Bacteriophages M13 and T4 Increase the Expression of Anchorage-Dependent Survival Pathway Genes and Down Regulate Androgen Receptor Expression in LNCaP Prostate Cell Line
Abstract
:1. Introduction
2. Materials and Methods
2.1. LNCaP Cell Culture
2.2. LNCaP Cell Exposure to Bacteriophages M13 and T4
2.3. MTT Reduction Assay
2.4. Hematoxylin and Eosin Staining
2.5. RNA Extraction and cDNA Synthesis for qPCR Studies
2.6. Statistical Analysis
3. Results
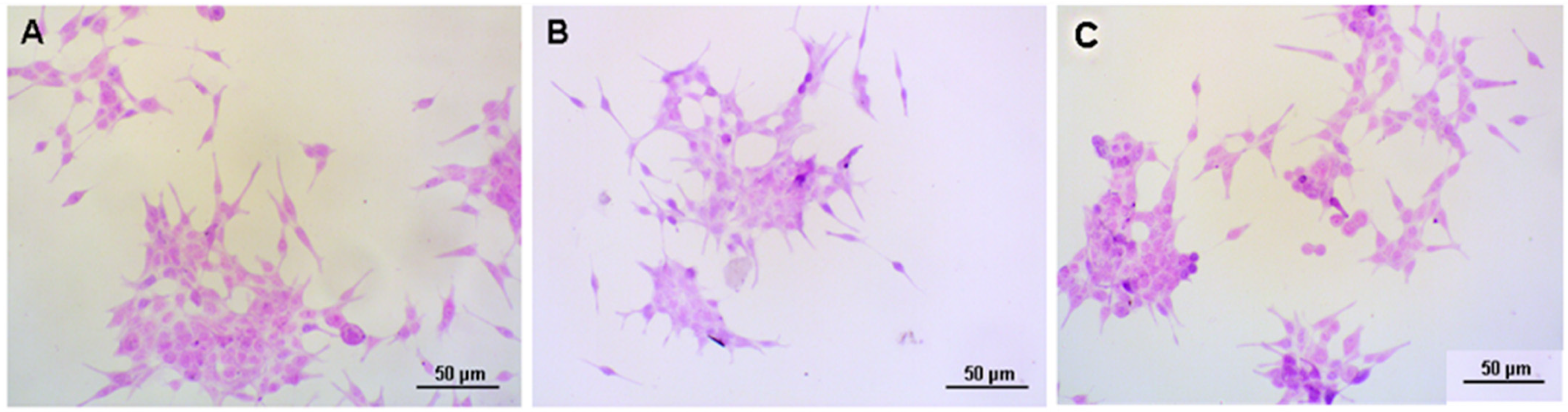
3.1. Cell Morphology
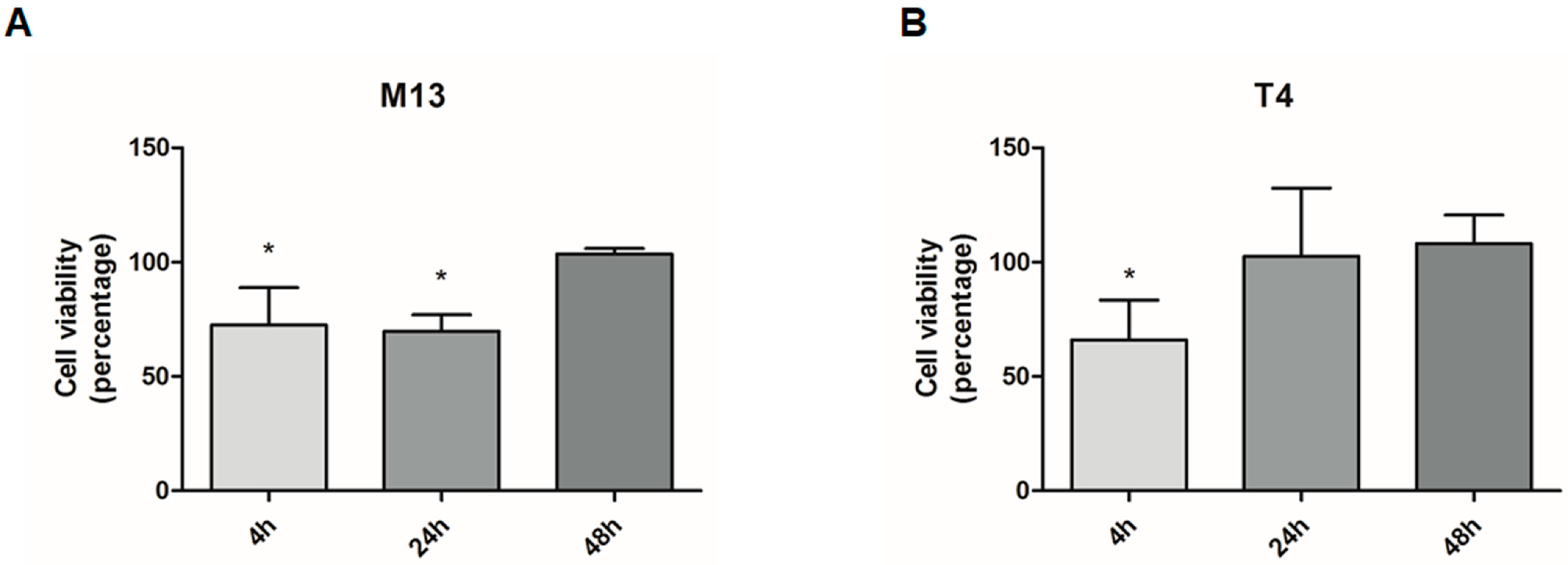
3.2. MTT Reduction-Cell Viability
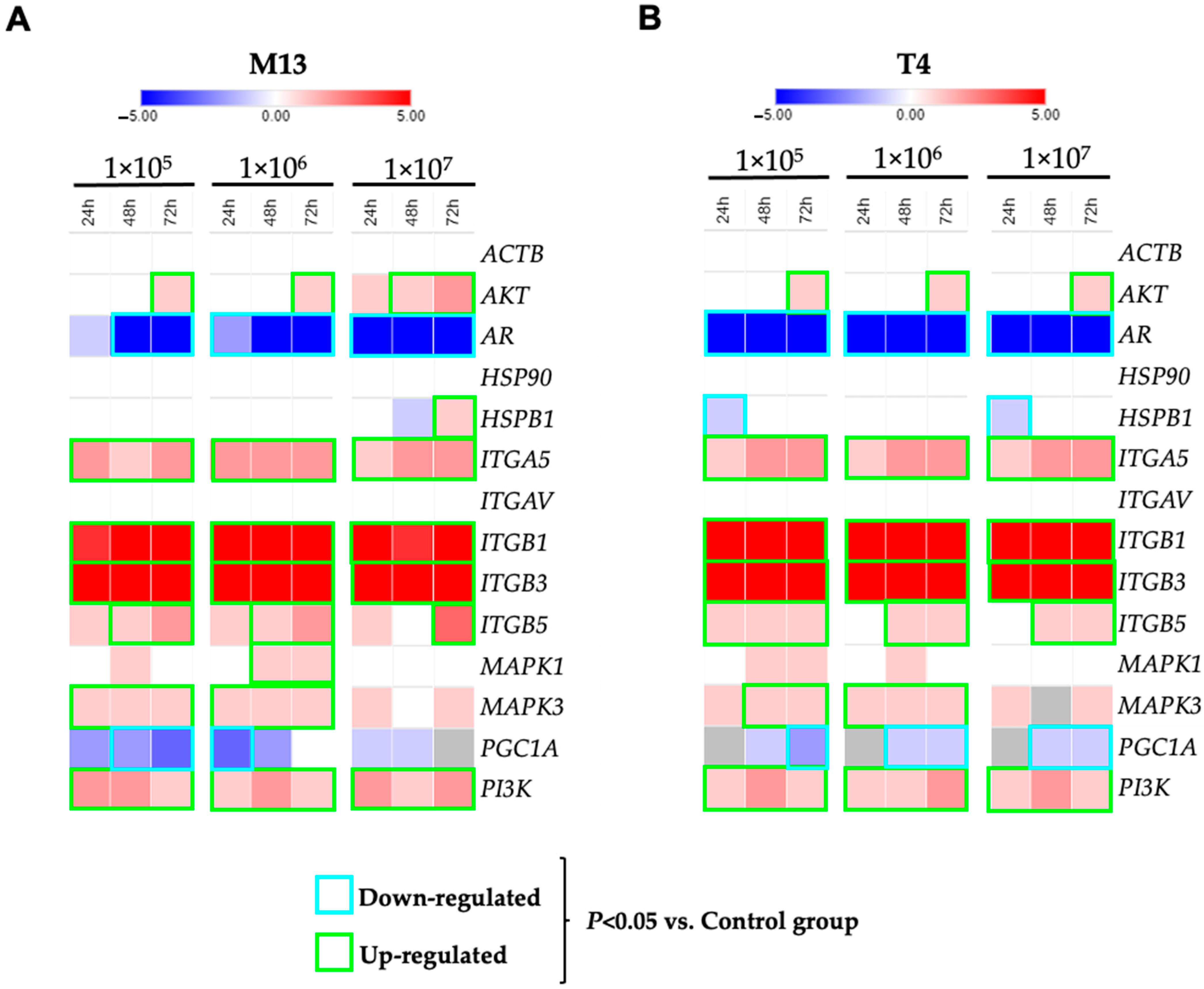
3.3. Gene Expression Profiles after Exposure to Bacteriophages M13 and T4
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sakr, W.A.; Grignon, D.J.; Crissman, J.D.; Heilbrun, L.K.; Cassin, B.J.; Pontes, J.J.; Haas, G.P. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: An autopsy study of 249 cases. In Vivo 1994, 8, 439–443. [Google Scholar] [PubMed]
- Nelson, W.G.; De Marzo, A.M.; Isaacs, W.B. Prostate cancer. N. Engl. J. Med. 2003, 349, 366–381. [Google Scholar] [CrossRef]
- Nuhn, P.; De Bono, J.S.; Fizazi, K.; Freedland, S.J.; Grilli, M.; Kantoff, P.W.; Sonpavde, G.; Sternberg, C.N.; Yegnasubramanian, S.; Antonarakis, E.S. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-resistant Prostate Cancer in the Era of Precision Oncology. Eur. Urol. 2019, 75, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Sumanasuriya, S.; De Bono, J. Treatment of Advanced Prostate Cancer-A Review of Current Therapies and Future Promise. Cold Spring Harb. Perspect. Med. 2018, 8, a030635. [Google Scholar] [CrossRef] [PubMed]
- Barquilha, C.N.; Santos, N.J.; Monção, C.C.D.; Barbosa, I.C.; Lima, F.O.; Justulin, L.A.; Pértega-Gomes, N.; Felisbino, S.L. Sulfiredoxin as a Potential Therapeutic Target for Advanced and Metastatic Prostate Cancer. Oxidative Med. Cell. Longev. 2020, 2020, 2148562. [Google Scholar] [CrossRef]
- Dabrowska, K.; Opolski, A.; Wietrzyk, J.; Switala-Jelen, K.; Godlewska, J.; Boratynski, J.; Syper, D.; Weber-Dabrowska, B.; Gorski, A. Anticancer activity of bacteriophage T4 and its mutant HAP1 in mouse experimental tumour models. Anticancer Res. 2004, 24, 3991–3995. [Google Scholar]
- Dabrowska, K.; Skaradziński, G.; Jończyk, P.; Kurzepa, A.; Wietrzyk, J.; Owczarek, B.; Zaczek, M.; Switała-Jeleń, K.; Boratyński, J.; Poźniak, G.; et al. The effect of bacteriophages T4 and HAP1 on in vitro melanoma migration. BMC Microbiol. 2009, 9, 13. [Google Scholar] [CrossRef] [Green Version]
