Long-Term Wastewater Surveillance for SARS-CoV-2: One-Year Study in Brazil
Abstract
:1. Introduction
2. Methodology
2.1. Sampling
2.2. RNA Extraction and Quantitative PCR
2.3. Detection Limit and qPCR Inhibition Control
2.4. Data Analysis
3. Results and Discussion
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pacca, C.C.; Zini, N.; Versiani, A.F.; Edoardo, E.D.O.; Milhim, B.H.G.A.; Campos, G.R.F.; Moraes, M.M.; dos Santos, T.M.I.L.; Dourado, F.S.; Marques, B.C.; et al. Follow-up of a hospital cohort during the first 3530 suspected cases of COVID-19 in Sao Jose do Rio Preto, Sao Paulo, Brazil. bioRxiv 2021. [Google Scholar] [CrossRef]
- Banho, C.A.; Sacchetto, L.; Campos, G.R.F.; Bittar, C.; Possebon, F.S.; Ullmann, L.S.; Marques, B.D.C.; da Silva, G.C.D.; Moraes, M.M.; Parra, M.C.P.; et al. Impact of SARS-CoV-2 Gamma lineage introduction and COVID-19 vaccination on the epidemiological landscape of a Brazilian city. Commun. Med. 2022, 2, 1–11. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples From a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Wu, Y.; Guo, C.; Tang, L.; Hong, Z.; Zhou, J.; Dong, X.; Yin, H.; Xiao, Q.; Tang, Y.; Qu, X.; et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol. Hepatol. 2020, 5, 434–435. [Google Scholar] [CrossRef]
- Xu, Y.; Li, X.; Zhu, B.; Liang, H.; Fang, C.; Gong, Y.; Guo, Q.; Sun, X.; Zhao, D.; Shen, J.; et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat. Med. 2020, 26, 502–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.J.; Wang, Y.Y.; Ji, M.Y.; Pei, F.Y.; Zhao, Q.Q.; Zhou, Y.Y.; Hong, Y.T.; Han, S.Y.; Wang, J.; Wang, Q.X.; et al. Transmission Routes Analysis of SARS-CoV-2: A Systematic Review and Case Report. Front. Cell Dev. Biol. 2020, 8, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Weber, F.A.; Schiwy, S.; Linnemann, V.; Brinkmann, M.; Widera, M.; Greve, C.; Janke, A.; Hollert, H.; Wintgens, T.; et al. Detection of SARS-CoV-2 in raw and treated wastewater in Germany—Suitability for COVID-19 surveillance and potential transmission risks. Sci. Total Environ. 2021, 751, 141750. [Google Scholar] [CrossRef]
- Santiso-Bellon, C.; Randazzo, W.; Perez-Cataluna, A.; Vila-Vicent, S.; Gozalbo-Rovira, R.; Munoz, C.; Buesa, J.; Sanchez, G.; Rodriguez Diaz, J. Epidemiological Surveillance of Norovirus and Rotavirus in Sewage (2016–2017) in Valencia (Spain). Microorganisms 2020, 8, 458. [Google Scholar] [CrossRef] [Green Version]
- Cuevas-Ferrando, E.; Randazzo, W.; Perez-Cataluna, A.; Sanchez, G. HEV Occurrence in Waste and Drinking Water Treatment Plants. Front. Microbiol. 2019, 10, 2937. [Google Scholar] [CrossRef]
- Asghar, H.; Diop, O.M.; Weldegebriel, G.; Malik, F.; Shetty, S.; El Bassioni, L.; Akande, A.O.; Al Maamoun, E.; Zaidi, S.; Adeniji, A.J.; et al. Environmental surveillance for polioviruses in the Global Polio Eradication Initiative. J. Infect. Dis. 2014, 210 (Suppl. S1), S294–S303. [Google Scholar] [CrossRef] [PubMed]
- Zahedi, A.; Monis, P.; Deere, D.; Ryan, U. Wastewater-based epidemiology-surveillance and early detection of waterborne pathogens with a focus on SARS-CoV-2, Cryptosporidium and Giardia. Parasitol. Res. 2021, 120, 4167–4188. [Google Scholar] [CrossRef]
- Randazzo, W.; Truchado, P.; Cuevas-Ferrando, E.; Simon, P.; Allende, A.; Sanchez, G. SARS-CoV-2 RNA in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020, 181, 115942. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020, 728, 138764. [Google Scholar] [CrossRef] [PubMed]
- Coronado, Y.; Navarro, R.; Mosqueda, C.; Valenzuela, V.; Prez, J.P.; Gonzlez-Mendoza, V.; de la Torre, M.; Rocha, J. SARS-CoV-2 in wastewater from Mexico City used for irrigation in the Mezquital Valley: Quantification and modeling of geographic dispersion. Environ. Manag. 2021, 68, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Hara-Yamamura, H.; Meuchi, Y.; Imai, S.; Honda, R. Detection of SARS-CoV-2 in wastewater in Japan during a COVID-19 outbreak. Sci. Total Environ. 2021, 758, 143578. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Zhang, K.; Du, W.; Ali, W.; Feng, X.; Zhang, H. The potential of wastewater-based epidemiology as surveillance and early warning of infectious disease outbreaks. Curr. Opin. Environ. Sci. Health 2020, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Patel, A.K.; Shah, A.V.; Raval, J.; Rajpara, N.; Joshi, M.; Joshi, C.G. First proof of the capability of wastewater surveillance for COVID-19 in India through detection of genetic material of SARS-CoV-2. Sci. Total Environ. 2020, 746, 141326. [Google Scholar] [CrossRef] [PubMed]
- CDC. Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-Time RT-PCR Primers and Probes. 2020. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/virus-requests.html (accessed on 22 August 2020).
