Molecular Insights into the Flavivirus Replication Complex
Abstract
:1. Introduction
2. The Flavivirus Replication Cycle
3. Function and Formation of the Replication Organelle
3.1. Function
3.2. Biogenesis
3.2.1. Replication Organelle Curvature
3.2.2. Host Factors in Assembly
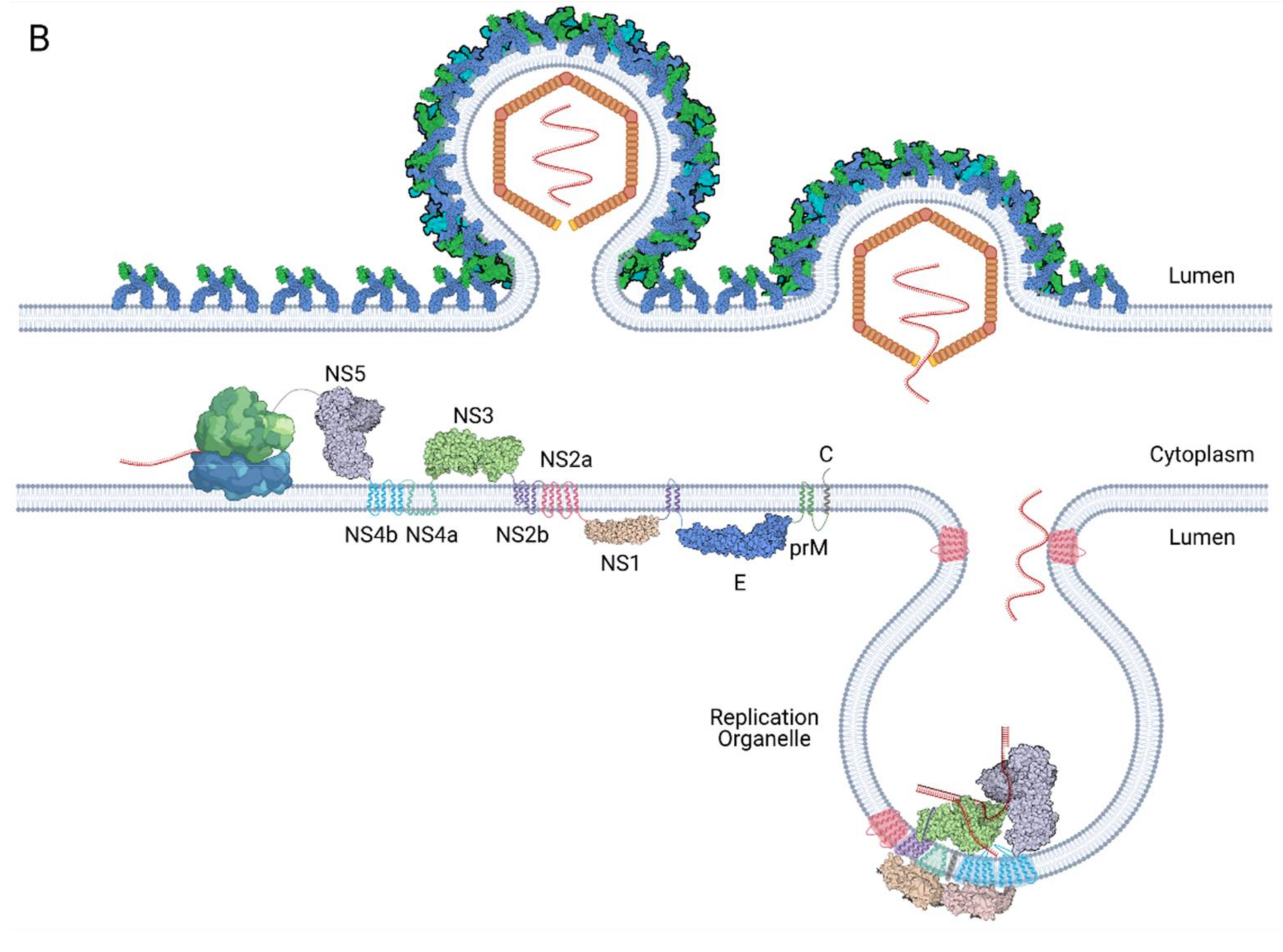
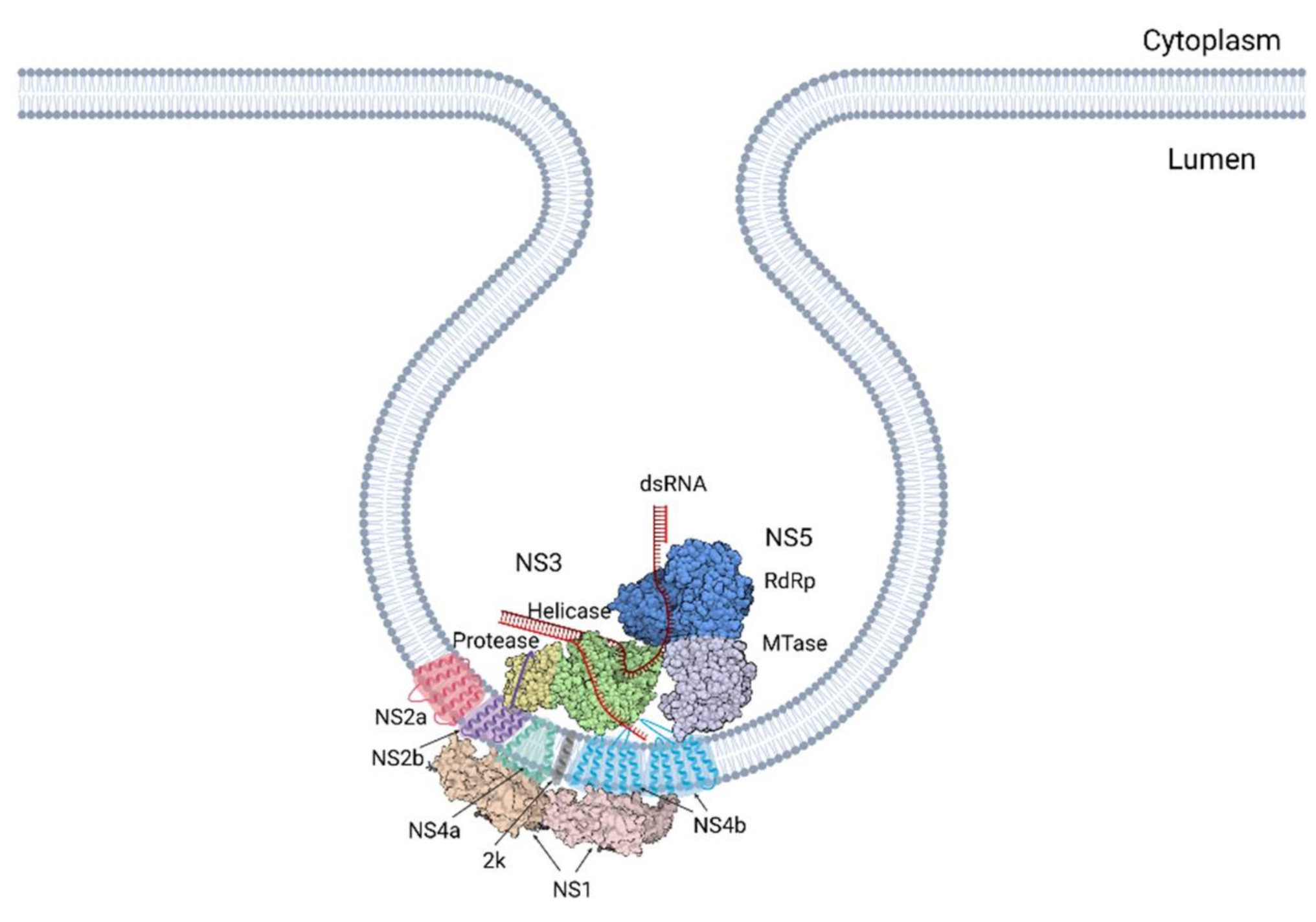
4. Architecture of the Replication Organelle
4.1. Structure, Composition and Arrangement
4.2. Convoluted Membranes
4.3. Host Factors in RC Function
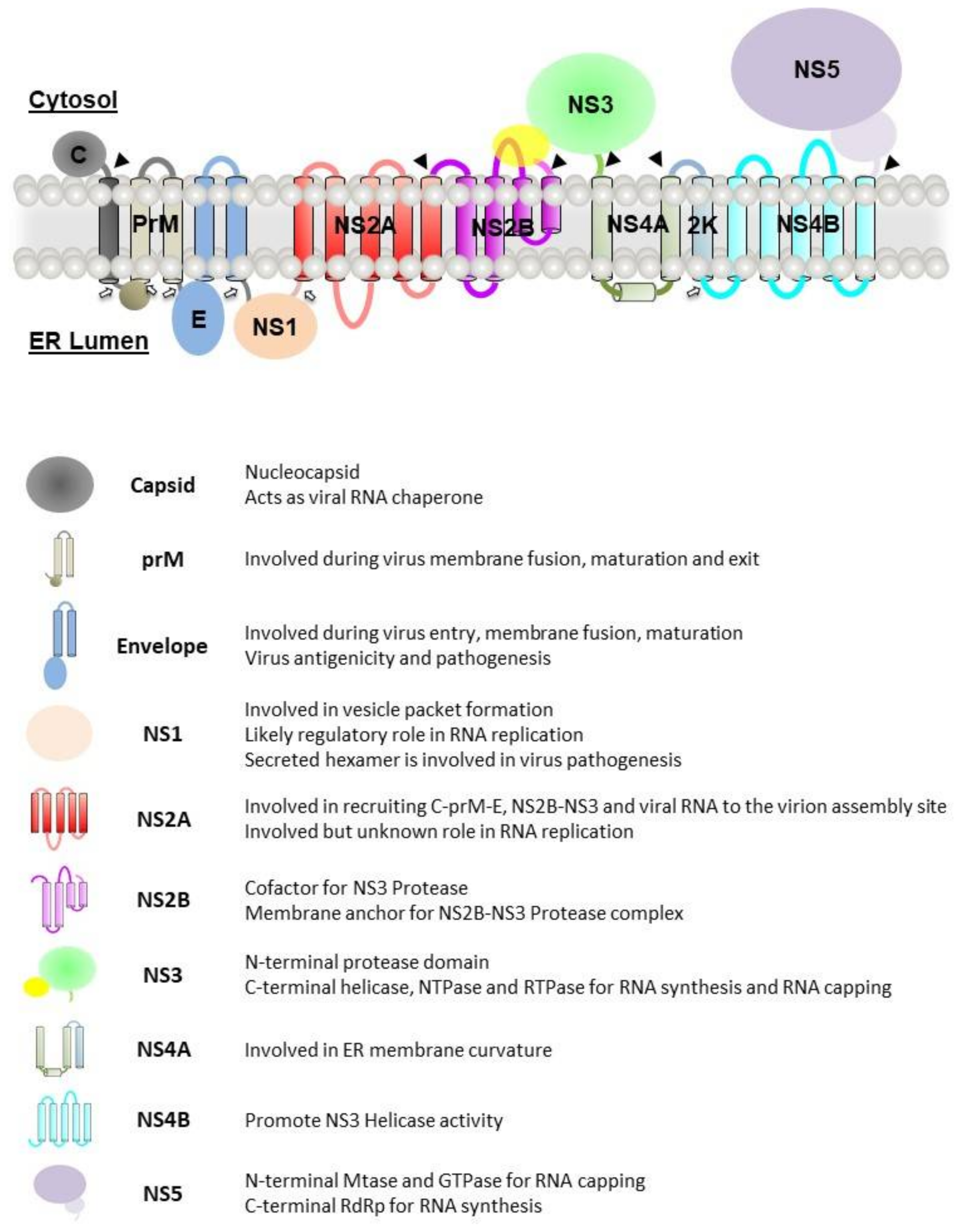
5. Nonstructural Proteins
5.1. NS1
| Viruses | PDB IDs | Remarks | Space Group | Ref. |
|---|---|---|---|---|
| DENV-1 | 4OIG | NS1 | C121 | [18] |
| 6WEQ | NS1 in complex with neutralising 2B7 Fab | P1211 | [58] | |
| DENV-2 | 4O6B | NS1 | H3 | [49] |
| 6WER | NS1 in complex with neutralising 2B7 Fab | I4122 | [58] | |
| 7K93 | NS1 in complex with neutralising 2B7 single chain Fab variable region (scFv) | P212121 | ||
| 7BSC | C-terminal NS1 in complex with 1G5.3 Fab | P21221 | [59] | |
| JEV | 5O19 | C-terminal NS1 | I212121 | [60] |
| WNV | 4OIE | NS1 | P3121 | [18] |
| 4OII | NS1 In complex with neutralising 22NS1 Fab | C121 | ||
| 4O6C | NS1 | P3 | [49] | |
| 4O6D | NS1 | P321 | ||
| YFV | 5YXA | C-terminal NS1 | P212121 | [61] |
| ZIKV | 5GS6 | NS1 from 2015 Brazil strain | P4122 | [48] |
| 5IY3 | NS1 | C2221 | [62] | |
| 5K6K | NS1 | I222 | [17] | |
| 5X8Y | NS1 | C2221 | [63] | |
| 7BSD | C-terminal NS1 in complex with 1G5.3 Fab | P212121 | [59] |
5.2. NS2A
5.3. NS2B
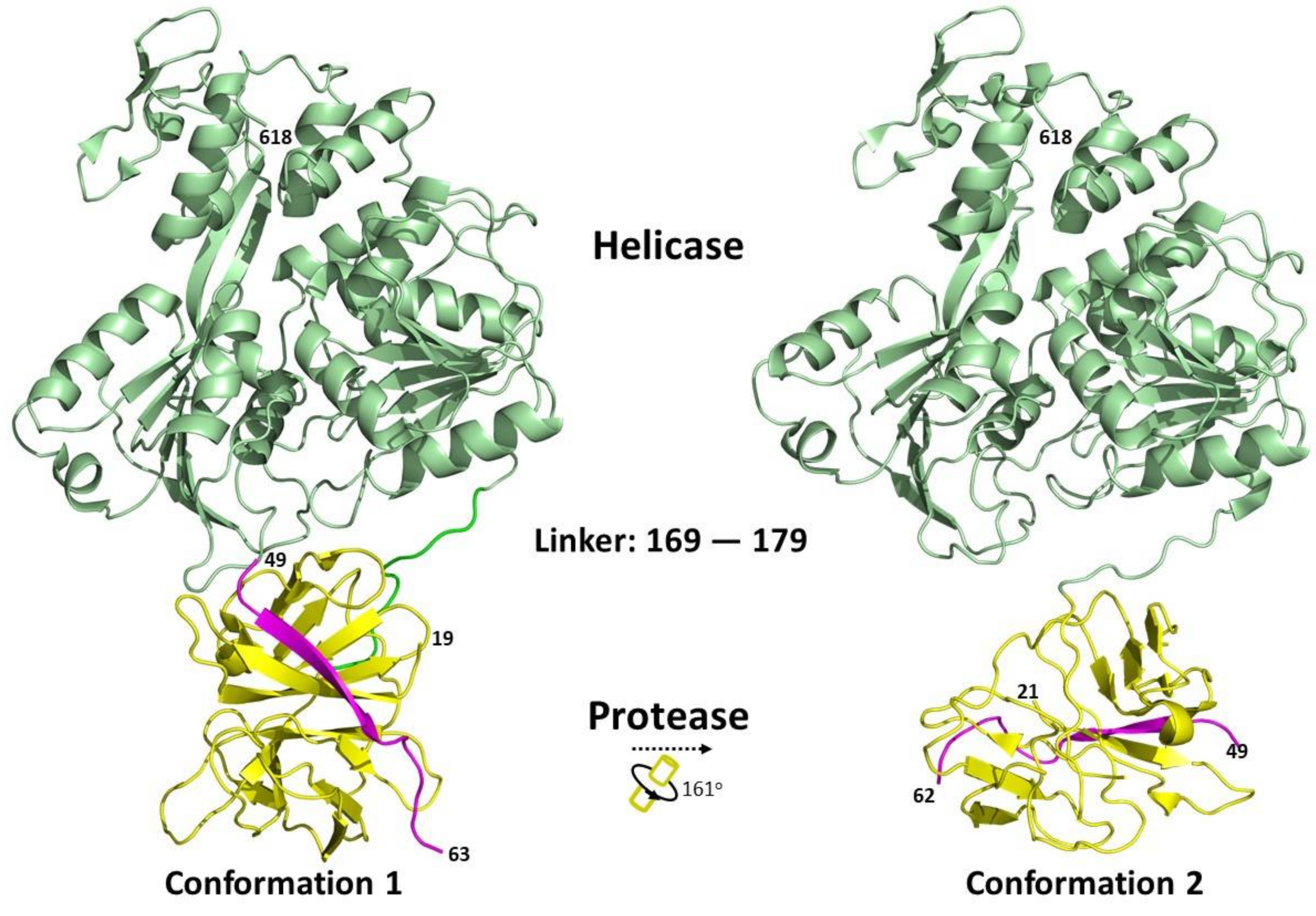
5.4. NS3
| Viruses | PDB IDs | Protease Active Site | Remarks | Ref. |
|---|---|---|---|---|
| DENV-4 | 2VBC | Open | Partial NS2B cofactor linked to NS3 via a G4SG4 linker | [16] |
| 2WHX | Open | Partial NS2B cofactor linked to NS3 via a G4SG4 linker | [86] | |
| 2WZQ | Open | Partial NS2B cofactor linked to NS3 via a G4SG4 linker, glycine insertion mutant between E173 and P174 | ||
| 5YVJ | Open | Partial NS2B cofactor linked to NS3 via a G4SG4 linker | [94] | |
| 5YVU | Closed | NS2B cofactor linked to NS3 via a G4SG4 linker, in complex with BPTI | ||
| 5YVV | Closed | NS2B cofactor linked to NS3 via a G4SG4 linker | ||
| 5YVW | Closed | Unlinked NS2B cofactor | ||
| 5YVY | Closed | Unlinked NS2B cofactor | ||
| 5YW1 | Closed | Unlinked NS2B cofactor, in complex with BPTI | ||
| MVEV | 2WV9 | Open | Partial NS2B cofactor linked to NS3 via a G4SG4 linker | [87] |
5.5. NS4A
5.6. NS4B
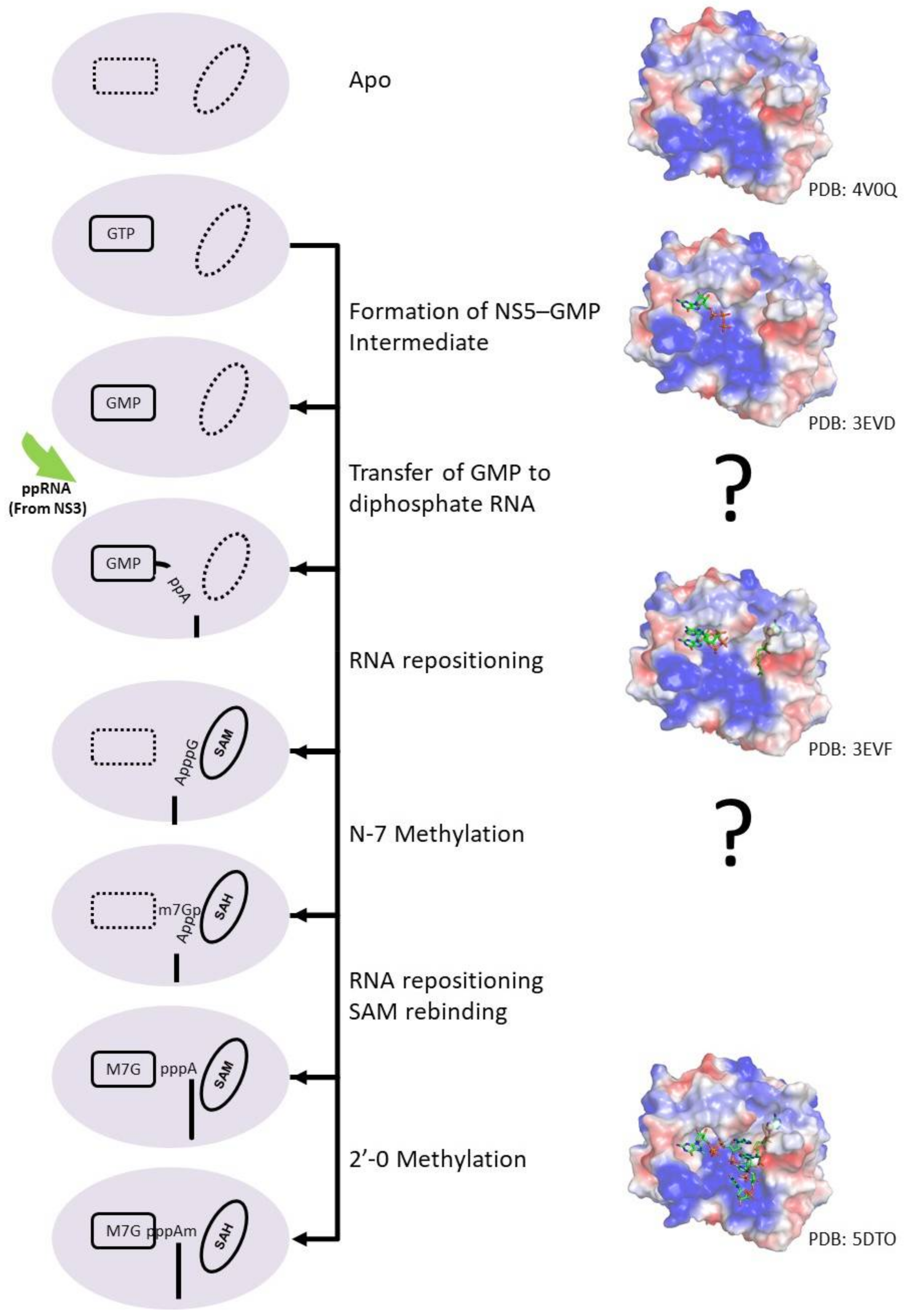
5.7. NS5
| Viruses | PDB IDs | Ligands | Remarks | Space Group | Ref. |
|---|---|---|---|---|---|
| DENV-3 | 4V0Q | SAH and Zn | Conformation 5 | P21212 | [115] |
| 4V0R | GTP, Mg, SAH and Zn | ||||
| 5DTO | M7G, Mg, SAH and Zn | [111] | |||
| 5CCV | SAH and Zn | [123] | |||
| 5JJR | Compound 29, SAH, Mg and Zn | [124] | |||
| 5JJS | Compound 27, SAH, Mg and Zn | ||||
| DENV-2 | 5ZQK | SAM, Mg and Zn | Conformation 3 | P1211 | [15] |
| 6KR2 | SAH and Zn | Conformation 3 | P1211 | [118] | |
| 6KR3 | SAH and Zn | Conformation 4 | C2221 | ||
| JEV | 4K6M | SAH and Zn | Conformation 1 | H3 | [119] |
| YFV | 6QSN | SAH and Zn | Conformation 2 | P212121 | [121] |
| ZIKV | 5TFR | SAH and Zn | Conformation 1 | P21212 | [125] |
| 5TMH | P22121 | [116] | |||
| 5U0B | P21212 | [117] | |||
| 5M2X | P212121 | [126] | |||
| 5M2Z | P65 | ||||
| 6I7P | P212121 |

6. Interaction Network within the Replication Complex
| Virus | Protein: Residues | Protein: Residues | References |
|---|---|---|---|
| DENV | NS1 (G161, W168) | NS4A-2K-NS4B | [54] |
| NS1: 30–180 | NS4A-2K-NS4B: 35–61 (Y41) | [57] | |
| NS2B | NS3 | ||
| NS3: Helicase Domain II and III | NS4B: 129–165 (Q134) | [106,134,135] | |
| NS3: 566–585 (N570) | NS5: 320–341 (K330) | [138,139] | |
| NS4A: 40–76 (L48, T54, L60) | NS4B: 84–146 | [140] | |
| NS4B | NS4B | [102] | |
| WNV | NS1 | NS4B | [55] |
| NS2A | NS2B | [74] | |
| NS2A | NS4A | [74] | |
| NS2B | NS3 | [74] | |
| NS2B | NS4A | [74] | |
| NS2B | NS4B | [74] | |
| NS3 | NS5 | [74] | |
| NS4A | NS4B | [74] | |
| JEV | NS2A | NS2B | [75] |
| NS3 | NS5 | [141] | |
| YFV | NS1 | NS4A | [56] |
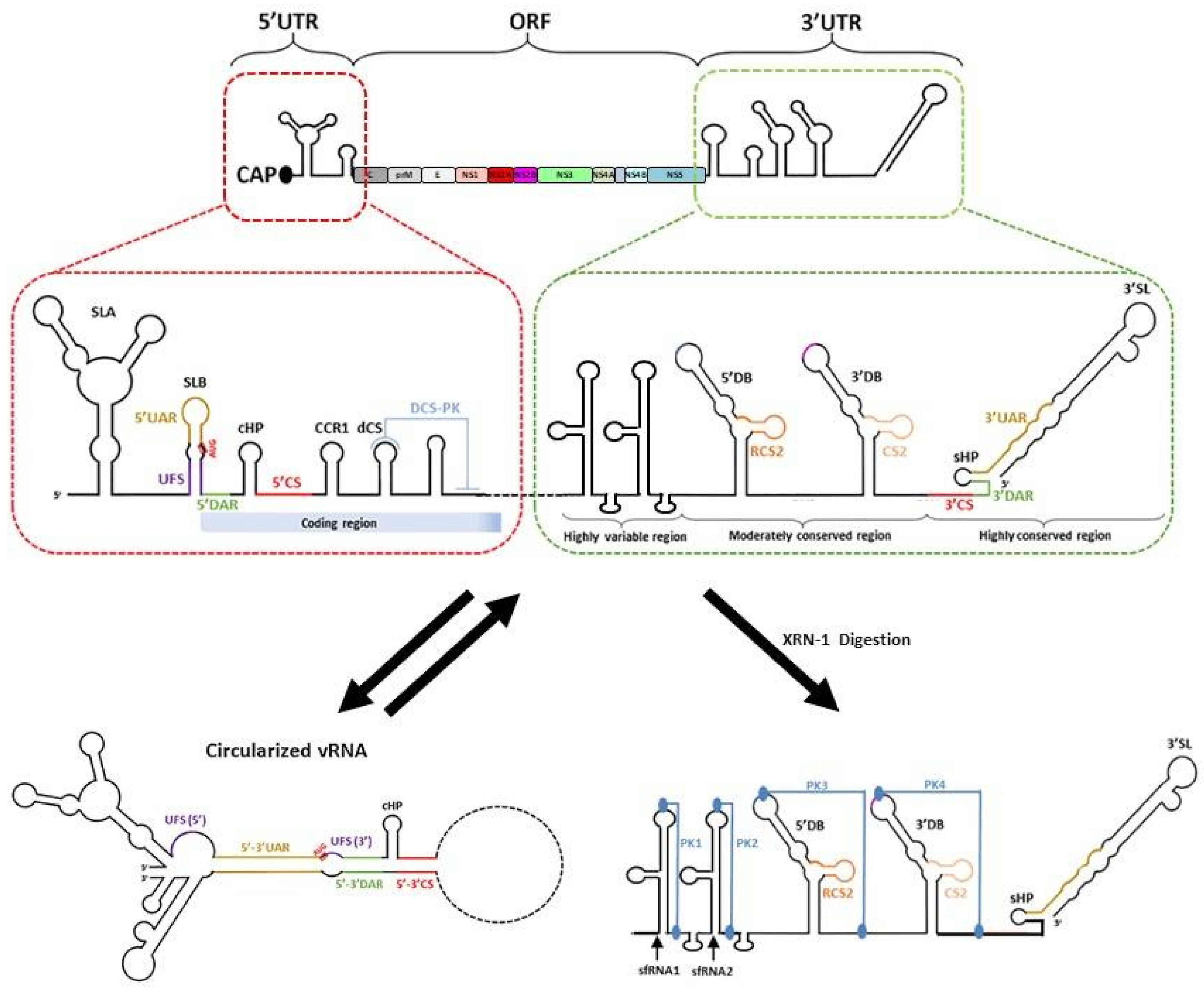
7. The Viral RNA
7.1. The 5′ UTR
7.2. The 3′UTR
7.3. Genome Cyclisation and Long-Range RNA–RNA Interactions
7.4. Subgenomic Flaviviral RNA (sfRNA)
8. Future Work
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baltimore, D. Expression of animal virus genomes. Bacteriol. Rev. 1971, 35, 235–241. [Google Scholar] [CrossRef] [PubMed]
- Paul, D.; Bartenschlager, R. Architecture and biogenesis of plus-strand RNA virus replication factories. World J. Virol. 2013, 2, 32–48. [Google Scholar] [CrossRef] [PubMed]
- Denison, M.R. Seeking membranes: Positive-strand RNA virus replication complexes. PLoS Biol. 2008, 6, e270. [Google Scholar] [CrossRef] [Green Version]
- Kato, H.; Takahasi, K.; Fujita, T. RIG-I-like receptors: Cytoplasmic sensors for non-self RNA. Immunol. Rev. 2011, 243, 91–98. [Google Scholar] [CrossRef] [Green Version]
- Pierson, T.C.; Diamond, M.S. The continued threat of emerging flaviviruses. Nat. Microbiol. 2020. [Google Scholar] [CrossRef]
- Harrington, J.; Kroeger, A.; Runge-Ranzinger, S.; O’Dempsey, T. Detecting and responding to a dengue outbreak: Evaluation of existing strategies in country outbreak response planning. J. Trop. Med. 2013, 2013, 756832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- WHO. Fact Sheet: Dengue and Severe Dengue; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Smit, J.M.; Moesker, B.; Rodenhuis-Zybert, I.; Wilschut, J. Flavivirus cell entry and membrane fusion. Viruses 2011, 3, 160–171. [Google Scholar] [CrossRef] [Green Version]
- Westaway, E.G.; Brinton, M.A.; Gaidamovich, S.; Horzinek, M.C.; Igarashi, A.; Kaariainen, L.; Lvov, D.K.; Porterfield, J.S.; Russell, P.K.; Trent, D.W. Flaviviridae. Intervirology 1985, 24, 183–192. [Google Scholar] [CrossRef]