- DePorter, S.M.; McNaughton, B.R. Engineered M13 bacteriophage nanocarriers for intracellular delivery of exogenous proteins to human prostate cancer cells. Bioconjug. Chem. 2014, 25, 1620–1625. [Google Scholar] [CrossRef]
- Gogokhia, L.; Buhrke, K.; Bell, R.; Hoffman, B.; Brown, D.G.; Hanke-Gogokhia, C.; Ajami, N.J.; Wong, M.C.; Ghazaryan, A.; Valentine, J.F.; et al. Expansion of Bacteriophages Is Linked to Aggravated Intestinal Inflammation and Colitis. Cell Host Microbe 2019, 25, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Kolesanova, E.F.; Melnikova, M.V.; Bolshakova, T.N.; Rybalkina, E.Y.; Sivov, I.G. Bacteriophage MS2 As a Tool for Targeted Delivery in Solid Tumor Chemotherapy. Acta Nat. 2019, 11, 98–101. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Cleto, S.; Sillankorva, S.; Azeredo, J.; Lu, T.K. Genetically Engineered Phages: A Review of Advances over the Last Decade. Microbiol. Mol. Biol. Rev. 2016, 80, 523–543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoo, S.Y.; Jin, H.E.; Choi, D.S.; Kobayashi, M.; Farouz, Y.; Wang, S.; Lee, S.W. M13 Bacteriophage and Adeno-Associated Virus Hybrid for Novel Tissue Engineering Material with Gene Delivery Functions. Adv. Healthc. Mater. 2016, 5, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Mirshekari, H.; Moosavi Basri, S.M.; Bahrami, S.; Moghoofei, M.; Hamblin, M.R. Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 2016, 106, 45–62. [Google Scholar] [CrossRef] [Green Version]
- Bodner, K.; Melkonian, A.L.; Covert, M.W. The Enemy of My Enemy: New Insights Regarding Bacteriophage-Mammalian Cell Interactions. Trends Microbiol. 2021, 29, 528–541. [Google Scholar] [CrossRef]
- Bichet, M.C.; Chin, W.H.; Richards, W.; Lin, Y.W.; Avellaneda-Franco, L.; Hernandez, C.A.; Oddo, A.; Chernyavskiy, O.; Hilsenstein, V.; Neild, A.; et al. Bacteriophage uptake by mammalian cell layers represents a potential sink that may impact phage therapy. iScience 2021, 24, 102287. [Google Scholar] [CrossRef]
- Møller-Olsen, C.; Ho, S.F.S.; Shukla, R.D.; Feher, T.; Sagona, A.P. Engineered K1F bacteriophages kill intracellular Escherichia coli K1 in human epithelial cells. Sci. Rep. 2018, 8, 17559. [Google Scholar] [CrossRef] [Green Version]