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamza, I.A.; Jurzik, L.; Stang, A.; Sure, K.; Uberla, K.; Wilhelm, M. Detection of human viruses in rivers of a densly-populated area in Germany using a virus adsorption elution method optimized for PCR analyses. Water Res. 2009, 43, 2657–2668. [Google Scholar] [CrossRef] [PubMed]
- Nagarkar, M.; Keely, S.P.; Jahne, M.; Wheaton, E.; Hart, C.; Smith, B.; Garland, J.; Varughese, E.A.; Braam, A.; Wiechman, B.; et al. SARS-CoV-2 monitoring at three sewersheds of different scales and complexity demonstrates distinctive relationships between wastewater measurements and COVID-19 case data. Sci. Total Environ. 2022, 816, 151534. [Google Scholar] [CrossRef] [PubMed]
- Lazuka, A.; Arnal, C.; Soyeux, E.; Sampson, M.; Lepeuple, A.S.; Deleuze, Y.; Pouradier Duteil, S.; Lacroix, S. COVID-19 wastewater based epidemiology: Long-term monitoring of 10 WWTP in France reveals the importance of the sampling context. Water Sci. Technol. 2021, 84, 1997–2013. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Xiao, A.; Zhang, J.; Moniz, K.; Endo, N.; Armas, F.; Bushman, M.; Chai, P.R.; Duvallet, C.; Erickson, T.B.; et al. Wastewater surveillance of SARS-CoV-2 across 40 U.S. states from February to June 2020. Water Res. 2021, 202, 117400. [Google Scholar] [CrossRef]
- Weidhaas, J.; Aanderud, Z.T.; Roper, D.K.; VanDerslice, J.; Gaddis, E.B.; Ostermiller, J.; Hoffman, K.; Jamal, R.; Heck, P.; Zhang, Y.; et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021, 775, 145790. [Google Scholar] [CrossRef]
- Melvin, R.G.; Chaudhry, N.; Georgewill, O.; Freese, R.; Simmons, G.E. Predictive power of SARS-CoV-2 wastewater surveillance for diverse populations across a large geographical range. medRxiv 2021. [Google Scholar] [CrossRef]
- Johnson, R.; Muller, C.J.F.; Ghoor, S.; Louw, J.; Archer, E.; Surujlal-Naicker, S.; Berkowitz, N.; Volschenk, M.; Brocker, L.H.L.; Wolfaardt, G.; et al. Qualitative and quantitative detection of SARS-CoV-2 RNA in untreated wastewater in Western Cape Province, South Africa. S. Afr. Med. J. 2021, 111, 198–202. [Google Scholar] [CrossRef]