- Westaway, E.G.; Mackenzie, J.M.; Kenney, M.T.; Jones, M.K.; Khromykh, A.A. Ultrastructure of Kunjin virus-infected cells: Colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J. Virol. 1997, 71, 6650–6661. [Google Scholar] [CrossRef] [Green Version]
- Leary, K.; Blair, C.D. Sequential events in the morphogenesis of japanese Encephalitis virus. J. Ultrastruct. Res. 1980, 72, 123–129. [Google Scholar] [CrossRef]
- Allison, S.L.; Schalich, J.; Stiasny, K.; Mandl, C.W.; Kunz, C.; Heinz, F.X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J. Virol. 1995, 69, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Stadler, K.; Allison, S.L.; Schalich, J.; Heinz, F.X. Proteolytic activation of tick-borne encephalitis virus by furin. J. Virol. 1997, 71, 8475–8481. [Google Scholar] [CrossRef] [Green Version]
- El Sahili, A.; Soh, T.S.; Schiltz, J.; Gharbi-Ayachi, A.; Seh, C.C.; Shi, P.Y.; Lim, S.P.; Lescar, J. NS5 from Dengue Virus Serotype 2 Can Adopt a Conformation Analogous to That of Its Zika Virus and Japanese Encephalitis Virus Homologues. J. Virol. 2019, 94. [Google Scholar] [CrossRef]
- Luo, D.; Xu, T.; Hunke, C.; Gruber, G.; Vasudevan, S.G.; Lescar, J. Crystal structure of the NS3 protease-helicase from dengue virus. J. Virol. 2008, 82, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Brown, W.C.; Akey, D.L.; Konwerski, J.R.; Tarrasch, J.T.; Skiniotis, G.; Kuhn, R.J.; Smith, J.L. Extended surface for membrane association in Zika virus NS1 structure. Nat. Struct. Mol. Biol. 2016, 23, 865–867. [Google Scholar] [CrossRef]
- Edeling, M.A.; Diamond, M.S.; Fremont, D.H. Structural basis of Flavivirus NS1 assembly and antibody recognition. Proc. Natl. Acad. Sci. USA 2014, 111, 4285–4290. [Google Scholar] [CrossRef] [Green Version]
- Rouvinski, A.; Guardado-Calvo, P.; Barba-Spaeth, G.; Duquerroy, S.; Vaney, M.C.; Kikuti, C.M.; Navarro Sanchez, M.E.; Dejnirattisai, W.; Wongwiwat, W.; Haouz, A.; et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 2015, 520, 109–113. [Google Scholar] [CrossRef]
- Fibriansah, G.; Tan, J.L.; Smith, S.A.; de Alwis, R.; Ng, T.S.; Kostyuchenko, V.A.; Jadi, R.S.; Kukkaro, P.; de Silva, A.M.; Crowe, J.E.; et al. A highly potent human antibody neutralizes dengue virus serotype 3 by binding across three surface proteins. Nat. Commun. 2015, 6, 6341. [Google Scholar] [CrossRef]
- Li, L.; Lok, S.M.; Yu, I.M.; Zhang, Y.; Kuhn, R.J.; Chen, J.; Rossmann, M.G. The flavivirus precursor membrane-envelope protein complex: Structure and maturation. Science 2008, 319, 1830–1834. [Google Scholar] [CrossRef] [Green Version]
- Paul, D.; Bartenschlager, R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu. Rev. Virol. 2015, 2, 289–310. [Google Scholar] [CrossRef] [PubMed]
- Kikkert, M. Innate Immune Evasion by Human Respiratory RNA Viruses. J. Innate Immunity 2020, 12, 4–20. [Google Scholar] [CrossRef]
- Welsch, S.; Miller, S.; Romero-Brey, I.; Merz, A.; Bleck, C.K.; Walther, P.; Fuller, S.D.; Antony, C.; Krijnse-Locker, J.; Bartenschlager, R. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 2009, 5, 365–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bollati, M.; Alvarez, K.; Assenberg, R.; Baronti, C.; Canard, B.; Cook, S.; Coutard, B.; Decroly, E.; de Lamballerie, X.; Gould, E.A.; et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antivir. Res. 2010, 87, 125–148. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Kastner, S.; Krijnse-Locker, J.; Buhler, S.; Bartenschlager, R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2007, 282, 8873–8882. [Google Scholar] [CrossRef] [Green Version]
- Roosendaal, J.; Westaway, E.G.; Khromykh, A.; Mackenzie, J.M. Regulated cleavages at the West Nile virus NS4A-2K-NS4B junctions play a major role in rearranging cytoplasmic membranes and Golgi trafficking of the NS4A protein. J. Virol. 2006, 80, 4623–4632. [Google Scholar] [CrossRef] [Green Version]
- Egger, D.; Wolk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef] [Green Version]
- Ci, Y.; Liu, Z.Y.; Zhang, N.N.; Niu, Y.; Yang, Y.; Xu, C.; Yang, W.; Qin, C.F.; Shi, L. Zika NS1-induced ER remodeling is essential for viral replication. J. Cell Biol. 2020, 219. [Google Scholar] [CrossRef]
- Cerikan, B.; Goellner, S.; Neufeldt, C.J.; Haselmann, U.; Mulder, K.; Chatel-Chaix, L.; Cortese, M.; Bartenschlager, R. A Non-Replicative Role of the 3′ Terminal Sequence of the Dengue Virus Genome in Membranous Replication Organelle Formation. Cell Rep. 2020, 32, 107859. [Google Scholar] [CrossRef]
- Heaton, N.S.; Perera, R.; Berger, K.L.; Khadka, S.; Lacount, D.J.; Kuhn, R.J.; Randall, G. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA 2010, 107, 17345–17350. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.M.; Khromykh, A.A.; Parton, R.G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2007, 2, 229–239. [Google Scholar] [CrossRef] [Green Version]
- Perera, R.; Riley, C.; Isaac, G.; Hopf-Jannasch, A.S.; Moore, R.J.; Weitz, K.W.; Pasa-Tolic, L.; Metz, T.O.; Adamec, J.; Kuhn, R.J. Dengue virus infection perturbs lipid homeostasis in infected mosquito cells. PLoS Pathog. 2012, 8, e1002584. [Google Scholar] [CrossRef]
- Tani, H.; Shiokawa, M.; Kaname, Y.; Kambara, H.; Mori, Y.; Abe, T.; Moriishi, K.; Matsuura, Y. Involvement of ceramide in the propagation of Japanese encephalitis virus. J. Virol. 2010, 84, 2798–2807. [Google Scholar] [CrossRef] [Green Version]
- Aktepe, T.E.; Liebscher, S.; Prier, J.E.; Simmons, C.P.; Mackenzie, J.M. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017, 21, 1639–1654. [Google Scholar] [CrossRef] [Green Version]
- Aktepe, T.E.; Mackenzie, J.M. Shaping the flavivirus replication complex: It is curvaceous! Cell. Microbiol. 2018, 20, e12884. [Google Scholar] [CrossRef] [Green Version]
