cell Res. 2011, 21, 1271–1275. [Google Scholar] [CrossRef]
- Griffin, S.D.C. Plugging the holes in hepatitis C virus antiviral therapy. Proc. Natl. Acad. Sci. USA 2009, 106, 12567–12568. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.; Beales, L.P.; Clarke, D.S.; Worsfold, O.; Evans, S.; Jaeger, J.; Harris, M.P.; Rowlands, D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003, 535, 34–38. [Google Scholar] [CrossRef]
- Cady, S.; Schmidt-Rohr, K.; Wang, J.; Soto, C.S.; DeGrado, W.F.; Hong, M. Structure of the amantadine binding site of influenza M2 proton channels in lipid bilayers. Nat. Cell Biol. 2010, 463, 689–692. [Google Scholar] [CrossRef] [PubMed]
- Ewart, G.D.; Mills, K.; Cox, G.B.; Gage, P.W. Amiloride derivatives block ion channel activity and enhancement of virus-like particle budding caused by HIV-1 protein Vpu. Eur. Biophys. J. 2002, 31, 26–35. [Google Scholar] [CrossRef]
- Watanabe, S.; Watanabe, T.; Kawaoka, Y. Influenza A Virus Lacking M2 Protein as a Live Attenuated Vaccine. J. Virol. 2009, 83, 5947–5950. [Google Scholar] [CrossRef]
- Haijema, B.J.; Volders, H.; Rottier, P.J.M. Live, Attenuated Coronavirus Vaccines through the Directed Deletion of Group-Specific Genes Provide Protection against Feline Infectious Peritonitis. J. Virol. 2004, 78, 3863–3871. [Google Scholar] [CrossRef] [PubMed]
- Dedeurwaerder, A.; Desmarets, L.M.; Olyslaegers, D.A.; Vermeulen, B.L.; Dewerchin, H.L.; Nauwynck, H.J. The role of accessory proteins in the replication of feline infectious peritonitis virus in peripheral blood monocytes. Vet. Microbiol. 2013, 162, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Sola, I.; Alonso, S.; Zúñiga, S.; Balasch, M.; Plana-Durán, J.; Enjuanes, L. Engineering the Transmissible Gastroenteritis Virus Genome as an Expression Vector Inducing Lactogenic Immunity. J. Virol. 2003, 77, 4357–4369. [Google Scholar] [CrossRef] [PubMed]
- Pewe, L.; Zhou, H.; Netland, J.; Tangudu, C.; Olivares, H.; Shi, L.; Look, D.; Gallagher, T.; Perlman, S. A Severe Acute Respiratory Syndrome-Associated Coronavirus-Specific Protein Enhances Virulence of an Attenuated Murine Coronavirus. J. Virol. 2005, 79, 11335–11342. [Google Scholar] [CrossRef][Green Version]
- Menachery, V.D.; Mitchell, H.D.; Cockrell, A.S.; Gralinski, L.E.; Yount, B.L., Jr.; Graham, R.L.; McAnarney, E.T.; Douglas, M.G.; Scobey, T.; Beall, A.; et al. MERS-CoV Accessory ORFs Play Key Role for Infection and Pathogenesis. mBio 2017, 8, e00665-17. [Google Scholar] [CrossRef]
- De Haan, C.A.; Masters, P.S.; Shen, X.; Weiss, S.; Rottier, P.J. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 2002, 296, 177–189. [Google Scholar] [CrossRef]
- Van Beurden, S.J.; Berends, A.J.; Kramer-Kuhl, A.; Spekreijse, D.; Chenard, G.; Philipp, H.C.; Mundt, E.; Rottier, P.J.M.; Verheije, M.H. Recombinant live attenuated avian coronavirus vaccines with deletions in the accessory genes 3ab and/or 5ab protect against infectious bronchitis in chickens. Vaccine 2018, 36, 1085–1092. [Google Scholar] [CrossRef]
- Zhao, X.; Jiang, Y.; Cheng, X.; Yu, Y.; Gao, M.; Zhou, S. Pathogenicity of a QX-like strain of infectious bronchitis virus and effects of accessory proteins 3a and 3b in chickens. Vet. Microbiol. 2019, 239, 108464. [Google Scholar] [CrossRef]
- Kint, J.; Fernandez-Gutierrez, M.; Maier, H.J.; Britton, P.; Langereis, M.A.; Koumans, J.; Wiegertjes, G.F.; Forlenza, M. Activation of the Chicken Type I Interferon Response by Infectious Bronchitis Coronavirus. J. Virol. 2015, 89, 1156–1167. [Google Scholar] [CrossRef]