- Kopperi, H.; Tharak, A.; Hemalatha, M.; Kiran, U.; Gokulan, C.G.; Mishra, R.K.; Mohan, S.V. Defining the methodological approach for wastewater-based epidemiological studies-Surveillance of SARS-CoV-2. Environ. Technol. Innov. 2021, 23, 101696. [Google Scholar] [CrossRef]
- Haramoto, E.; Malla, B.; Thakali, O.; Kitajima, M. First environmental surveillance for the presence of SARS-CoV-2 RNA in wastewater and river water in Japan. Sci. Total Environ. 2020, 737, 140405. [Google Scholar] [CrossRef]
- Hasan, S.W.; Ibrahim, Y.; Daou, M.; Kannout, H.; Jan, N.; Lopes, A.; Alsafar, H.; Yousef, A.F. Detection and quantification of SARS-CoV-2 RNA in wastewater and treated effluents: Surveillance of COVID-19 epidemic in the United Arab Emirates. Sci. Total Environ. 2021, 764, 142929. [Google Scholar] [CrossRef] [PubMed]
- Balboa, S.; Mauricio-Iglesias, M.; Rodriguez, S.; Martinez-Lamas, L.; Vasallo, F.J.; Regueiro, B.; Lema, J.M. The fate of SARS-COV-2 in WWTPS points out the sludge line as a suitable spot for detection of COVID-19. Sci. Total Environ. 2021, 772, 145268. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, W.; Bertsch, P.M.; Angel, N.; Bibby, K.; Bivins, A.; Dierens, L.; Edson, J.; Ehret, J.; Gyawali, P.; Hamilton, K.A.; et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: A surveillance tool for assessing the presence of COVID-19 infected travellers. J. Travel Med. 2020, 27, 1–11. [Google Scholar] [CrossRef]
- Cavany, S.; Bivins, A.; Wu, Z.; North, D.; Bibby, K.; Perkins, T.A. Inferring SARS-CoV-2 RNA shedding into wastewater relative to the time of infection. Epidemiol. Infect. 2022, 150, 1–23. [Google Scholar] [CrossRef]
- D’Aoust, P.M.; Mercier, E.; Montpetit, D.; Jia, J.J.; Alexandrov, I.; Neault, N.; Baig, A.T.; Mayne, J.; Zhang, X.; Alain, T.; et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021, 188, 116560. [Google Scholar] [CrossRef] [PubMed]
- Nemudryi, A.; Nemudraia, A.; Wiegand, T.; Surya, K.; Buyukyoruk, M.; Cicha, C.; Vanderwood, K.K.; Wilkinson, R.; Wiedenheft, B. Temporal Detection and Phylogenetic Assessment of SARS-CoV-2 in Municipal Wastewater. Cell Rep. Med. 2020, 1, 100098. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Joshi, M.; Patel, A.K.; Joshi, C.G. Unravelling the early warning capability of wastewater surveillance for COVID-19: A temporal study on SARS-CoV-2 RNA detection and need for the escalation. Environ. Res. 2021, 196, 110946. [Google Scholar] [CrossRef]
