Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions
Abstract
:1. Introduction
1.1. Native Prions and Chaperone Proteins
1.2. Amino Acid Composition of Prion-Forming Domains
1.3. Synthetic Polyglutamine Models of Prion Propagation and Human Diseases
2. Materials and Methods
2.1. Yeast Strains and Plasmids
2.2. SDD-AGE, SDS-PAGE, and Immunoblot Analyses
2.3. SIS1 Repression and Plasmid-Shuffling
3. Results
3.1. Confirming PolyQX Aggregation and Hsp104-Dependent Propagation
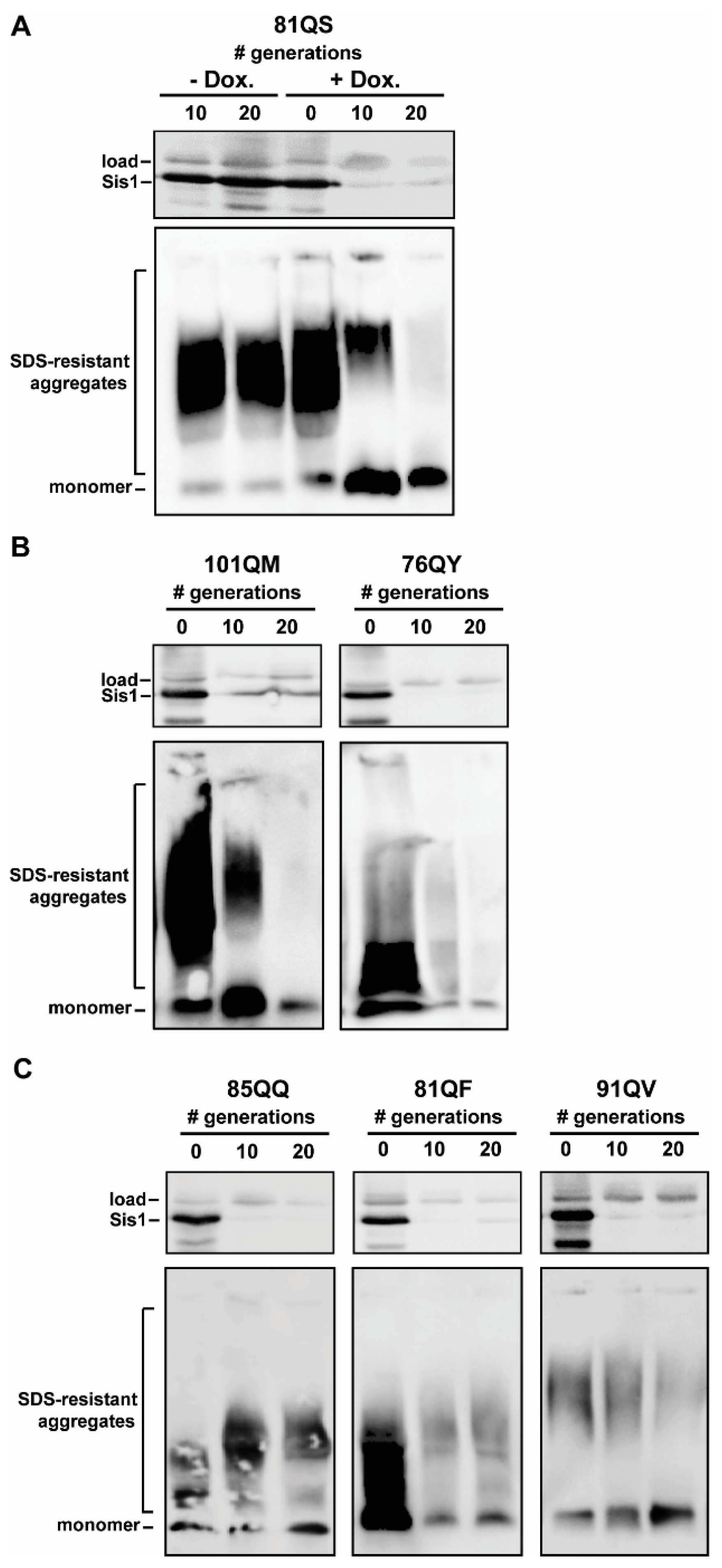
3.2. Assaying for Sis1-Depdendence
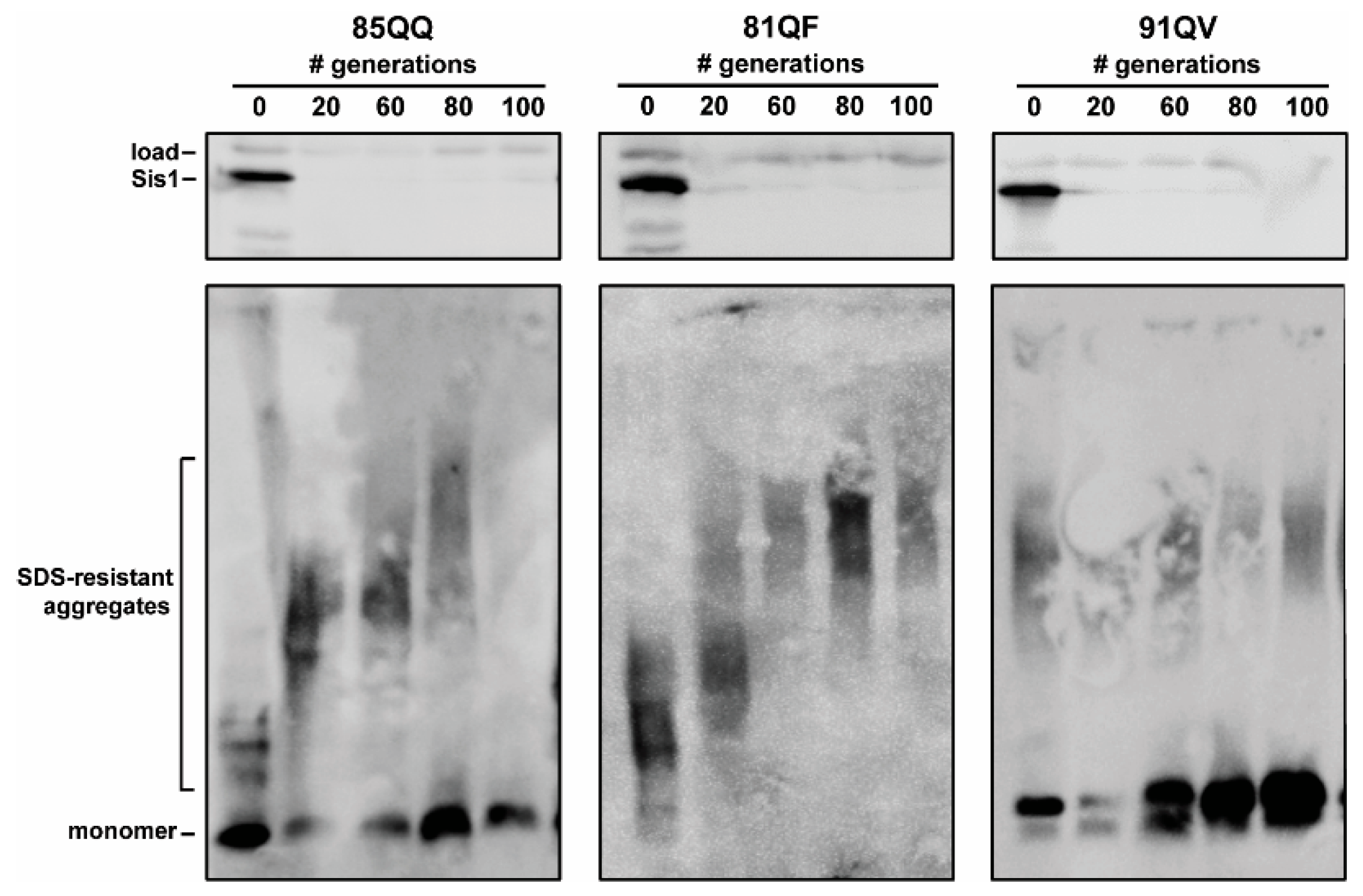
3.3. Pilot Work on Sis1-Domain Dependence and Secondary JDP Requirements
4. Discussion
4.1. Impact of SIS1 Repression
4.2. Sis1 Domain Requirements
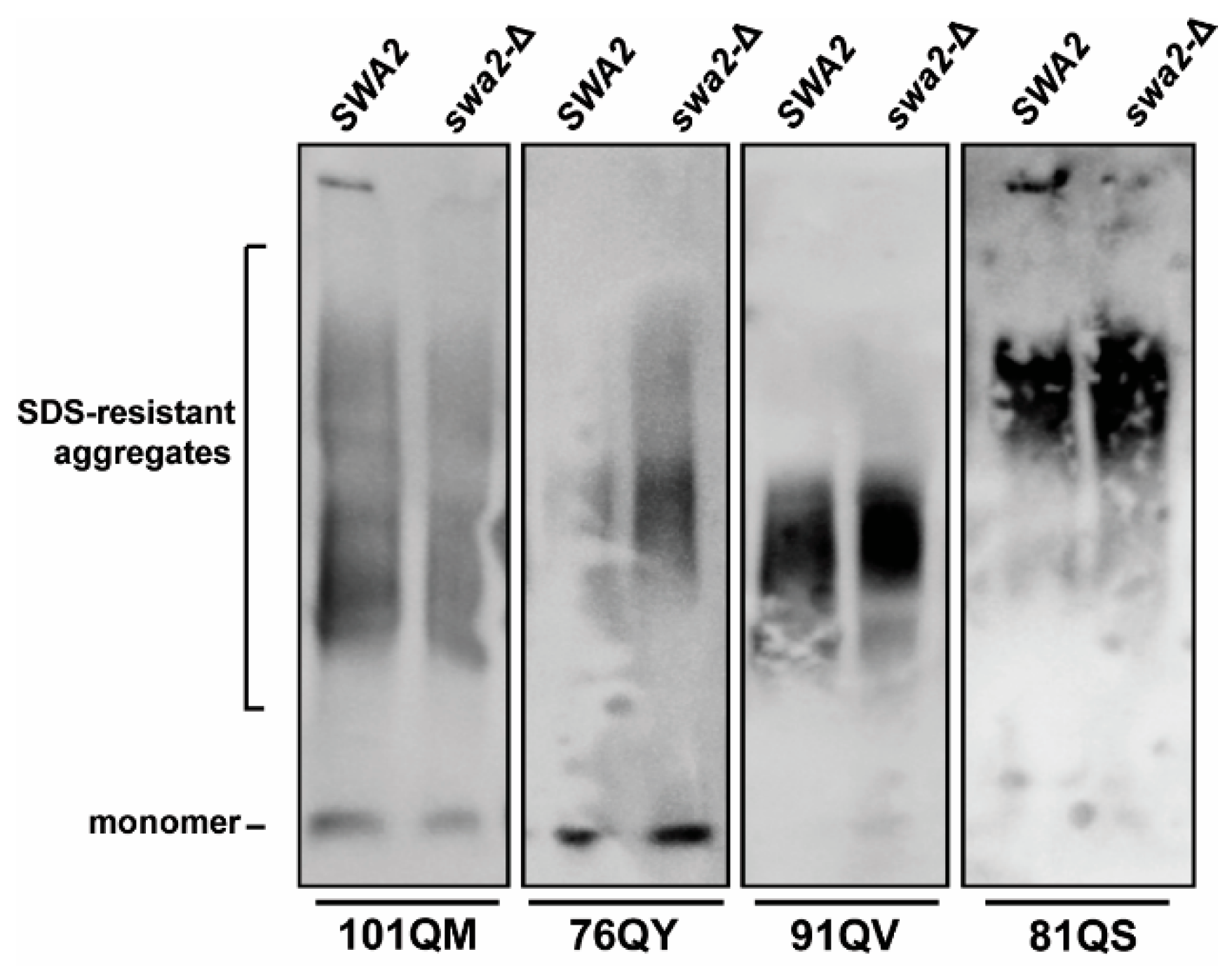
4.3. Secondary JDP Requirements
4.4. Amino Acid Composition, Current Limitations, and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prusiner, S.B. Biology and Genetics of Prions Causing Neurodegeneration. Annu. Rev. Genet. 2013, 47, 601–623. [Google Scholar] [CrossRef]
- Wickner, R.B. [URE3] as an Altered URE2 Protein: Evidence for a Prion Analog in Saccharomyces cerevisiae. Science 1994, 264, 566–569. [Google Scholar] [CrossRef] [PubMed]
- Wickner, R.B.; Masison, D.C.; Edskes, H.K. [PSI] and [URE3] as yeast prions. Yeast 1995, 11, 1671–1685. [Google Scholar] [CrossRef]
- Derkatch, I.L.; O Chernoff, Y.; Kushnirov, V.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics 1996, 144, 1375–1386. [Google Scholar] [CrossRef]
- Derkatch, I.L.; E Bradley, M.; Hong, J.Y.; Liebman, S.W. Prions Affect the Appearance of Other Prions: The Story of [PIN+]. Cell 2001, 106, 171–182. [Google Scholar] [CrossRef]
- Alberti, S.; Halfmann, R.; King, O.; Kapila, A.; Lindquist, S. A Systematic Survey Identifies Prions and Illuminates Sequence Features of Prionogenic Proteins. Cell 2009, 137, 146–158. [Google Scholar] [CrossRef]