- Zhang, M.; Li, W.; Zhou, P.; Liu, D.; Luo, R.; Jongkaewwattana, A.; He, Q. Genetic manipulation of porcine deltacoronavirus reveals insights into NS6 and NS7 functions: A novel strategy for vaccine design. Emerg. Microbes Infect. 2019, 9, 20–31. [Google Scholar] [CrossRef]
- Ye, C.; Chiem, K.; Park, J.G.; Oladunni, F.; Platt, R.N., 2nd; Anderson, T.; Almazan, F.; de la Torre, J.C.; Martinez-Sobrido, L. Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome. mBio 2020, 11, e02168-20. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Muruato, A.; Lokugamage, K.G.; Narayanan, K.; Zhang, X.; Zou, J.; Liu, J.; Schindewolf, C.; Bopp, N.E.; Aguilar, P.V.; et al. An Infectious cDNA Clone of SARS-CoV-2. Cell Host Microbe 2020, 27, 841–848.e3. [Google Scholar] [CrossRef] [PubMed]
- Chiem, K.; Morales Vasquez, D.; Park, J.G.; Platt, R.N.; Anderson, T.; Walter, M.R.; Kobie, J.J.; Ye, C.; Martinez-Sobrido, L. Generation and Characterization of recombinant SARS-CoV-2 expressing reporter genes. J. Virol. 2021, 95, e02209-20. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Muruato, A.E.; Zhang, X.; Lokugamage, K.G.; Fontes-Garfias, C.R.; Zou, J.; Liu, J.; Ren, P.; Balakrishnan, M.; Cihlar, T.; et al. A nanoluciferase SARS-CoV-2 for rapid neutralization testing and screening of anti-infective drugs for COVID-19. Nat. Commun. 2020, 11, 5214. [Google Scholar] [CrossRef] [PubMed]
- Silvas, J.; Morales-Vasquez, D.; Park, J.G.; Chiem, K.; Torrelles, J.; Platt, R.N.; Anderson, T.; Ye, C.; Martinez-Sobrido, L. Contribution of SARS-CoV-2 accessory proteins to viral pathogenicity in K18 hACE2 transgenic mice. bioRxiv 2021. [Google Scholar] [CrossRef]
- Keep, S.; Oade, M.S.; Lidzbarski-Silvestre, F.; Bentley, K.; Stevenson-Leggett, P.; Freimanis, G.L.; Tennakoon, C.; Sanderson, N.; Hammond, J.A.; Jones, R.C.; et al. Multiple novel non-canonically transcribed sub-genomic mRNAs produced by avian coronavirus infectious bronchitis virus. J. Gen. Virol. 2020, 101, 1103–1118. [Google Scholar] [CrossRef]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef]
- Nelson, C.W.; Ardern, Z.; Goldberg, T.L.; Meng, C.; Kuo, C.H.; Ludwig, C.; Kolokotronis, S.O.; Wei, X. Dynamically evolving novel overlapping gene as a factor in the SARS-CoV-2 pandemic. Elife 2020, 9, e59633. [Google Scholar] [CrossRef]
- Peng, Q.; Fang, L.; Ding, Z.; Wang, D.; Peng, G.; Xiao, S. Rapid manipulation of the porcine epidemic diarrhea virus genome by CRISPR/Cas9 technology. J. Virol. Methods 2020, 276, 113772. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, P.; Fang, L.; Zhang, H.; Xia, S.; Xiao, S. Functions of Coronavirus Accessory Proteins: Overview of the State of the Art. Viruses 2021, 13, 1139. https://doi.org/10.3390/v13061139
Fang P, Fang L, Zhang H, Xia S, Xiao S. Functions of Coronavirus Accessory Proteins: Overview of the State of the Art. Viruses. 2021; 13(6):1139. https://doi.org/10.3390/v13061139
Chicago/Turabian StyleFang, Puxian, Liurong Fang, Huichang Zhang, Sijin Xia, and Shaobo Xiao. 2021. "Functions of Coronavirus Accessory Proteins: Overview of the State of the Art" Viruses 13, no. 6: 1139. https://doi.org/10.3390/v13061139
APA StyleFang, P., Fang, L., Zhang, H., Xia, S., & Xiao, S. (2021). Functions of Coronavirus Accessory Proteins: Overview of the State of the Art. Viruses, 13(6), 1139. https://doi.org/10.3390/v13061139