- Koureas, M.; Amoutzias, G.D.; Vontas, A.; Kyritsi, M.; Pinaka, O.; Papakonstantinou, A.; Dadouli, K.; Hatzinikou, M.; Koutsolioutsou, A.; Mouchtouri, V.A.; et al. Wastewater monitoring as a supplementary surveillance tool for capturing SARS-CoV-2 community spread. A case study in two Greek municipalities. Environ. Res. 2021, 200, 111749. [Google Scholar] [CrossRef]
- Petala, M.; Kostoglou, M.; Karapantsios, T.; Dovas, C.I.; Lytras, T.; Paraskevis, D.; Roilides, E.; Koutsolioutsou-Benaki, A.; Panagiotakopoulos, G.; Sypsa, V.; et al. Relating SARS-CoV-2 shedding rate in wastewater to daily positive tests data: A consistent model based approach. Sci. Total Environ. 2022, 807 Pt 2, 150838. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Modeling wastewater temperature and attenuation of sewage-borne biomarkers globally. Water Res. 2020, 172, 115473. [Google Scholar] [CrossRef]
- Kevill, J.L.; Pellett, C.; Farkas, K.; Brown, M.R.; Bassano, I.; Denise, H.; McDonald, J.E.; Malham, S.K.; Porter, J.; Warren, J.; et al. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Sci. Total Environ. 2022, 808, 151916. [Google Scholar] [CrossRef]
- Burra, P.; Soto-Diaz, K.; Chalen, I.; Gonzalez-Ricon, R.J.; Istanto, D.; Caetano-Anolles, G. Temperature and Latitude Correlate with SARS-CoV-2 Epidemiological Variables but not with Genomic Change Worldwide. Evol. Bioinform. 2021, 17, 1176934321989695. [Google Scholar] [CrossRef]
- Tran, H.N.; Le, G.T.; Nguyen, D.T.; Juang, R.S.; Rinklebe, J.; Bhatnagar, A.; Lima, E.C.; Iqbal, H.M.N.; Sarmah, A.K.; Chao, H.P. SARS-CoV-2 coronavirus in water and wastewater: A critical review about presence and concern. Environ. Res. 2021, 193, 110265. [Google Scholar] [CrossRef]