- Yi, Z.; Yuan, Z.; Rice, C.M.; MacDonald, M.R. Flavivirus replication complex assembly revealed by DNAJC14 functional mapping. J. Virol. 2012, 86, 11815–11832. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, H.H.; Schneider, W.M.; Rozen-Gagnon, K.; Miles, L.A.; Schuster, F.; Razooky, B.; Jacobson, E.; Wu, X.; Yi, S.; Rudin, C.M.; et al. TMEM41B Is a Pan-flavivirus Host Factor. Cell 2021, 184, 133–148 e120. [Google Scholar] [CrossRef]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar] [CrossRef] [Green Version]
- Chatel-Chaix, L.; Cortese, M.; Romero-Brey, I.; Bender, S.; Neufeldt, C.J.; Fischl, W.; Scaturro, P.; Schieber, N.; Schwab, Y.; Fischer, B.; et al. Dengue Virus Perturbs Mitochondrial Morphodynamics to Dampen Innate Immune Responses. Cell Host Microbe 2016, 20, 342–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortese, M.; Goellner, S.; Acosta, E.G.; Neufeldt, C.J.; Oleksiuk, O.; Lampe, M.; Haselmann, U.; Funaya, C.; Schieber, N.; Ronchi, P.; et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017, 18, 2113–2123. [Google Scholar] [CrossRef] [Green Version]
- Junjhon, J.; Pennington, J.G.; Edwards, T.J.; Perera, R.; Lanman, J.; Kuhn, R.J. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 2014, 88, 4687–4697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shah, P.S.; Link, N.; Jang, G.M.; Sharp, P.P.; Zhu, T.; Swaney, D.L.; Johnson, J.R.; Von Dollen, J.; Ramage, H.R.; Satkamp, L.; et al. Comparative Flavivirus-Host Protein Interaction Mapping Reveals Mechanisms of Dengue and Zika Virus Pathogenesis. Cell 2018, 175, 1931–1945 e1918. [Google Scholar] [CrossRef] [Green Version]
- Ooi, Y.S.; Majzoub, K.; Flynn, R.A.; Mata, M.A.; Diep, J.; Li, J.K.; van Buuren, N.; Rumachik, N.; Johnson, A.G.; Puschnik, A.S.; et al. An RNA-centric dissection of host complexes controlling flavivirus infection. Nat. Microbiol. 2019, 4, 2369–2382. [Google Scholar] [CrossRef] [PubMed]
- Phillips, S.L.; Soderblom, E.J.; Bradrick, S.S.; Garcia-Blanco, M.A. Identification of Proteins Bound to Dengue Viral RNA In Vivo Reveals New Host Proteins Important for Virus Replication. mBio 2016, 7, e01865-15. [Google Scholar] [CrossRef] [Green Version]
- Marceau, C.D.; Puschnik, A.S.; Majzoub, K.; Ooi, Y.S.; Brewer, S.M.; Fuchs, G.; Swaminathan, K.; Mata, M.A.; Elias, J.E.; Sarnow, P.; et al. Genetic dissection of Flaviviridae host factors through genome-scale CRISPR screens. Nature 2016, 535, 159–163. [Google Scholar] [CrossRef] [Green Version]
- Fons, R.D.; Bogert, B.A.; Hegde, R.S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003, 160, 529–539. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Song, H.; Qi, J.; Liu, Y.; Wang, H.; Su, C.; Shi, Y.; Gao, G.F. Contribution of intertwined loop to membrane association revealed by Zika virus full-length NS1 structure. EMBO J. 2016, 35, 2170–2178. [Google Scholar] [CrossRef] [Green Version]
- Akey, D.L.; Brown, W.C.; Dutta, S.; Konwerski, J.; Jose, J.; Jurkiw, T.J.; DelProposto, J.; Ogata, C.M.; Skiniotis, G.; Kuhn, R.J.; et al. Flavivirus NS1 structures reveal surfaces for associations with membranes and the immune system. Science 2014, 343, 881–885. [Google Scholar] [CrossRef] [Green Version]
- Mackenzie, J.M.; Jones, M.K.; Young, P.R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology 1996, 220, 232–240. [Google Scholar] [CrossRef] [Green Version]
- Youn, S.; Ambrose, R.L.; Mackenzie, J.M.; Diamond, M.S. Non-structural protein-1 is required for West Nile virus replication complex formation and viral RNA synthesis. Virol. J. 2013, 10, 339. [Google Scholar] [CrossRef] [Green Version]
- Lindenbach, B.D.; Rice, C.M. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 1997, 71, 9608–9617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khromykh, A.A.; Sedlak, P.L.; Guyatt, K.J.; Hall, R.A.; Westaway, E.G. Efficient trans-complementation of the flavivirus kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J. Virol. 1999, 73, 10272–10280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaszczyca, A.; Scaturro, P.; Neufeldt, C.J.; Cortese, M.; Cerikan, B.; Ferla, S.; Brancale, A.; Pichlmair, A.; Bartenschlager, R. A novel interaction between dengue virus nonstructural protein 1 and the NS4A-2K-4B precursor is required for viral RNA replication but not for formation of the membranous replication organelle. PLoS Pathog. 2019, 15, e1007736. [Google Scholar] [CrossRef]
- Youn, S.; Li, T.; McCune, B.T.; Edeling, M.A.; Fremont, D.H.; Cristea, I.M.; Diamond, M.S. Evidence for a genetic and physical interaction between nonstructural proteins NS1 and NS4B that modulates replication of West Nile virus. J. Virol. 2012, 86, 7360–7371. [Google Scholar] [CrossRef] [Green Version]
- Lindenbach, B.D.; Rice, C.M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 1999, 73, 4611–4621. [Google Scholar] [CrossRef] [Green Version]
- Tan, M.J.A.; Brown, N.G.; Chan, K.W.K.; Jin, J.Y.; Zu Kong, S.Y.; Vasudevan, S.G. Mutations in the cytoplasmic domain of dengue virus NS4A affect virus fitness and interactions with other non-structural proteins. J. Gen. Virol. 2020. [Google Scholar] [CrossRef]
- Biering, S.B.; Akey, D.L.; Wong, M.P.; Brown, W.C.; Lo, N.T.N.; Puerta-Guardo, H.; Tramontini Gomes de Sousa, F.; Wang, C.; Konwerski, J.R.; Espinosa, D.A.; et al. Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction. Science 2021, 371, 194–200. [Google Scholar] [CrossRef]
- Modhiran, N.; Song, H.; Liu, L.; Bletchly, C.; Brillault, L.; Amarilla, A.A.; Xu, X.; Qi, J.; Chai, Y.; Cheung, S.T.M.; et al. A broadly protective antibody that targets the flavivirus NS1 protein. Science 2021, 371, 190–194. [Google Scholar] [CrossRef]