- Du, Z.; Park, K.-W.; Yu, H.; Fan, Q.; Li, L. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat. Genet. 2008, 40, 460–465. [Google Scholar] [CrossRef]
- Liebman, S.W.; Chernoff, Y.O. Prions in yeast. Genetics 2012, 191, 1041–1072. [Google Scholar] [CrossRef]
- Verma, A.K.; Diwan, D.; Raut, S.; Dobriyal, N.; Brown, R.E.; Gowda, V.; Hines, J.K.; Sahi, C. Evolutionary Conservation and Emerging Functional Diversity of the Cytosolic Hsp70:J Protein Chaperone Network of Arabidopsis thaliana. G3 Genes Genomes Genet. 2017, 7, 1941–1954. [Google Scholar] [CrossRef]
- Verma, A.K.; Tamadaddi, C.; Tak, Y.; Lal, S.S.; Cole, S.J.; Hines, J.K.; Sahi, C. The expanding world of plant J-domain proteins. Crit. Rev. Plant Sci. 2019, 38, 382–400. [Google Scholar] [CrossRef]
- Stein, K.C.; Bengoechea, R.; Harms, M.B.; Weihl, C.C.; True, H.L. Myopathy-causing Mutations in an HSP40 Chaperone Disrupt Processing of Specific Client Conformers. J. Biol. Chem. 2014, 289, 21120–21130. [Google Scholar] [CrossRef]
- Lopez, N.; Aron, R.; Craig, E.A. Specificity of class II Hsp40 Sis1 in maintenance of yeast prion [RNQ+]. Mol. Biol. Cell 2003, 14, 1172–1181. [Google Scholar] [CrossRef]
- Reidy, M.; Masison, D.C. Yeast prions help identify and define chaperone interaction networks. Curr. Pharm. Biotechnol. 2014, 15, 1008–1018. [Google Scholar] [CrossRef]
- Reidy, M.; Sharma, R.; Roberts, B.-L.; Masison, D.C. Human J-protein DnaJB6b Cures a Subset of Saccharomyces cerevisiae Prions and Selectively Blocks Assembly of Structurally Related Amyloids. J. Biol. Chem. 2016, 291, 4035–4047. [Google Scholar] [CrossRef]
- Sporn, Z.A.; Hines, J.K. Hsp40 function in yeast prion propagation: Amyloid diversity necessitates chaperone functional complexity. Prion 2015, 9, 80–89. [Google Scholar] [CrossRef]
