Identification of Key Genes Involved in Resistance to Early Stage of BmNPV Infection in Silkworms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of BmNPV and Silkworm Midgut Samples
2.2. RNA Extraction and RNA-Seq
2.3. RNA-Seq Data Analysis
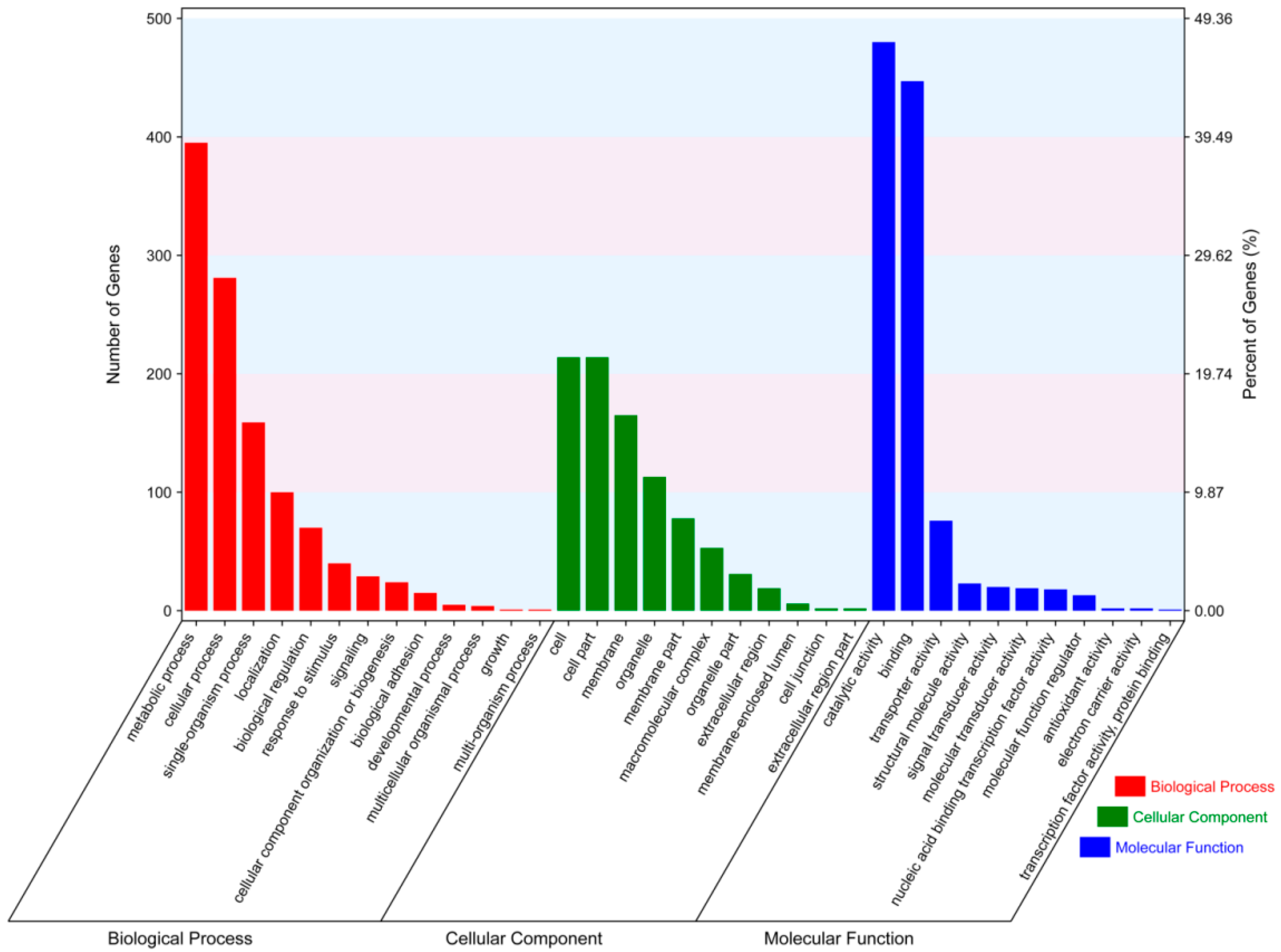
2.4. Functional Annotation
2.5. Verification of DEGs by qRT-PCR
2.6. Data Statistics
3. Results
3.1. Resistance of the Two Silkworm Varieties to BmNPV Infection