- Poonsiri, T.; Wright, G.S.A.; Diamond, M.S.; Turtle, L.; Solomon, T.; Antonyuk, S.V. Structural Study of the C-Terminal Domain of Nonstructural Protein 1 from Japanese Encephalitis Virus. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Han, M.; Qi, J.; Hilgenfeld, R.; Luo, T.; Shi, Y.; Gao, G.F.; Song, H. Crystal structure of the C-terminal fragment of NS1 protein from yellow fever virus. Sci. China Life Sci. 2017, 60, 1403–1406. [Google Scholar] [CrossRef]
- Song, H.; Qi, J.; Haywood, J.; Shi, Y.; Gao, G.F. Zika virus NS1 structure reveals diversity of electrostatic surfaces among flaviviruses. Nat. Struct. Mol. Biol. 2016, 23, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Chen, C.; Liu, S.; Zhou, H.; Yang, K.; Zhao, Q.; Ji, X.; Chen, C.; Xie, W.; Wang, Z.; et al. A Mutation Identified in Neonatal Microcephaly Destabilizes Zika Virus NS1 Assembly in Vitro. Sci. Rep. 2017, 7, 42580. [Google Scholar] [CrossRef]
- Mackenzie, J.M.; Khromykh, A.A.; Jones, M.K.; Westaway, E.G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology 1998, 245, 203–215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Xie, X.; Zou, J.; Xia, H.; Shan, C.; Chen, X.; Shi, P.Y. Genetic and biochemical characterizations of Zika virus NS2A protein. Emerg. Microbes Infect. 2019, 8, 585–602. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Gayen, S.; Kang, C.; Yuan, Z.; Shi, P.Y. Membrane topology and function of dengue virus NS2A protein. J. Virol. 2013, 87, 4609–4622. [Google Scholar] [CrossRef] [Green Version]
- Xie, X.; Zou, J.; Puttikhunt, C.; Yuan, Z.; Shi, P.Y. Two distinct sets of NS2A molecules are responsible for dengue virus RNA synthesis and virion assembly. J. Virol. 2015, 89, 1298–1313. [Google Scholar] [CrossRef] [Green Version]
- Barnard, T.R.; Abram, Q.H.; Lin, Q.F.; Wang, A.B.; Sagan, S.M. Molecular Determinants of Flavivirus Virion Assembly. Trends Biochem. Sci. 2021. [Google Scholar] [CrossRef]
- Ng, E.Y.; Loh, Y.R.; Li, Y.; Li, Q.; Kang, C. Expression, purification of Zika virus membrane protein-NS2B in detergent micelles for NMR studies. Protein Expr. Purif. 2019, 154, 1–6. [Google Scholar] [CrossRef]
- Li, Y.; Li, Q.; Wong, Y.L.; Liew, L.S.; Kang, C. Membrane topology of NS2B of dengue virus revealed by NMR spectroscopy. Biochim. Biophys. Acta 2015, 1848, 2244–2252. [Google Scholar] [CrossRef] [Green Version]
- Kang, C.; Keller, T.H.; Luo, D. Zika Virus Protease: An Antiviral Drug Target. Trends Microbiol. 2017, 25, 797–808. [Google Scholar] [CrossRef]
- Hill, M.E.; Kumar, A.; Wells, J.A.; Hobman, T.C.; Julien, O.; Hardy, J.A. The Unique Cofactor Region of Zika Virus NS2B-NS3 Protease Facilitates Cleavage of Key Host Proteins. ACS Chem. Biol. 2018, 13, 2398–2405. [Google Scholar] [CrossRef]
- Clum, S.; Ebner, K.E.; Padmanabhan, R. Cotranslational membrane insertion of the serine proteinase precursor NS2B-NS3(Pro) of dengue virus type 2 is required for efficient in vitro processing and is mediated through the hydrophobic regions of NS2B. J. Biol. Chem. 1997, 272, 30715–30723. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Takeda, K.; Markoff, L. Protein-protein interactions among West Nile non-structural proteins and transmembrane complex formation in mammalian cells. Virology 2013, 446, 365–377. [Google Scholar] [CrossRef] [Green Version]
- Li, X.D.; Deng, C.L.; Ye, H.Q.; Zhang, H.L.; Zhang, Q.Y.; Chen, D.D.; Zhang, P.T.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Transmembrane Domains of NS2B Contribute to both Viral RNA Replication and Particle Formation in Japanese Encephalitis Virus. J. Virol. 2016, 90, 5735–5749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Erbel, P.; Schiering, N.; D’Arcy, A.; Renatus, M.; Kroemer, M.; Lim, S.P.; Yin, Z.; Keller, T.H.; Vasudevan, S.G.; Hommel, U. Structural basis for the activation of flaviviral NS3 proteases from dengue and West Nile virus. Nat. Struct. Mol. Biol. 2006, 13, 372–373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, Y.; Loh, Y.R.; Phoo, W.W.; Hung, A.W.; Kang, C.; Luo, D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. [Google Scholar] [CrossRef]
- Luo, D.; Vasudevan, S.G.; Lescar, J. The flavivirus NS2B-NS3 protease-helicase as a target for antiviral drug development. Antivir. Res. 2015, 118, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Phoo, W.W.; Luo, D. Functional interplay among the flavivirus NS3 protease, helicase, and cofactors. Virol. Sin. 2014, 29, 74–85. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Coloma, J.; Garcia-Sastre, A.; Aggarwal, A.K. Structure of the NS3 helicase from Zika virus. Nat. Struct. Mol. Biol. 2016, 23, 752–754. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Ji, X.; Yang, X.; Xie, W.; Yang, K.; Chen, C.; Wu, C.; Chi, H.; Mu, Z.; Wang, Z.; et al. The crystal structure of Zika virus helicase: Basis for antiviral drug design. Protein Cell 2016, 7, 450–454. [Google Scholar] [CrossRef] [Green Version]
- Xu, T.; Sampath, A.; Chao, A.; Wen, D.; Nanao, M.; Chene, P.; Vasudevan, S.G.; Lescar, J. Structure of the Dengue virus helicase/nucleoside triphosphatase catalytic domain at a resolution of 2.4 A. J. Virol. 2005, 79, 10278–10288. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Xu, T.; Watson, R.P.; Scherer-Becker, D.; Sampath, A.; Jahnke, W.; Yeong, S.S.; Wang, C.H.; Lim, S.P.; Strongin, A.; et al. Insights into RNA unwinding and ATP hydrolysis by the flavivirus NS3 protein. EMBO J. 2008, 27, 3209–3219. [Google Scholar] [CrossRef] [Green Version]
- Tian, H.; Ji, X.; Yang, X.; Zhang, Z.; Lu, Z.; Yang, K.; Chen, C.; Zhao, Q.; Chi, H.; Mu, Z.; et al. Structural basis of Zika virus helicase in recognizing its substrates. Protein Cell 2016, 7, 562–570. [Google Scholar] [CrossRef] [Green Version]
- Swarbrick, C.M.D.; Basavannacharya, C.; Chan, K.W.K.; Chan, S.A.; Singh, D.; Wei, N.; Phoo, W.W.; Luo, D.; Lescar, J.; Vasudevan, S.G. NS3 helicase from dengue virus specifically recognizes viral RNA sequence to ensure optimal replication. Nucleic Acids Res. 2017, 45, 12904–12920. [Google Scholar] [CrossRef] [Green Version]