- Kampinga, H.H.; Andreasson, C.; Barducci, A.; Cheetham, M.E.; Cyr, D.; Emanuelsson, C.; Genevaux, P.; Gestwicki, J.E.; Goloubinoff, P.; Huerta-Cepas, J.; et al. Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 2018, 24, 7–15. [Google Scholar] [CrossRef]
- Tuite, M.F.; Mundy, C.R.; Cox, B.S. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics 1981, 98, 691–711. [Google Scholar] [CrossRef]
- Grimminger, V.; Richter, K.; Imhof, A.; Buchner, J.; Walter, S. The Prion Curing Agent Guanidinium Chloride Specifically Inhibits ATP Hydrolysis by Hsp104. J. Biol. Chem. 2004, 279, 7378–7383. [Google Scholar] [CrossRef]
- Wickner, R.B.; Edskes, H.K.; Shewmaker, F.; Nakayashiki, T. Prions of fungi: Inherited structures and biological roles. Nat. Rev. Genet. 2007, 5, 611–618. [Google Scholar] [CrossRef]
- Winkler, J.; Tyedmers, J.; Bukau, B.; Mogk, A. Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation. J. Cell Biol. 2012, 198, 387–404. [Google Scholar] [CrossRef] [Green Version]
- Satpute-Krishnan, P.; Langseth, S.X.; Serio, T.R. Hsp104-Dependent Remodeling of Prion Complexes Mediates Protein-Only Inheritance. PLoS Biol. 2007, 5, e24. [Google Scholar] [CrossRef]
- Killian, A.N.; Hines, J.K. Chaperone functional specificity promotes yeast prion diversity. PLoS Pathog. 2018, 14, e1006695. [Google Scholar] [CrossRef]
- Sahi, C.; Craig, E.A. Network of general and specialty J protein chaperones of the yeast cytosol. Proc. Natl. Acad. Sci. USA 2007, 104, 7163–7168. [Google Scholar] [CrossRef]
- Higurashi, T.; Hines, J.K.; Sahi, C.; Aron, R.; Craig, E.A. Specificity of the J-protein Sis1 in the propagation of 3 yeast prions. Proc. Natl. Acad. Sci. USA 2008, 105, 16596–16601. [Google Scholar] [CrossRef]
- Hines, J.K.; Li, X.; Du, Z.; Higurashi, T.; Li, L.; Craig, E.A. [SWI], the prion formed by the chromatin remodeling factor Swi1, is highly sensitive to alterations in Hsp70 chaperone system activity. PLoS Genet. 2011, 7, e1001309. [Google Scholar] [CrossRef]