- Van Belleghem, J.D.; Dąbrowska, K.; Vaneechoutte, M.; Barr, J.J.; Bollyky, P.L. Interactions between Bacteriophage, Bacteria, and the Mammalian Immune System. Viruses 2018, 11, 10. [Google Scholar] [CrossRef] [Green Version]
- Hodyra-Stefaniak, K.; Miernikiewicz, P.; Drapała, J.; Drab, M.; Jończyk-Matysiak, E.; Lecion, D.; Kaźmierczak, Z.; Beta, W.; Majewska, J.; Harhala, M.; et al. Mammalian Host-Versus-Phage immune response determines phage fate in vivo. Sci. Rep. 2015, 5, 14802. [Google Scholar] [CrossRef] [Green Version]
- Merril, C.R.; Geier, M.R.; Petricciani, J.C. Bacterial virus gene expression in human cells. Nature 1971, 233, 398–400. [Google Scholar] [CrossRef]
- Geier, M.R.; Merril, C.R. Lambda phage transcription in human fibroblasts. Virology 1972, 47, 638–643. [Google Scholar] [CrossRef]
- Wenger, S.L.; Steele, M.W.; Turner, J.H. Incorporation of bacteriophage DNA into the genome of cultured human lymphocytes. In Vitro 1981, 17, 695–700. [Google Scholar] [CrossRef]
- Nguyen, S.; Baker, K.; Padman, B.S.; Patwa, R.; Dunstan, R.A.; Weston, T.A.; Schlosser, K.; Bailey, B.; Lithgow, T.; Lazarou, M.; et al. Bacteriophage Transcytosis Provides a Mechanism To Cross Epithelial Cell Layers. mBio 2017, 8, e01874-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Wu, M.; Liu, X.; Liu, Z.; Zhou, Q.; Niu, Z.; Huang, Y. Probing the endocytic pathways of the filamentous bacteriophage in live cells using ratiometric pH fluorescent indicator. Adv. Healthc. Mater. 2015, 4, 413–419. [Google Scholar] [CrossRef]
- Ivanenkov, V.; Felici, F.; Menon, A.G. Uptake and intracellular fate of phage display vectors in mammalian cells. Biochim. Biophys. Acta 1999, 1448, 450–462. [Google Scholar] [CrossRef] [Green Version]
- Øie, C.I.; Wolfson, D.L.; Yasunori, T.; Dumitriu, G.; Sørensen, K.K.; McCourt, P.A.; Ahluwalia, B.S.; Smedsrød, B. Liver sinusoidal endothelial cells contribute to the uptake and degradation of entero bacterial viruses. Sci. Rep. 2020, 10, 898. [Google Scholar] [CrossRef]
- Dedrick, R.M.; Guerrero-Bustamante, C.A.; Garlena, R.A.; Russell, D.A.; Ford, K.; Harris, K.; Gilmour, K.C.; Soothill, J.; Jacobs-Sera, D.; Schooley, R.T.; et al. Engineered bacteriophages for treatment of a patient with a disseminated drug-resistant Mycobacterium abscessus. Nat. Med. 2019, 25, 730–733. [Google Scholar] [CrossRef]