- Luo, D.; Wei, N.; Doan, D.N.; Paradkar, P.N.; Chong, Y.; Davidson, A.D.; Kotaka, M.; Lescar, J.; Vasudevan, S.G. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J. Biol. Chem. 2010, 285, 18817–18827. [Google Scholar] [CrossRef] [Green Version]
- Assenberg, R.; Mastrangelo, E.; Walter, T.S.; Verma, A.; Milani, M.; Owens, R.J.; Stuart, D.I.; Grimes, J.M.; Mancini, E.J. Crystal structure of a novel conformational state of the flavivirus NS3 protein: Implications for polyprotein processing and viral replication. J. Virol. 2009, 83, 12895–12906. [Google Scholar] [CrossRef] [Green Version]
- Chernov, A.V.; Shiryaev, S.A.; Aleshin, A.E.; Ratnikov, B.I.; Smith, J.W.; Liddington, R.C.; Strongin, A.Y. The two-component NS2B-NS3 proteinase represses DNA unwinding activity of the West Nile virus NS3 helicase. J. Biol. Chem. 2008, 283, 17270–17278. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Ci, Y.; Wang, L.; Yang, Y.; Zhang, L.; Xu, C.; Qin, C.; Shi, L. Zika virus NS3 is a canonical RNA helicase stimulated by NS5 RNA polymerase. Nucleic Acids Res. 2019, 47, 8693–8707. [Google Scholar] [CrossRef]
- Mastrangelo, E.; Milani, M.; Bollati, M.; Selisko, B.; Peyrane, F.; Pandini, V.; Sorrentino, G.; Canard, B.; Konarev, P.V.; Svergun, D.I.; et al. Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full length NS3 protein. J. Mol. Biol. 2007, 372, 444–455. [Google Scholar] [CrossRef] [Green Version]
- Lei, J.; Hansen, G.; Nitsche, C.; Klein, C.D.; Zhang, L.; Hilgenfeld, R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 2016, 353, 503–505. [Google Scholar] [CrossRef] [Green Version]
- Leon-Juarez, M.; Martinez-Castillo, M.; Shrivastava, G.; Garcia-Cordero, J.; Villegas-Sepulveda, N.; Mondragon-Castelan, M.; Mondragon-Flores, R.; Cedillo-Barron, L. Recombinant Dengue virus protein NS2B alters membrane permeability in different membrane models. Virol. J. 2016, 13, 1. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.A.; Razaz, M.; Hayward, S. The DynDom database of protein domain motions. Bioinformatics 2003, 19, 1290–1291. [Google Scholar] [CrossRef] [Green Version]
- Phoo, W.W.; El Sahili, A.; Zhang, Z.; Chen, M.W.; Liew, C.W.; Lescar, J.; Vasudevan, S.G.; Luo, D. Crystal structures of full length DENV4 NS2B-NS3 reveal the dynamic interaction between NS2B and NS3. Antivir. Res. 2020, 182, 104900. [Google Scholar] [CrossRef]
- Li, Y.; Lee, M.Y.; Loh, Y.R.; Kang, C. Secondary structure and membrane topology of dengue virus NS4A protein in micelles. Biochim. Biophys. Acta Biomembr. 2018, 1860, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Stern, O.; Hung, Y.F.; Valdau, O.; Yaffe, Y.; Harris, E.; Hoffmann, S.; Willbold, D.; Sklan, E.H. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J. Virol. 2013, 87, 4080–4085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.M.; Xie, X.; Zou, J.; Li, S.H.; Lee, M.Y.; Dong, H.; Qin, C.F.; Kang, C.; Shi, P.Y. Determinants of Dengue Virus NS4A Protein Oligomerization. J. Virol. 2015, 89, 6171–6183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiryaev, S.A.; Chernov, A.V.; Aleshin, A.E.; Shiryaeva, T.N.; Strongin, A.Y. NS4A regulates the ATPase activity of the NS3 helicase: A novel cofactor role of the non-structural protein NS4A from West Nile virus. J. Gen. Virol. 2009, 90, 2081–2085. [Google Scholar] [CrossRef]
- Li, Y.; Wong, Y.L.; Lee, M.Y.; Li, Q.; Wang, Q.Y.; Lescar, J.; Shi, P.Y.; Kang, C. Secondary Structure and Membrane Topology of the Full-Length Dengue Virus NS4B in Micelles. Angew. Chem. Int. Ed. Engl. 2016, 55, 12068–12072. [Google Scholar] [CrossRef]
- Miller, S.; Sparacio, S.; Bartenschlager, R. Subcellular localization and membrane topology of the Dengue virus type 2 Non-structural protein 4B. J. Biol. Chem. 2006, 281, 8854–8863. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kim, Y.M.; Zou, J.; Wang, Q.Y.; Gayen, S.; Wong, Y.L.; Lee, L.T.; Xie, X.; Huang, Q.; Lescar, J.; et al. Secondary structure and membrane topology of dengue virus NS4B N-terminal 125 amino acids. Biochim. Biophys. Acta 2015, 1848, 3150–3157. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Xie, X.; Lee, L.T.; Chandrasekaran, R.; Reynaud, A.; Yap, L.; Wang, Q.Y.; Dong, H.; Kang, C.; Yuan, Z.; et al. Dimerization of flavivirus NS4B protein. J. Virol. 2014, 88, 3379–3391. [Google Scholar] [CrossRef] [Green Version]
- Ngo, A.M.; Shurtleff, M.J.; Popova, K.D.; Kulsuptrakul, J.; Weissman, J.S.; Puschnik, A.S. The ER membrane protein complex is required to ensure correct topology and stable expression of flavivirus polyproteins. Elife 2019, 8. [Google Scholar] [CrossRef]
- Lin, D.L.; Inoue, T.; Chen, Y.J.; Chang, A.; Tsai, B.; Tai, A.W. The ER Membrane Protein Complex Promotes Biogenesis of Dengue and Zika Virus Non-structural Multi-pass Transmembrane Proteins to Support Infection. Cell Rep. 2019, 27, 1666–1674 e1664. [Google Scholar] [CrossRef] [Green Version]
- Barrows, N.J.; Anglero-Rodriguez, Y.; Kim, B.; Jamison, S.F.; Le Sommer, C.; McGee, C.E.; Pearson, J.L.; Dimopoulos, G.; Ascano, M.; Bradrick, S.S.; et al. Dual roles for the ER membrane protein complex in flavivirus infection: Viral entry and protein biogenesis. Sci. Rep. 2019, 9, 9711. [Google Scholar] [CrossRef] [Green Version]
- Umareddy, I.; Chao, A.; Sampath, A.; Gu, F.; Vasudevan, S.G. Dengue virus NS4B interacts with NS3 and dissociates it from single-stranded RNA. J. Gen. Virol. 2006, 87, 2605–2614. [Google Scholar] [CrossRef]
- Dong, H.; Fink, K.; Zust, R.; Lim, S.P.; Qin, C.F.; Shi, P.Y. Flavivirus RNA methylation. J. Gen. Virol. 2014, 95, 763–778. [Google Scholar] [CrossRef] [Green Version]