- Sondheimer, N.; Lopez, N.; Craig, E.A.; Lindquist, S. The role of Sis1 in the maintenance of the [RNQ+] prion. EMBO J. 2001, 20, 2435–2442. [Google Scholar] [CrossRef]
- Troisi, E.M.; Rockman, M.E.; Nguyen, P.P.; Oliver, E.E.; Hines, J.K. Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1]. Mol. Microbiol. 2015, 97, 926–941. [Google Scholar] [CrossRef]
- Harris, J.M.; Nguyen, P.P.; Patel, M.J.; Sporn, Z.A.; Hines, J.K. Functional diversification of hsp40: Distinct j-protein functional requirements for two prions allow for chaper-one-dependent prion selection. PLoS Genet. 2014, 10, e1004510. [Google Scholar] [CrossRef]
- Stein, K.C.; True, H.L. Extensive diversity of prion strains is defined by differential chaperone interactions and distinct amyloi-dogenic regions. PLoS Genet. 2014, 10, e1004337. [Google Scholar] [CrossRef]
- Killian, A.N.; Miller, S.C.; Hines, J.K. Impact of Amyloid Polymorphism on Prion-Chaperone Interactions in Yeast. Viruses 2019, 11, 349. [Google Scholar] [CrossRef] [Green Version]
- Taylor, K.L.; Cheng, N.; Williams, R.W.; Steven, A.C.; Wickner, R.B. Prion Domain Initiation of Amyloid Formation in Vitro from Native Ure2p. Science 1999, 283, 1339–1343. [Google Scholar] [CrossRef]
- Glover, J.R.; Kowal, A.S.; Schirmer, E.C.; Patino, M.M.; Liu, J.-J.; Lindquist, S. Self-Seeded Fibers Formed by Sup35, the Protein Determinant of [PSI+], a Heritable Prion-like Factor of S. cerevisiae. Cell 1997, 89, 811–819. [Google Scholar] [CrossRef]
- Vitrenko, Y.A.; Pavon, M.E.; Stone, S.I.; Liebman, S.W. Propagation of the [PIN+] prion by fragments of Rnq1 fused to GFP. Curr. Genet. 2007, 51, 309–319. [Google Scholar] [CrossRef]
- Du, Z.; Crow, E.T.; Kang, H.S.; Li, L. Distinct Subregions of Swi1 Manifest Striking Differences in Prion Transmission and SWI/SNF Function. Mol. Cell. Biol. 2010, 30, 4644–4655. [Google Scholar] [CrossRef]