- Bachrach, G.; Leizerovici-Zigmond, M.; Zlotkin, A.; Naor, R.; Steinberg, D. Bacteriophage isolation from human saliva. Lett. Appl. Microbiol. 2003, 36, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Gorski, A.; Dabrowska, K.; Switala-Jeleń, K.; Nowaczyk, M.; Weber-Dabrowska, B.; Boratynski, J.; Wietrzyk, J.; Opolski, A. New insights into the possible role of bacteriophages in host defense and disease. Med. Immunol. 2003, 2, 2. [Google Scholar] [CrossRef] [Green Version]
- Huh, H.; Wong, S.; St Jean, J.; Slavcev, R. Bacteriophage interactions with mammalian tissue: Therapeutic applications. Adv. Drug Deliv. Rev. 2019, 145, 4–17. [Google Scholar] [CrossRef]
- Dabrowska, K.; Opolski, A.; Wietrzyk, J.; Switala-Jelen, K.; Boratynski, J.; Nasulewicz, A.; Lipinska, L.; Chybicka, A.; Kujawa, M.; Zabel, M.; et al. Antitumor activity of bacteriophages in murine experimental cancer models caused possibly by inhibition of beta3 integrin signaling pathway. Acta Virol. 2004, 48, 241–248. [Google Scholar] [PubMed]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Berridge, M.V.; Tan, A.S. Characterization of the cellular reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT): Subcellular localization, substrate dependence, and involvement of mitochondrial electron transport in MTT reduction. Arch. Biochem. Biophys. 1993, 303, 474–482. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E.; Pierschbacher, M.D. New perspectives in cell adhesion: RGD and integrins. Science 1987, 238, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Pasqualini, R.; Koivunen, E.; Ruoslahti, E. A peptide isolated from phage display libraries is a structural and functional mimic of an RGD-binding site on integrins. J. Cell Biol. 1995, 130, 1189–1196. [Google Scholar] [CrossRef] [Green Version]
- Pasqualini, R.; Bourdoulous, S.; Koivunen, E.; Woods, V.L.; Ruoslahti, E. A polymeric form of fibronectin has antimetastatic effects against multiple tumor types. Nat. Med. 1996, 2, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Ruoslahti, E. Integrin signaling and matrix assembly. Tumor Biol. 1996, 17, 117–124. [Google Scholar] [CrossRef]
- Rivinoja, A.; Laakkonen, P. Identification of homing peptides using the in vivo phage display technology. Methods Mol. Biol. 2011, 683, 401–415. [Google Scholar] [CrossRef]
- Dąbrowska, K.; Miernikiewicz, P.; Piotrowicz, A.; Hodyra, K.; Owczarek, B.; Lecion, D.; Kaźmierczak, Z.; Letarov, A.; Górski, A. Immunogenicity studies of proteins forming the T4 phage head surface. J. Virol. 2014, 88, 12551–12557. [Google Scholar] [CrossRef] [Green Version]