- Issur, M.; Geiss, B.J.; Bougie, I.; Picard-Jean, F.; Despins, S.; Mayette, J.; Hobdey, S.E.; Bisaillon, M. The flavivirus NS5 protein is a true RNA guanylyltransferase that catalyzes a two-step reaction to form the RNA cap structure. RNA 2009, 15, 2340–2350. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Ray, D.; Zhao, Y.; Dong, H.; Ren, S.; Li, Z.; Guo, Y.; Bernard, K.A.; Shi, P.Y.; Li, H. Structure and function of flavivirus NS5 methyltransferase. J. Virol. 2007, 81, 3891–3903. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Ren, S.; Zhang, B.; Zhou, Y.; Puig-Basagoiti, F.; Li, H.; Shi, P.Y. West Nile virus methyltransferase catalyzes two methylations of the viral RNA cap through a substrate-repositioning mechanism. J. Virol. 2008, 82, 4295–4307. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Soh, T.S.; Lim, S.P.; Chung, K.Y.; Swaminathan, K.; Vasudevan, S.G.; Shi, P.Y.; Lescar, J.; Luo, D. Molecular basis for specific viral RNA recognition and 2′-O-ribose methylation by the dengue virus nonstructural protein 5 (NS5). Proc. Natl. Acad. Sci. USA 2015, 112, 14834–14839. [Google Scholar] [CrossRef] [Green Version]
- Coloma, J.; Jain, R.; Rajashankar, K.R.; Garcia-Sastre, A.; Aggarwal, A.K. Structures of NS5 Methyltransferase from Zika Virus. Cell Rep. 2016, 16, 3097–3102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Sahili, A.; Lescar, J. Dengue Virus Non-Structural Protein 5. Viruses 2017, 9, 91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yap, T.L.; Xu, T.; Chen, Y.L.; Malet, H.; Egloff, M.P.; Canard, B.; Vasudevan, S.G.; Lescar, J. Crystal structure of the dengue virus RNA-dependent RNA polymerase catalytic domain at 1.85-angstrom resolution. J. Virol. 2007, 81, 4753–4765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Soh, T.S.; Zheng, J.; Chan, K.W.; Phoo, W.W.; Lee, C.C.; Tay, M.Y.; Swaminathan, K.; Cornvik, T.C.; Lim, S.P.; et al. A crystal structure of the Dengue virus NS5 protein reveals a novel inter-domain interface essential for protein flexibility and virus replication. PLoS Pathog. 2015, 11, e1004682. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Tan, X.F.; Thurmond, S.; Zhang, Z.M.; Lin, A.; Hai, R.; Song, J. The structure of Zika virus NS5 reveals a conserved domain conformation. Nat. Commun. 2017, 8, 14763. [Google Scholar] [CrossRef]
- Zhao, B.; Yi, G.; Du, F.; Chuang, Y.C.; Vaughan, R.C.; Sankaran, B.; Kao, C.C.; Li, P. Structure and function of the Zika virus full-length NS5 protein. Nat. Commun. 2017, 8, 14762. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Ye, H.Q.; Zhang, Q.Y.; Lu, G.; Zhang, B.; Gong, P. A conformation-based intra-molecular initiation factor identified in the flavivirus RNA-dependent RNA polymerase. PLoS Pathog. 2020, 16, e1008484. [Google Scholar] [CrossRef]
- Lu, G.; Gong, P. Crystal Structure of the full-length Japanese encephalitis virus NS5 reveals a conserved methyltransferase-polymerase interface. PLoS Pathog. 2013, 9, e1003549. [Google Scholar] [CrossRef] [Green Version]
- Saw, W.G.; Tria, G.; Gruber, A.; Subramanian Manimekalai, M.S.; Zhao, Y.; Chandramohan, A.; Srinivasan Anand, G.; Matsui, T.; Weiss, T.M.; Vasudevan, S.G.; et al. Structural insight and flexible features of NS5 proteins from all four serotypes of Dengue virus in solution. Acta Cryst. D Biol. Cryst. 2015, 71, 2309–2327. [Google Scholar] [CrossRef] [Green Version]
- Dubankova, A.; Boura, E. Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses. Antivir. Res. 2019, 169, 104536. [Google Scholar] [CrossRef]
- Zhao, Y.; Soh, T.S.; Chan, K.W.; Fung, S.S.; Swaminathan, K.; Lim, S.P.; Shi, P.Y.; Huber, T.; Lescar, J.; Luo, D.; et al. Flexibility of NS5 Methyltransferase-Polymerase Linker Region Is Essential for Dengue Virus Replication. J. Virol. 2015, 89, 10717–10721. [Google Scholar] [CrossRef] [Green Version]
- Klema, V.J.; Ye, M.; Hindupur, A.; Teramoto, T.; Gottipati, K.; Padmanabhan, R.; Choi, K.H. Dengue Virus Nonstructural Protein 5 (NS5) Assembles into a Dimer with a Unique Methyltransferase and Polymerase Interface. PLoS Pathog. 2016, 12, e1005451. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.P.; Noble, C.G.; Seh, C.C.; Soh, T.S.; El Sahili, A.; Chan, G.K.; Lescar, J.; Arora, R.; Benson, T.; Nilar, S.; et al. Potent Allosteric Dengue Virus NS5 Polymerase Inhibitors: Mechanism of Action and Resistance Profiling. PLoS Pathog. 2016, 12, e1005737. [Google Scholar] [CrossRef] [Green Version]
- Upadhyay, A.K.; Cyr, M.; Longenecker, K.; Tripathi, R.; Sun, C.; Kempf, D.J. Crystal structure of full-length Zika virus NS5 protein reveals a conformation similar to Japanese encephalitis virus NS5. Acta Cryst. F Struct. Biol. Commun. 2017, 73, 116–122. [Google Scholar] [CrossRef] [Green Version]
- Ferrero, D.S.; Ruiz-Arroyo, V.M.; Soler, N.; Uson, I.; Guarne, A.; Verdaguer, N. Supramolecular arrangement of the full-length Zika virus NS5. PLoS Pathog. 2019, 15, e1007656. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.P.; Koh, J.H.; Seh, C.C.; Liew, C.W.; Davidson, A.D.; Chua, L.S.; Chandrasekaran, R.; Cornvik, T.C.; Shi, P.Y.; Lescar, J. A crystal structure of the dengue virus non-structural protein 5 (NS5) polymerase delineates interdomain amino acid residues that enhance its thermostability and de novo initiation activities. J. Biol. Chem. 2013, 288, 31105–31114. [Google Scholar] [CrossRef] [Green Version]
- Potisopon, S.; Priet, S.; Collet, A.; Decroly, E.; Canard, B.; Selisko, B. The methyltransferase domain of dengue virus protein NS5 ensures efficient RNA synthesis initiation and elongation by the polymerase domain. Nucleic Acids Res. 2014, 42, 11642–11656. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Lu, G.; Zhang, B.; Gong, P. Perturbation in the conserved methyltransferase-polymerase interface of flavivirus NS5 differentially affects polymerase initiation and elongation. J. Virol. 2015, 89, 249–261. [Google Scholar] [CrossRef] [Green Version]