3.2. Overview of the RNA-Seq Data
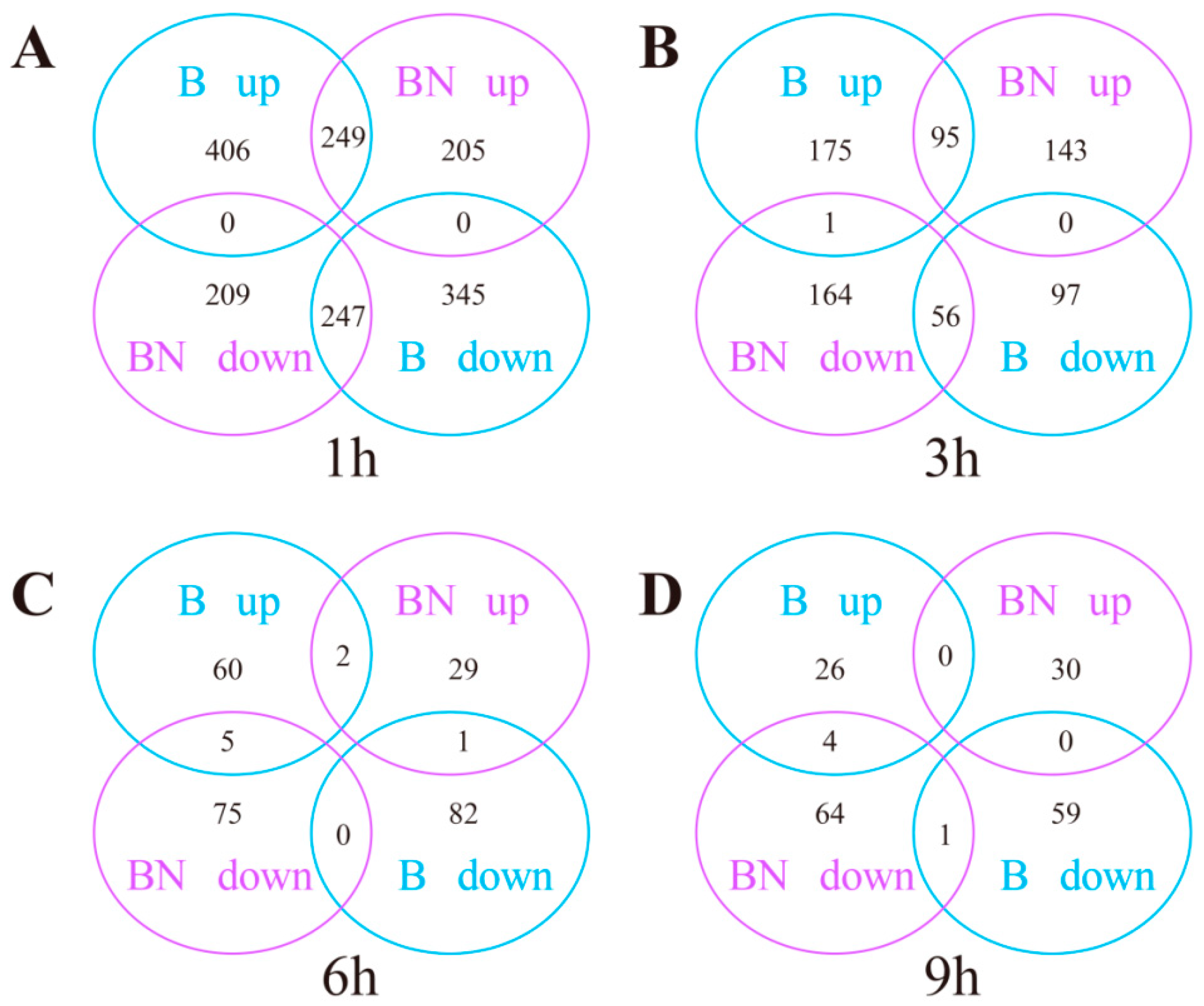
3.3. Overall Analyses of DEGs at Different Time Points Post-BmNPV Infection
3.4. Transcriptome Changes at Different