- Streuli, C.H. Integrins as architects of cell behavior. Mol. Biol. Cell 2016, 27, 2885–2888. [Google Scholar] [CrossRef] [PubMed]
- Lehti, T.A.; Pajunen, M.I.; Skog, M.S.; Finne, J. Internalization of a polysialic acid-binding Escherichia coli bacteriophage into eukaryotic neuroblastoma cells. Nat. Commun. 2017, 8, 1915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaźmierczak, Z.; Majewska, J.; Milczarek, M.; Owczarek, B.; Dąbrowska, K. Circulation of Fluorescently Labelled Phage in a Murine Model. Viruses 2021, 13, 297. [Google Scholar] [CrossRef]
- Chen, H.; Zhou, L.; Wu, X.; Li, R.; Wen, J.; Sha, J.; Wen, X. The PI3K/AKT pathway in the pathogenesis of prostate cancer. Front. Biosci. 2016, 21, 1084–1091. [Google Scholar] [CrossRef] [Green Version]
- Noorolyai, S.; Shajari, N.; Baghbani, E.; Sadreddini, S.; Baradaran, B. The relation between PI3K/AKT signalling pathway and cancer. Gene 2019, 698, 120–128. [Google Scholar] [CrossRef]
- Jurmeister, S.; Ramos-Montoya, A.; Sandi, C.; Pertega-Gomes, N.; Wadhwa, K.; Lamb, A.D.; Dunning, M.J.; Attig, J.; Carroll, J.S.; Fryer, L.G.; et al. Identification of potential therapeutic targets in prostate cancer through a cross-species approach. EMBO Mol. Med. 2018, 3, e8274. [Google Scholar] [CrossRef] [PubMed]
- Barra, F.; Evangelisti, G.; Ferro Desideri, L.; Di Domenico, S.; Ferraioli, D.; Vellone, V.G.; De Cian, F.; Ferrero, S. Investigational PI3K/AKT/mTOR inhibitors in development for endometrial cancer. Expert Opin. Investig. Drugs 2019, 28, 131–142. [Google Scholar] [CrossRef] [PubMed]
- du Rusquec, P.; Blonz, C.; Frenel, J.S.; Campone, M. Targeting the PI3K/Akt/mTOR pathway in estrogen-receptor positive HER2 negative advanced breast cancer. Adv. Med. Oncol. 2020, 12. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, O.; Brunner, S.F.; Yaeger, R.; Ly, P.; Nechemia-Arbely, Y.; Kim, D.H.; Fang, R.; Castillon, G.A.; Yu, M.; Li, J.S.Z.; et al. Publisher Correction: Chromothripsis drives the evolution of gene amplification in cancer. Nature 2021, 591, E19. [Google Scholar] [CrossRef]