- Ross, E.D.; Baxa, U.; Wickner, R.B. Scrambled Prion Domains Form Prions and Amyloid. Mol. Cell. Biol. 2004, 24, 7206–7213. [Google Scholar] [CrossRef]
- Ross, E.D.; Edskes, H.K.; Terry, M.J.; Wickner, R.B. Primary sequence independence for prion formation. Proc. Natl. Acad. Sci. USA 2005, 102, 12825–12830. [Google Scholar] [CrossRef]
- Ross, E.D.; Toombs, J.A. The effects of amino acid composition on yeast prion formation and prion domain interactions. Prion 2010, 4, 60–65. [Google Scholar] [CrossRef]
- Cascarina, S.M.; Paul, K.R.; Machihara, S.; Ross, E.D. Sequence features governing aggregation or degradation of prion-like proteins. PLoS Genet. 2018, 14, e1007517. [Google Scholar] [CrossRef]
- Toombs, J.; McCarty, B.R.; Ross, E.D. Compositional Determinants of Prion Formation in Yeast. Mol. Cell. Biol. 2010, 30, 319–332. [Google Scholar] [CrossRef]
- Alexandrov, I.M.; Vishnevskaya, A.B.; Ter-Avanesyan, M.D.; Kushnirov, V.V. Appearance and Propagation of Polyglutamine-based Amyloids in Yeast: TYROSINE RESIDUES ENABLE POLYMER FRAGMENTATION. J. Biol. Chem. 2008, 283, 15185–15192. [Google Scholar] [CrossRef] [Green Version]
- Alexandrov, A.I.; Polyanskaya, A.B.; Serpionov, G.V.; Ter-Avanesyan, M.D.; Kushnirov, V.V. The Effects of Amino Acid Composition of Glutamine-Rich Domains on Amyloid Formation and Fragmentation. PLoS ONE 2012, 7, e46458. [Google Scholar] [CrossRef]
- MacLea, K.S.; Paul, K.R.; Ben-Musa, Z.; Waechter, A.; Shattuck, J.E.; Gruca, M.; Ross, E.D. Distinct Amino Acid Compositional Requirements for Formation and Maintenance of the [PSI+] Prion in Yeast. Mol. Cell. Biol. 2015, 35, 899–911. [Google Scholar] [CrossRef]
- Hines, J.K.; Craig, E.A. The sensitive [SWI(+)] prion: New perspectives on yeast prion diversity. Prion 2011, 5, 164–168. [Google Scholar] [CrossRef]