Time Points Post-BmNPV Infection
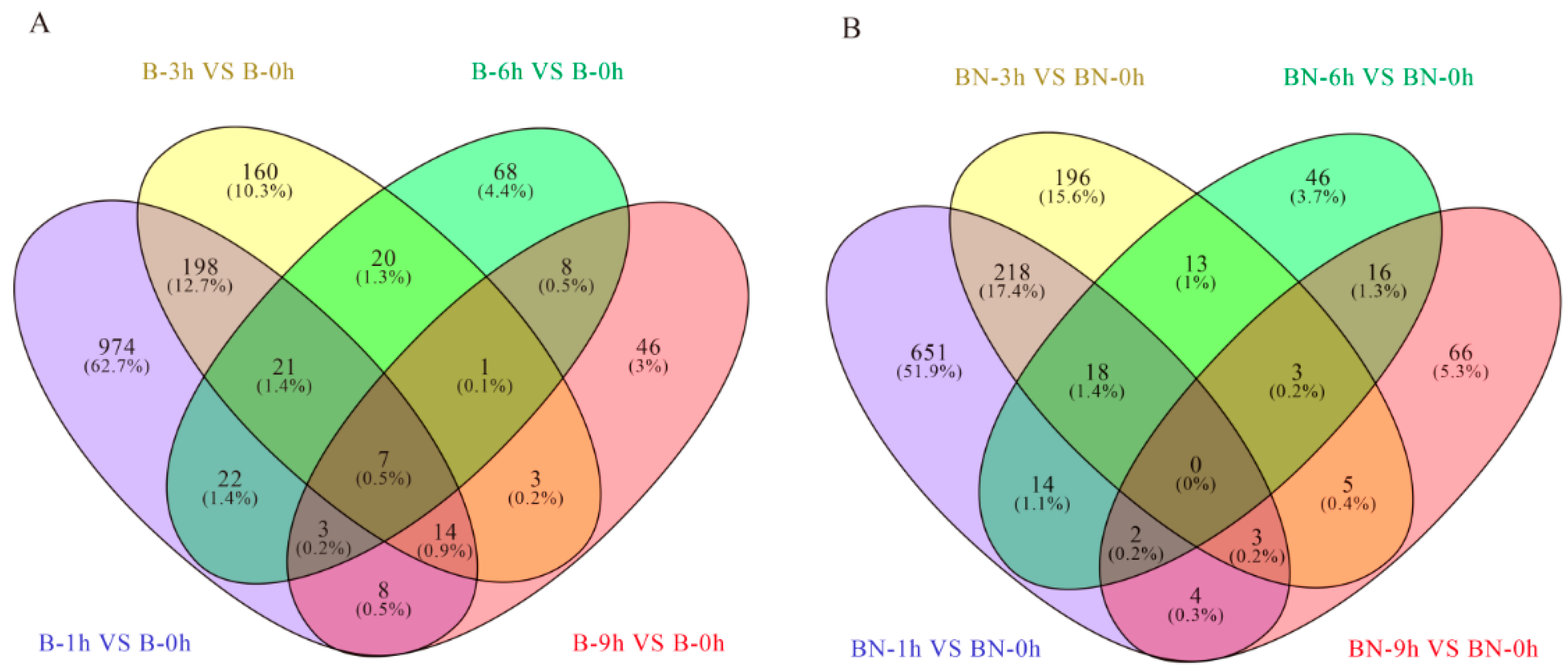
3.4.1. Transcriptome Changes at Different Time Points in the Susceptible (B) Variety
3.4.2. Transcriptome Changes at Different Time