- Choi, S.K.; Kam, H.; Kim, K.Y.; Park, S.I.; Lee, Y.S. Targeting Heat Shock Protein 27 in Cancer: A Druggable Target for Cancer Treatment? Cancers 2019, 11, 1195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cayado-Gutiérrez, N.; Moncalero, V.L.; Rosales, E.M.; Berón, W.; Salvatierra, E.E.; Alvarez-Olmedo, D.; Radrizzani, M.; Ciocca, D.R. Downregulation of Hsp27 (HSPB1) in MCF-7 human breast cancer cells induces upregulation of PTEN. Cell Stress Chaperones 2013, 18, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, J.C.; Tuukkanen, A.; Schroeder, M.; Fahrig, T.; Fahrig, R. RP101 (brivudine) binds to heat shock protein HSP27 (HSPB1) and enhances survival in animals and pancreatic cancer patients. J. Cancer Res. Clin. Oncol. 2011, 137, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- Torrano, V.; Valcarcel-Jimenez, L.; Cortazar, A.R.; Liu, X.; Urosevic, J.; Castillo-Martin, M.; Fernández-Ruiz, S.; Morciano, G.; Caro-Maldonado, A.; Guiu, M.; et al. The metabolic co-regulator PGC1α suppresses prostate cancer metastasis. Nat. Cell Biol. 2016, 18, 645–656. [Google Scholar] [CrossRef] [Green Version]
- Tennakoon, J.B.; Shi, Y.; Han, J.J.; Tsouko, E.; White, M.A.; Burns, A.R.; Zhang, A.; Xia, X.; Ilkayeva, O.R.; Xin, L.; et al. Androgens regulate prostate cancer cell growth via an AMPK-PGC-1α-mediated metabolic switch. Oncogene 2014, 33, 5251–5261. [Google Scholar] [CrossRef] [Green Version]
- Sanmukh, S.G.; Felisbino, S.L. Development of pipette tip gap closure migration assay (s-ARU method) for studying semi-adherent cell lines. Cytotechnology 2018, 70, 1685–1695. [Google Scholar] [CrossRef] [Green Version]
- Coumar, M.S.; Tsai, F.Y.; Kanwar, J.R.; Sarvagalla, S.; Cheung, C.H. Treat cancers by targeting survivin: Just a dream or future reality? Cancer Treat. Rev. 2013, 39, 802–811. [Google Scholar] [CrossRef]
- Solit, D.B.; Scher, H.I.; Rosen, N. Hsp90 as a therapeutic target in prostate cancer. Semin. Oncol. 2003, 30, 709–716. [Google Scholar] [CrossRef]
- Centenera, M.M.; Carter, S.L.; Gillis, J.L.; Marrocco-Tallarigo, D.L.; Grose, R.H.; Tilley, W.D.; Butler, L.M. Co-targeting AR and HSP90 suppresses prostate cancer cell growth and prevents resistance mechanisms. Endocr. Relat. Cancer 2015, 22, 805–818. [Google Scholar] [CrossRef] [Green Version]
- Arrigo, A.P.; Gibert, B. HspB1, HspB5 and HspB4 in Human Cancers: Potent Oncogenic Role of Some of Their Client Proteins. Cancers 2014, 6, 333–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Genes | Primer Sense | Primer Anti-Sense |
|---|---|---|
| ACTB | GATTCCTATGTGGGCGACGA | TGTAGAAGGTGTGGTGCCAG |
| AKT | CATCGCTTCTTTGCCGGTATC | ACTCCATGCTGTCATCTTGGTC |