- Takahashi, T.; Katada, S.; Onodera, O. Polyglutamine Diseases: Where does Toxicity Come from? What is Toxicity? Where are We Going? J. Mol. Cell Biol. 2010, 2, 180–191. [Google Scholar] [CrossRef]
- Krobitsch, S.; Lindquist, S. Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 1589–1594. [Google Scholar] [CrossRef]
- DePace, A.H.; Santoso, A.; Hillner, P.; Weissman, J.S. A Critical Role for Amino-Terminal Glutamine/Asparagine Repeats in the Formation and Propagation of a Yeast Prion. Cell 1998, 93, 1241–1252. [Google Scholar] [CrossRef]
- Osherovich, L.Z.; Cox, B.S.; Tuite, M.F.; Weissman, J.S. Dissection and Design of Yeast Prions. PLoS Biol. 2004, 2, e86. [Google Scholar] [CrossRef]
- Yan, W.; Craig, E.A. The Glycine-Phenylalanine-Rich Region Determines the Specificity of the Yeast Hsp40 Sis1. Mol. Cell. Biol. 1999, 19, 7751–7758. [Google Scholar] [CrossRef]
- Aron, R.; Higurashi, T.; Sahi, C.; Craig, E.A. J-protein co-chaperone Sis1 required for generation of [RNQ+] seeds necessary for prion propagation. EMBO J. 2007, 26, 3794–3803. [Google Scholar] [CrossRef]
- Hines, J.K.; Higurashi, T.; Srinivasan, M.; Craig, E.A. Influence of prion variant and yeast strain variation on prion-molecular chaperone requirements. Prion 2011, 5, 238–244. [Google Scholar] [CrossRef] [Green Version]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Derkatch, I.L.; Bradley, M.E.; Masse, S.V.; Zadorsky, S.P.; Polozkov, G.V.; Inge-Vechtomov, S.G.; Liebman, S.W. Dependence and independence of [PSI(+)] and [PIN(+)]: A two-prion system in yeast? EMBO J. 2000, 19, 1942–1952. [Google Scholar] [CrossRef] [PubMed]
- Derkatch, I.L.; Uptain, S.M.; Outeiro, T.F.; Krishnan, R.; Lindquist, S.L.; Liebman, S.W. Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc. Natl. Acad. Sci. USA 2004, 101, 12934–12939. [Google Scholar] [CrossRef]