- Kampf, G.; Todt, D.; Pfaender, S.; Steinmann, E. Persistence of coronaviruses on inanimate surfaces and their inactivation with biocidal agents. J. Hosp. Infect. 2020, 104, 246–251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henwood, A.F. Coronavirus disinfection in histopathology. J. Histotechnol. 2020, 43, 102–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ort, C.; Lawrence, M.G.; Rieckermann, J.; Joss, A. Sampling for pharmaceuticals and personal care products (PPCPs) and illicit drugs in wastewater systems: Are your conclusions valid? A critical review. Environ. Sci. Technol. 2010, 44, 6024–6035. [Google Scholar] [CrossRef] [PubMed]
- Ort, C.; Lawrence, M.G.; Reungoat, J.; Mueller, J.F. Sampling for PPCPs in wastewater systems: Comparison of different sampling modes and optimization strategies. Environ. Sci. Technol. 2010, 44, 6289–6296. [Google Scholar] [CrossRef]
- Urdan, T.C. Statistics in Plain English, 4th ed.; Routledge: London, UK, 2016; p. 286. [Google Scholar]
- Hair, J.F.B., Jr.; Black, W.C.; Babin, B.J.; Anderson, R.E. Multivariate Data Analysis, 7th ed.; Pearson: London, UK, 2009; p. 816. [Google Scholar]
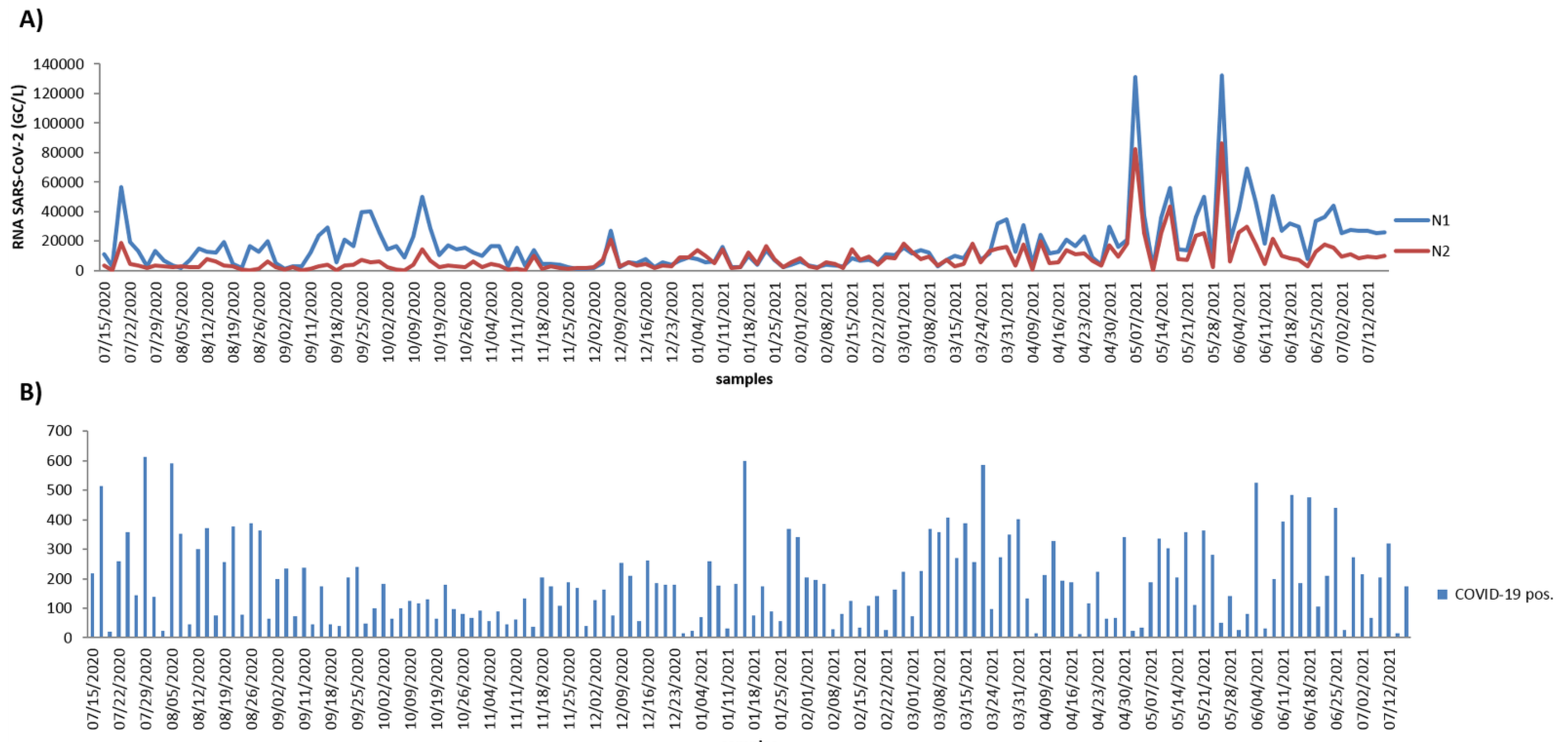
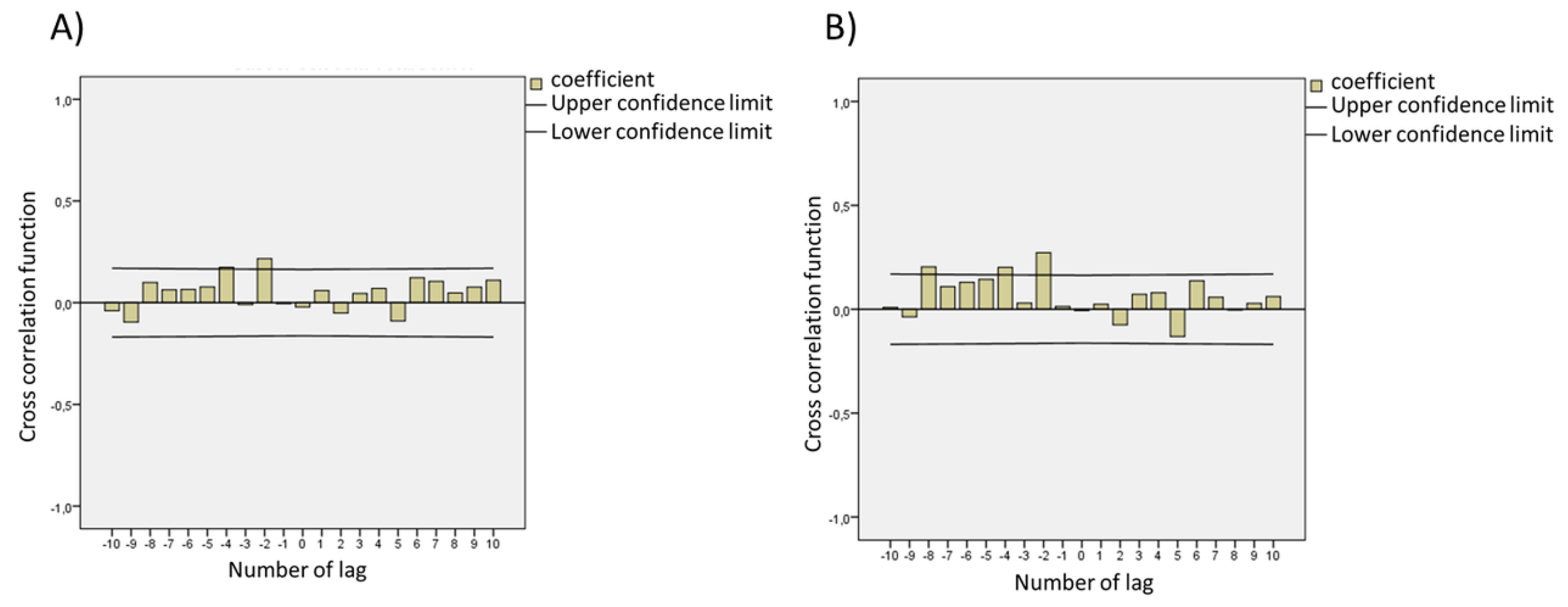
| Parameters | Minimum | Maximum | Average | Standard Deviation | Median (Md) |
|---|---|---|---|---|---|
| N1 RNA copies: normalized to flow | 10.7 × 1010 | 1333.2 × 1010 | 169.6 × 1010 | 183.7 × 1010 | 125.9 × 1010 |
| N2 RNA copies: normalized to flow | 0.00 | 837.4 × 1010 | 85.7 × 1010 | 110.6 × 1010 | 54.4 × 1010 |
| SARS-CoV-2-positive cases | 12.00 | 613.00 | 188.34 | 140.11 | 174 |
| Air temperature (°C) | 0.00 | 34.30 | 24.41 | 3.70 | 24.5 |
| Average flow (L/s) | 1019.00 | 1394.00 | 1169.55 | 71.62 | 1162 |
| Total flow (m3/day) | 87,876.00 | 121,542.00 | 100,826.41 | 6150.29 | 100,067 |
| pH | 7.04 | 7.81 | 7.51 | 0.11 | 7.5 |
| Wastewater temperature (°C) | 18.00 | 30.80 | 26.03 | 2.37 | 26.4 |
| Chemical oxygen demand (mg/L) | 411.00 | 2039.00 | 725.60 | 283.56 | 622.5 |
| N1 | N2 | ||
|---|---|---|---|
| SARS-CoV-2-Positive cases | Correlation Coefficient | −0.033 | 0.000 |
| Sig. (bilateral) | 0.69 | 0.999 | |
| N | 150 | 150 | |
| Air Temperature (°C) | Correlation Coefficient | −0.361 ** | −0.300 ** |
| Sig. (bilateral) | >0.001 | >0.001 | |