| AR | GACATGCGTTTGGAGACTGC | CAATCATTTCTGCTGGCGCA |
| GAPDH | GAATGGGCAGCCGTTAGGAA | ATCACCCGGAGGAGAAATCG |
| HSP90 | AGGGGGAAAGGGGAGTATCT | ATGTCAACCCTTGGAGCAGC |
| HSPB1 | CGCGGAAATACACGCTGCC | GACTCGAAGGTGACTGGGATG |
| ITGA5 | GGGTGGTGCTGTCTACCTC | GTGGAGCGCATGCCAAGATG |
| ITGAV | AGGCACCCTCCTTCTGATCC | CTTGGCATAATCTCTATTGCCTGT |
| ITGB1 | GCCAAATGGGACACGCAAGA | GTGTTGTGGGATTTGCACGG |
| ITGB3 | CTGCCGTGACGAGATTGAGT | CCTTGGGACACTCTGGCTCT |
| ITGB5 | GGGCTCTACTCAGTGGTTTCG | GGCTTCCGAAGTCCTCTTTG |
| MAPK1 | TCAGCTAACGTTCTGCACCG | ACTTGGTGTAGCCCTTGGA |
| MAPK3 | ATCTTCCAGGAGACAGCACG | TTCTAACAGTCTGGCGGGAG |
| PGC1A | GAAGGGTACTTTTCTGCCCCT | CTTCTTCCAGCCTTGGGGAG |
| PI3K | AGAGCCCCGAGCGTTT | TCGTGGAGGCATTGTTCTGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanmukh, S.G.; dos Santos, N.J.; Barquilha, C.N.; Cucielo, M.S.; de Carvalho, M.; dos Reis, P.P.; Delella, F.K.; Carvalho, H.F.; Felisbino, S.L. Bacteriophages M13 and T4 Increase the Expression of Anchorage-Dependent Survival Pathway Genes and Down Regulate Androgen Receptor Expression in LNCaP Prostate Cell Line. Viruses 2021, 13, 1754. https://doi.org/10.3390/v13091754
Sanmukh SG, dos Santos NJ, Barquilha CN, Cucielo MS, de Carvalho M, dos Reis PP, Delella FK, Carvalho HF, Felisbino SL. Bacteriophages M13 and T4 Increase the Expression of Anchorage-Dependent Survival Pathway Genes and Down Regulate Androgen Receptor Expression in LNCaP Prostate Cell Line. Viruses. 2021; 13(9):1754. https://doi.org/10.3390/v13091754
Chicago/Turabian StyleSanmukh, Swapnil Ganesh, Nilton José dos Santos, Caroline Nascimento Barquilha, Maira Smaniotto Cucielo, Márcio de Carvalho, Patricia Pintor dos Reis, Flávia Karina Delella, Hernandes F. Carvalho, and Sérgio Luis Felisbino. 2021. "Bacteriophages M13 and T4 Increase the Expression of Anchorage-Dependent Survival Pathway Genes and Down Regulate Androgen Receptor Expression in LNCaP Prostate Cell Line" Viruses 13, no. 9: 1754. https://doi.org/10.3390/v13091754
APA StyleSanmukh, S. G., dos Santos, N. J., Barquilha, C. N., Cucielo, M. S., de Carvalho, M., dos Reis, P. P., Delella, F. K., Carvalho, H. F., & Felisbino, S. L. (2021). Bacteriophages M13 and T4 Increase the Expression of Anchorage-Dependent Survival Pathway Genes and Down Regulate Androgen Receptor Expression in LNCaP Prostate Cell Line. Viruses, 13(9), 1754. https://doi.org/10.3390/v13091754