- Rusanov, T.; Kent, T.; Saeed, M.; Hoang, T.M.; Thomas, C.; Rice, C.M.; Pomerantz, R.T. Identification of a Small Interface between the Methyltransferase and RNA Polymerase of NS5 that is Essential for Zika Virus Replication. Sci. Rep. 2018, 8, 17384. [Google Scholar] [CrossRef]
- Uchil, P.D.; Satchidanandam, V. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 2003, 278, 24388–24398. [Google Scholar] [CrossRef] [Green Version]
- Li, X.D.; Ye, H.Q.; Deng, C.L.; Liu, S.Q.; Zhang, H.L.; Shang, B.D.; Shi, P.Y.; Yuan, Z.M.; Zhang, B. Genetic interaction between NS4A and NS4B for replication of Japanese encephalitis virus. J. Gen. Virol. 2015, 96, 1264–1275. [Google Scholar] [CrossRef]
- Tajima, S.; Takasaki, T.; Kurane, I. Restoration of replication-defective dengue type 1 virus bearing mutations in the N-terminal cytoplasmic portion of NS4A by additional mutations in NS4B. Arch. Virol. 2011, 156, 63–69. [Google Scholar] [CrossRef]
- Chatel-Chaix, L.; Fischl, W.; Scaturro, P.; Cortese, M.; Kallis, S.; Bartenschlager, M.; Fischer, B.; Bartenschlager, R. A Combined Genetic-Proteomic Approach Identifies Residues within Dengue Virus NS4B Critical for Interaction with NS3 and Viral Replication. J. Virol. 2015, 89, 7170–7186. [Google Scholar] [CrossRef] [Green Version]
- Zou, J.; Lee, L.T.; Wang, Q.Y.; Xie, X.; Lu, S.; Yau, Y.H.; Yuan, Z.; Geifman Shochat, S.; Kang, C.; Lescar, J.; et al. Mapping the Interactions between the NS4B and NS3 proteins of dengue virus. J. Virol. 2015, 89, 3471–3483. [Google Scholar] [CrossRef] [Green Version]
- Yon, C.; Teramoto, T.; Mueller, N.; Phelan, J.; Ganesh, V.K.; Murthy, K.H.; Padmanabhan, R. Modulation of the nucleoside triphosphatase/RNA helicase and 5'-RNA triphosphatase activities of Dengue virus type 2 nonstructural protein 3 (NS3) by interaction with NS5, the RNA-dependent RNA polymerase. J. Biol. Chem. 2005, 280, 27412–27419. [Google Scholar] [CrossRef] [Green Version]
- Cui, T.; Sugrue, R.J.; Xu, Q.; Lee, A.K.; Chan, Y.C.; Fu, J. Recombinant dengue virus type 1 NS3 protein exhibits specific viral RNA binding and NTPase activity regulated by the NS5 protein. Virology 1998, 246, 409–417. [Google Scholar] [CrossRef] [Green Version]
- Zou, G.; Chen, Y.L.; Dong, H.; Lim, C.C.; Yap, L.J.; Yau, Y.H.; Shochat, S.G.; Lescar, J.; Shi, P.Y. Functional analysis of two cavities in flavivirus NS5 polymerase. J. Biol. Chem. 2011, 286, 14362–14372. [Google Scholar] [CrossRef] [Green Version]
- Tay, M.Y.; Saw, W.G.; Zhao, Y.; Chan, K.W.; Singh, D.; Chong, Y.; Forwood, J.K.; Ooi, E.E.; Gruber, G.; Lescar, J.; et al. The C-terminal 50 amino acid residues of dengue NS3 protein are important for NS3-NS5 interaction and viral replication. J. Biol. Chem. 2015, 290, 2379–2394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, J.; Xie, X.; Wang, Q.Y.; Dong, H.; Lee, M.Y.; Kang, C.; Yuan, Z.; Shi, P.Y. Characterization of dengue virus NS4A and NS4B protein interaction. J. Virol. 2015, 89, 3455–3470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.J.; Kuo, M.D.; Chien, L.J.; Hsu, S.L.; Wang, Y.M.; Lin, J.H. RNA-protein interactions: Involvement of NS3, NS5, and 3' noncoding regions of Japanese encephalitis virus genomic RNA. J. Virol. 1997, 71, 3466–3473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5' and 3' Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Gebhard, L.G.; Filomatori, C.V.; Gamarnik, A.V. Functional RNA Elements in the Dengue Virus Genome. Viruses 2011, 3, 1739–1756. [Google Scholar] [CrossRef]
- Iglesias, N.G.; Gamarnik, A.V. Dynamic RNA structures in the dengue virus genome. RNA Biol. 2011, 8, 249–257. [Google Scholar] [CrossRef] [Green Version]
- Mazeaud, C.; Freppel, W.; Chatel-Chaix, L. The Multiples Fates of the Flavivirus RNA Genome During Pathogenesis. Front. Genet. 2018, 9, 595. [Google Scholar] [CrossRef]
- Khromykh, A.A.; Kondratieva, N.; Sgro, J.Y.; Palmenberg, A.; Westaway, E.G. Significance in replication of the terminal nucleotides of the flavivirus genome. J. Virol. 2003, 77, 10623–10629. [Google Scholar] [CrossRef] [Green Version]
- Markoff, L. 5′- and 3′-noncoding regions in flavivirus RNA. Adv. Virus Res. 2003, 59, 177–228. [Google Scholar]
- Selisko, B.; Potisopon, S.; Agred, R.; Priet, S.; Varlet, I.; Thillier, Y.; Sallamand, C.; Debart, F.; Vasseur, J.J.; Canard, B. Molecular basis for nucleotide conservation at the ends of the dengue virus genome. PLoS Pathog. 2012, 8, e1002912. [Google Scholar] [CrossRef] [Green Version]
- Filomatori, C.V.; Iglesias, N.G.; Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. RNA Sequences and Structures Required for the Recruitment and Activity of the Dengue Virus Polymerase. J. Biol. Chem. 2011, 286, 6929–6939. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.Y.; Li, X.F.; Jiang, T.; Deng, Y.Q.; Ye, Q.; Zhao, H.; Yu, J.Y.; Qin, C.F. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 2016, 5. [Google Scholar] [CrossRef]
- Friebe, P.; Harris, E. Interplay of RNA elements in the dengue virus 5' and 3' ends required for viral RNA replication. J. Virol. 2010, 84, 6103–6118. [Google Scholar] [CrossRef] [Green Version]
- Clyde, K.; Harris, E. RNA secondary structure in the coding region of dengue virus type 2 directs translation start codon selection and is required for viral replication. J. Virol. 2006, 80, 2170–2182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clyde, K.; Barrera, J.; Harris, E. The capsid-coding region hairpin element (cHP) is a critical determinant of dengue virus and West Nile virus RNA synthesis. Virology 2008, 379, 314–323. [Google Scholar] [CrossRef] [Green Version]
- Akiyama, B.M.; Laurence, H.M.; Massey, A.R.; Costantino, D.A.; Xie, X.; Yang, Y.; Shi, P.Y.; Nix, J.C.; Beckham, J.D.; Kieft, J.S. Zika virus produces noncoding RNAs using a multi-pseudoknot structure that confounds a cellular exonuclease. Science 2016, 354, 1148–1152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slonchak, A.; Khromykh, A.A. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antivir. Res. 2018, 159, 13–25. [Google Scholar] [CrossRef] [Green Version]