- Yang, Z.; Hong, J.Y.; Derkatch, I.L.; Liebman, S.W. Heterologous Gln/Asn-Rich Proteins Impede the Propagation of Yeast Prions by Altering Chaperone Availability. PLoS Genet. 2013, 9, e1003236. [Google Scholar] [CrossRef] [PubMed]
- Kurahashi, H.; Ishiwata, M.; Shibata, S.; Nakamura, Y. A Regulatory Role of the Rnq1 Nonprion Domain for Prion Propagation and Polyglutamine Aggregates. Mol. Cell. Biol. 2008, 28, 3313–3323. [Google Scholar] [CrossRef]
- Meriin, A.B.; Zhang, X.; He, X.; Newnam, G.P.; Chernoff, Y.O.; Sherman, M.Y. Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J. Cell Biol. 2002, 157, 997–1004. [Google Scholar] [CrossRef]
- Osherovich, L.Z.; Weissman, J.S. Multiple Gln/Asn-Rich Prion Domains Confer Susceptibility to Induction of the Yeast [PSI] Prion. Cell 2001, 106, 183–194. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Boeke, J.D. In vitro mutagenesis and plasmid shuffling: From cloned gene to mutant yeast. Methods Enzymol. 1991, 194, 302–318. [Google Scholar] [CrossRef]
- Gall, W.E.; Higginbotham, M.A.; Chen, C.-Y.; Ingram, M.F.; Cyr, D.M.; Graham, T.R. The auxilin-like phosphoprotein Swa2p is required for clathrin function in yeast. Curr. Biol. 2000, 10, 1349–1358. [Google Scholar] [CrossRef]
- Xiao, J.; Kim, L.S.; Graham, T.R. Dissection of Swa2p/Auxilin Domain Requirements for Cochaperoning Hsp70 Clathrin-uncoating Activity In Vivo. Mol. Biol. Cell 2006, 17, 3281–3290. [Google Scholar] [CrossRef] [Green Version]
- Oliver, E.E.; Troisi, E.M.; Hines, J.K. Prion-specific Hsp40 function: The role of the auxilin homolog Swa2. Prion 2017, 11, 174–185. [Google Scholar] [CrossRef] [PubMed]
- Schilke, B.A.; Ciesielski, S.; Ziegelhoffer, T.; Kamiya, E.; Tonelli, M.; Lee, W.; Cornilescu, G.; Hines, J.K.; Markley, J.L.; Craig, E.A. Broadening the functionality of a J-protein/Hsp70 molecular chaperone system. PLoS Genet. 2017, 13, e1007084. [Google Scholar] [CrossRef] [PubMed]