Points in the Resistant (BN) Variety
3.5. Transcriptome Differences between the Susceptible (B) and Resistant (BN) Varieties
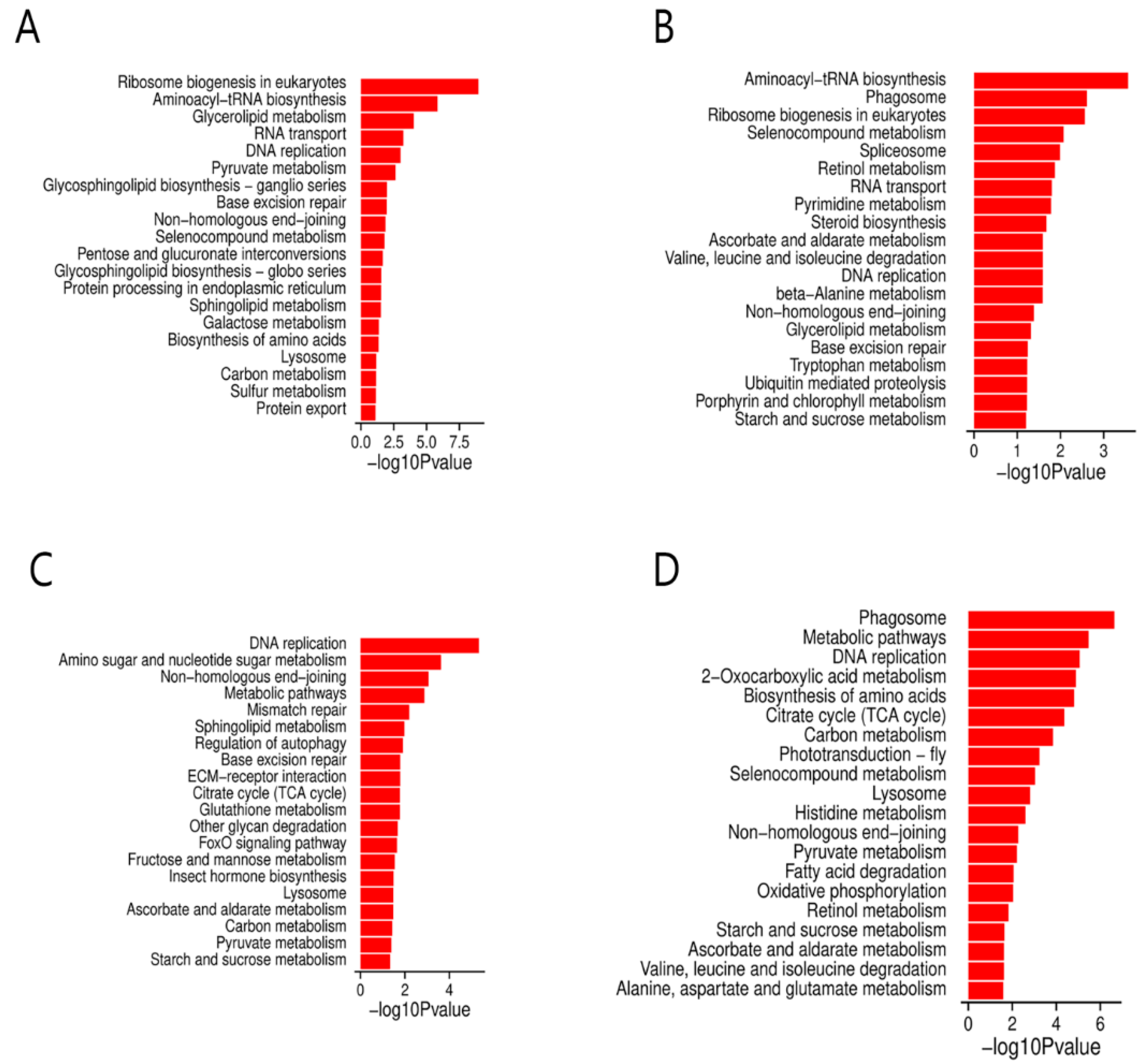
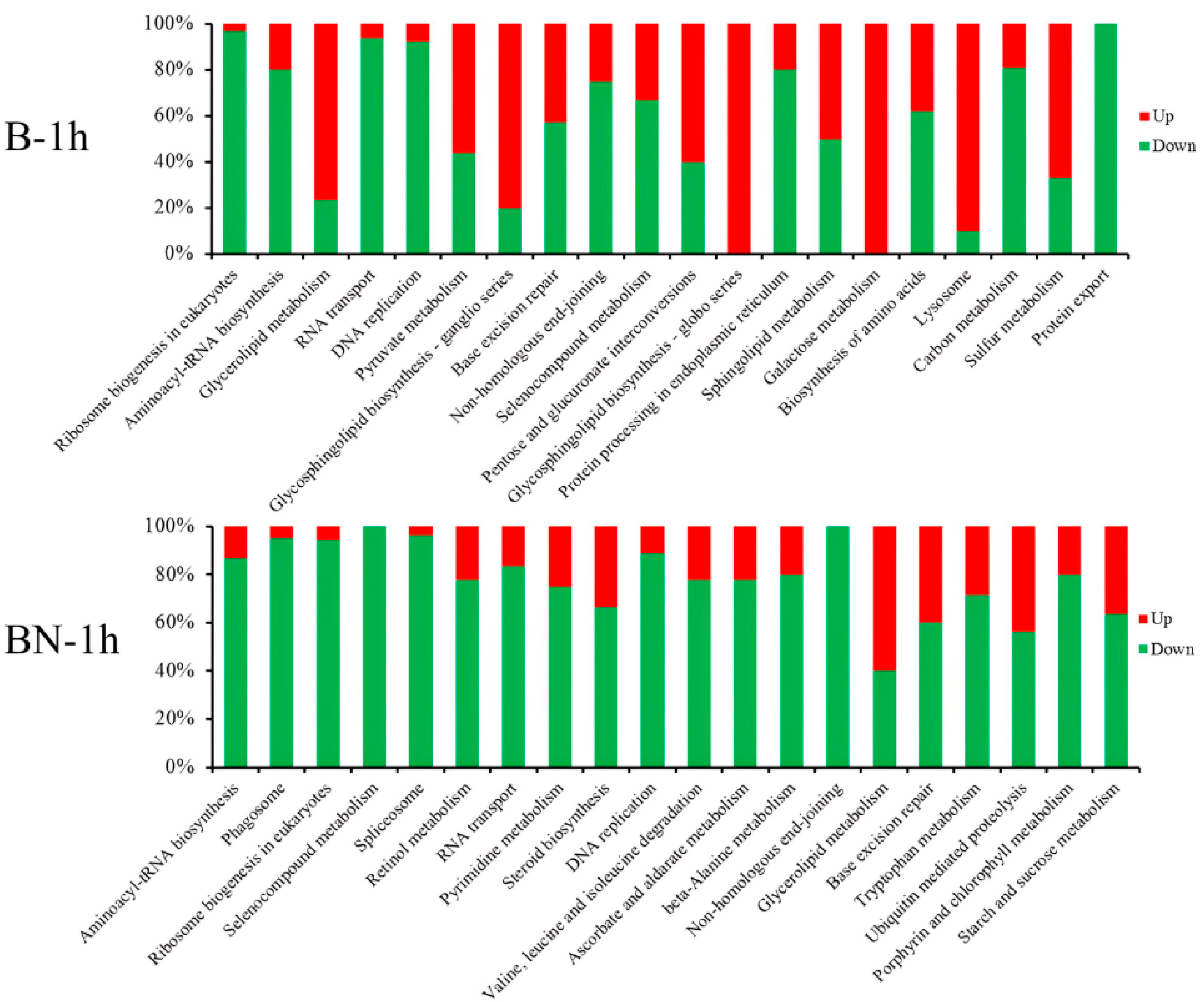