- Manzano, M.; Reichert, E.D.; Polo, S.; Falgout, B.; Kasprzak, W.; Shapiro, B.A.; Padmanabhan, R. Identification of cis-acting elements in the 3′-untranslated region of the dengue virus type 2 RNA that modulate translation and replication. J. Biol. Chem. 2011, 286, 22521–22534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villordo, S.M.; Alvarez, D.E.; Gamarnik, A.V. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA 2010, 16, 2325–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villordo, S.M.; Gamarnik, A.V. Genome cyclization as strategy for flavivirus RNA replication. Virus Res. 2009, 139, 230–239. [Google Scholar] [CrossRef] [Green Version]
- De Borba, L.; Villordo, S.M.; Iglesias, N.G.; Filomatori, C.V.; Gebhard, L.G.; Gamarnik, A.V. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J. Virol. 2015, 89, 3430–3437. [Google Scholar] [CrossRef] [Green Version]
- Vashist, S.; Bhullar, D.; Vrati, S. La protein can simultaneously bind to both 3′- and 5′-noncoding regions of Japanese encephalitis virus genome. DNA Cell Biol. 2011, 30, 339–346. [Google Scholar] [CrossRef]
- Friedrich, S.; Engelmann, S.; Schmidt, T.; Szczepankiewicz, G.; Bergs, S.; Liebert, U.G.; Kummerer, B.M.; Golbik, R.P.; Behrens, S.E. The Host Factor AUF1 p45 Supports Flavivirus Propagation by Triggering the RNA Switch Required for Viral Genome Cyclization. J. Virol. 2018, 92. [Google Scholar] [CrossRef] [Green Version]
- Friedrich, S.; Schmidt, T.; Geissler, R.; Lilie, H.; Chabierski, S.; Ulbert, S.; Liebert, U.G.; Golbik, R.P.; Behrens, S.E. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J. Virol. 2014, 88, 11586–11599. [Google Scholar] [CrossRef] [Green Version]
- De Nova-Ocampo, M.; Villegas-Sepulveda, N.; del Angel, R.M. Translation elongation factor-1alpha, La, and PTB interact with the 3’ untranslated region of dengue 4 virus RNA. Virology 2002, 295, 337–347. [Google Scholar] [CrossRef] [Green Version]
- Sanford, T.J.; Mears, H.V.; Fajardo, T.; Locker, N.; Sweeney, T.R. Circularization of flavivirus genomic RNA inhibits de novo translation initiation. Nucleic Acids Res. 2019, 47, 9789–9802. [Google Scholar] [CrossRef] [Green Version]
- Huber, R.G.; Lim, X.N.; Ng, W.C.; Sim, A.Y.L.; Poh, H.X.; Shen, Y.; Lim, S.Y.; Sundstrom, K.B.; Sun, X.; Aw, J.G.; et al. Structure mapping of dengue and Zika viruses reveals functional long-range interactions. Nat. Commun. 2019, 10, 1408. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wei, Y.; Mei, M.; Tang, L.; Sun, L.; Huang, W.; Zhou, J.; Zou, C.; Zhang, S.; Qin, C.F.; et al. Integrative Analysis of Zika Virus Genome RNA Structure Reveals Critical Determinants of Viral Infectivity. Cell Host Microbe 2018, 24, 875–886. [Google Scholar] [CrossRef] [Green Version]
- Dethoff, E.A.; Boerneke, M.A.; Gokhale, N.S.; Muhire, B.M.; Martin, D.P.; Sacco, M.T.; McFadden, M.J.; Weinstein, J.B.; Messer, W.B.; Horner, S.M.; et al. Pervasive tertiary structure in the dengue virus RNA genome. Proc. Natl. Acad. Sci. USA 2018, 115, 11513–11518. [Google Scholar] [CrossRef] [Green Version]
- Teo, C.S.; Chu, J.J. Cellular vimentin regulates construction of dengue virus replication complexes through interaction with NS4A protein. J. Virol. 2014, 88, 1897–1913. [Google Scholar] [CrossRef] [Green Version]
- Unchwaniwala, N.; Zhan, H.; Pennington, J.; Horswill, M.; den Boon, J.A.; Ahlquist, P. Subdomain cryo-EM structure of nodaviral replication protein A crown complex provides mechanistic insights into RNA genome replication. Proc. Natl. Acad. Sci. USA 2020, 117, 18680–18691. [Google Scholar] [CrossRef]
- Yan, L.; Ge, J.; Zheng, L.; Zhang, Y.; Gao, Y.; Wang, T.; Huang, Y.; Yang, Y.; Gao, S.; Li, M.; et al. Cryo-EM Structure of an Extended SARS-CoV-2 Replication and Transcription Complex Reveals an Intermediate State in Cap Synthesis. Cell 2021, 184, 184–193 e110. [Google Scholar] [CrossRef]
- Yan, L.; Zhang, Y.; Ge, J.; Zheng, L.; Gao, Y.; Wang, T.; Jia, Z.; Wang, H.; Huang, Y.; Li, M.; et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat. Commun. 2020, 11, 5874. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Malone, B.; Llewellyn, E.; Grasso, M.; Shelton, P.M.M.; Olinares, P.D.B.; Maruthi, K.; Eng, E.T.; Vatandaslar, H.; Chait, B.T.; et al. Structural Basis for Helicase-Polymerase Coupling in the SARS-CoV-2 Replication-Transcription Complex. Cell 2020, 182, 1560–1573 e1513. [Google Scholar] [CrossRef] [PubMed]
- Peng, Q.; Peng, R.; Yuan, B.; Zhao, J.; Wang, M.; Wang, X.; Wang, Q.; Sun, Y.; Fan, Z.; Qi, J.; et al. Structural and Biochemical Characterization of the nsp12-nsp7-nsp8 Core Polymerase Complex from SARS-CoV-2. Cell Rep. 2020, 31, 107774. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wu, J.; Wang, H.; Gao, Y.; Liu, Q.; Mu, A.; Ji, W.; Yan, L.; Zhu, Y.; Zhu, C.; et al. Structural Basis for RNA Replication by the SARS-CoV-2 Polymerase. Cell 2020, 182, 417–428 e413. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van den Elsen, K.; Quek, J.P.; Luo, D. Molecular Insights into the Flavivirus Replication Complex. Viruses 2021, 13, 956. https://doi.org/10.3390/v13060956
van den Elsen K, Quek JP, Luo D. Molecular Insights into the Flavivirus Replication Complex. Viruses. 2021; 13(6):956. https://doi.org/10.3390/v13060956
Chicago/Turabian Stylevan den Elsen, Kaïn, Jun Ping Quek, and Dahai Luo. 2021. "Molecular Insights into the Flavivirus Replication Complex" Viruses 13, no. 6: 956. https://doi.org/10.3390/v13060956
APA Stylevan den Elsen, K., Quek, J. P., & Luo, D. (2021). Molecular Insights into the Flavivirus Replication Complex. Viruses, 13(6), 956. https://doi.org/10.3390/v13060956