- Stein, K.C.; True, H.L. Structural variants of yeast prions show conformer-specific requirements for chaperone activity. Mol. Microbiol. 2014, 93, 1156–1171. [Google Scholar] [CrossRef] [PubMed]
- Moriyama, H.; Edskes, H.K.; Wickner, R.B. [URE3] Prion Propagation in Saccharomyces cerevisiae: Requirement for Chaperone Hsp104 and Curing by Overexpressed Chaperone Ydj1p. Mol. Cell. Biol. 2000, 20, 8916–8922. [Google Scholar] [CrossRef]
- Berger, S.E.; Nolte, A.M.; Kamiya, E.; Hines, J.K. Three J-proteins impact Hsp104-mediated variant-specific prion elimination: A new critical role for a low-complexity domain. Curr. Genet. 2019, 66, 51–58. [Google Scholar] [CrossRef]
- Astor, M.T.; Kamiya, E.; Sporn, Z.A.; Berger, S.; Hines, J.K. Variant-specific and reciprocal Hsp40 functions in Hsp104-mediated prion elimination. Mol. Microbiol. 2018, 109, 41–62. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miller, S.C.; Wegrzynowicz, A.K.; Cole, S.J.; Hayward, R.E.; Ganser, S.J.; Hines, J.K. Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions. Viruses 2022, 14, 2160. https://doi.org/10.3390/v14102160
Miller SC, Wegrzynowicz AK, Cole SJ, Hayward RE, Ganser SJ, Hines JK. Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions. Viruses. 2022; 14(10):2160. https://doi.org/10.3390/v14102160
Chicago/Turabian StyleMiller, Sarah C., Andrea K. Wegrzynowicz, Sierra J. Cole, Rachel E. Hayward, Samantha J. Ganser, and Justin K. Hines. 2022. "Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions" Viruses 14, no. 10: 2160. https://doi.org/10.3390/v14102160
APA StyleMiller, S. C., Wegrzynowicz, A. K., Cole, S. J., Hayward, R. E., Ganser, S. J., & Hines, J. K. (2022). Hsp40/JDP Requirements for the Propagation of Synthetic Yeast Prions. Viruses, 14(10), 2160. https://doi.org/10.3390/v14102160