| N | 150 | 150 | |
| pH | Correlation Coefficient | 0.215 ** | 0.281 ** |
| Sig. (bilateral) | 0.008 | >0.001 | |
| N | 150 | 150 | |
| Wastewater Temperature (°C) | Correlation Coefficient | −0.375 ** | −0.185 * |
| Sig. (bilateral) | >0.001 | 0.024 | |
| N | 150 | 150 | |
| Average Flow (L/s) | Correlation Coefficient | −0.298 ** | 0.063 |
| Sig. (bilateral) | >0.001 | 0.447 | |
| N | 150 | 150 | |
| Descriptive Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Rainfall | N | Mean | Standard Deviation | Minimum | Maximum | Percentiles | |||
| 25thg. | 50th (Median) | 75th | |||||||
| 0 | Total Flow | 110 | 99,792.7 | 5139.1 | 87,876.0 | 120,384.0 | 96,797.7 | 99,877.5 | 102,610.7 |
| CopiesN1 | 110 | 19,518.2 | 21,077.7 | 1076.0 | 132,348.0 | 6112.0 | 13,603.0 | 26,180.5 | |
| CopiesN2 | 110 | 9283.9 | 12,740.1 | 0 | 86,292.0 | 2422.8 | 5855.0 | 11,384.5 | |
| 1 | Total Flow | 40 | 103,668.9 | 7707.1 | 91,892.0 | 121,542.0 | 98,543.7 | 101,957.0 | 109,617.5 |
| CopiesN1 | 40 | 10,719.9 | 8621.5 | 1400.0 | 36,542.0 | 4246.5 | 7990.0 | 15,051.0 | |
| CopiesN2 | 40 | 6653.4 | 5646.2 | 1352.0 | 23,572.0 | 2756.0 | 4482.0 | 8792.5 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, R.M.; Carvalho, T.; Bittar, C.; Quevedo, D.M.; Miceli, R.N.; Nogueira, M.L.; Ferreira, H.L.; Costa, P.I.; Araújo, J.P., Jr.; Spilki, F.R.; et al. Long-Term Wastewater Surveillance for SARS-CoV-2: One-Year Study in Brazil. Viruses 2022, 14, 2333. https://doi.org/10.3390/v14112333
Martins RM, Carvalho T, Bittar C, Quevedo DM, Miceli RN, Nogueira ML, Ferreira HL, Costa PI, Araújo JP Jr., Spilki FR, et al. Long-Term Wastewater Surveillance for SARS-CoV-2: One-Year Study in Brazil. Viruses. 2022; 14(11):2333. https://doi.org/10.3390/v14112333
Chicago/Turabian StyleMartins, Renan Moura, Tamara Carvalho, Cintia Bittar, Daniela Muller Quevedo, Rafael Nava Miceli, Mauricio Lacerda Nogueira, Helena Lage Ferreira, Paulo Inácio Costa, João Pessoa Araújo, Jr., Fernando Rosado Spilki, and et al. 2022. "Long-Term Wastewater Surveillance for SARS-CoV-2: One-Year Study in Brazil" Viruses 14, no. 11: 2333. https://doi.org/10.3390/v14112333