3.6. KEGG Pathway Analysis of DEGs at Different Time Points
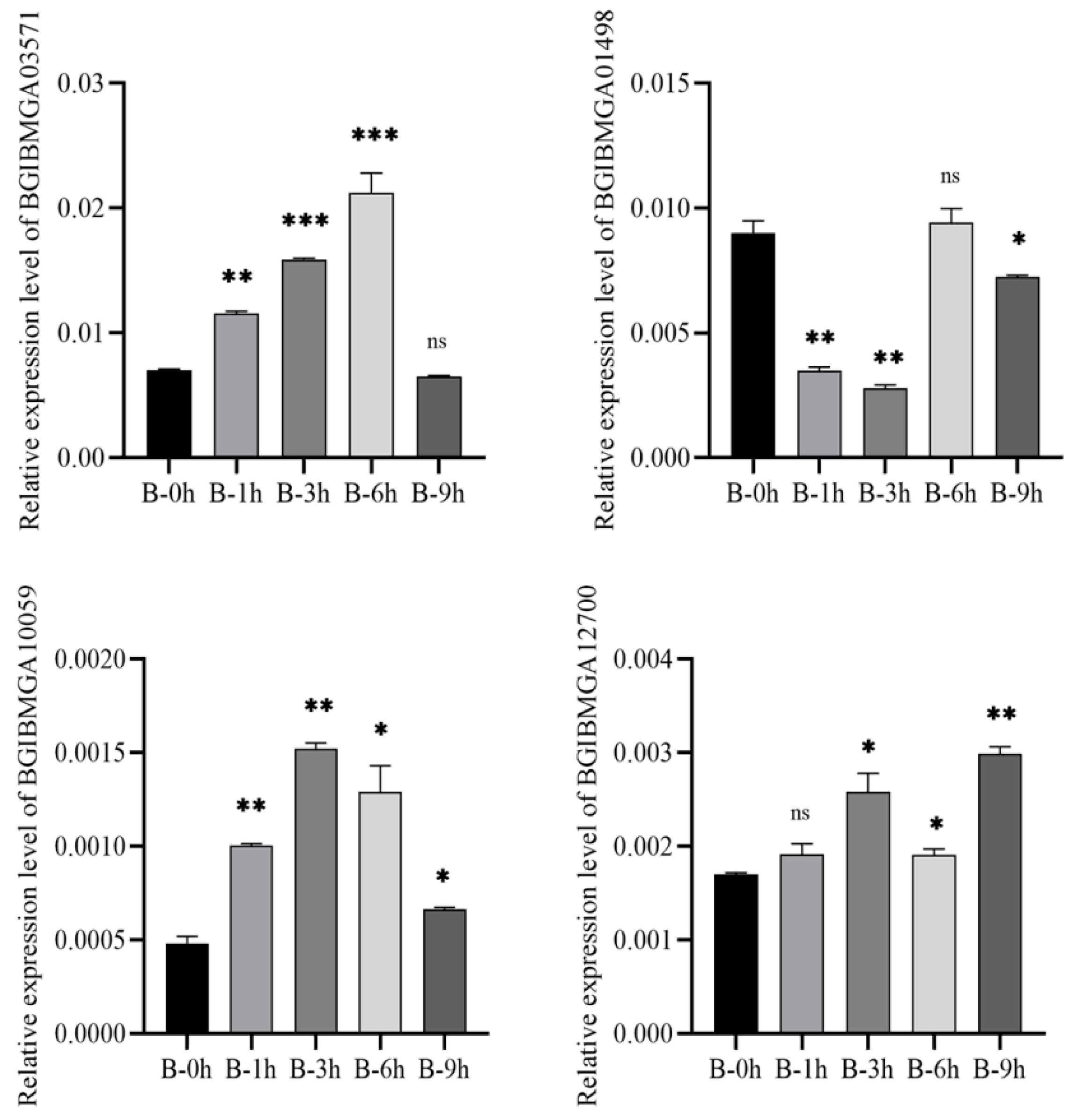
3.7. Analysis of DEGs Associated with the Carbon Metabolism, DNA Replication, Starch, and Sucrose Metabolism Pathways
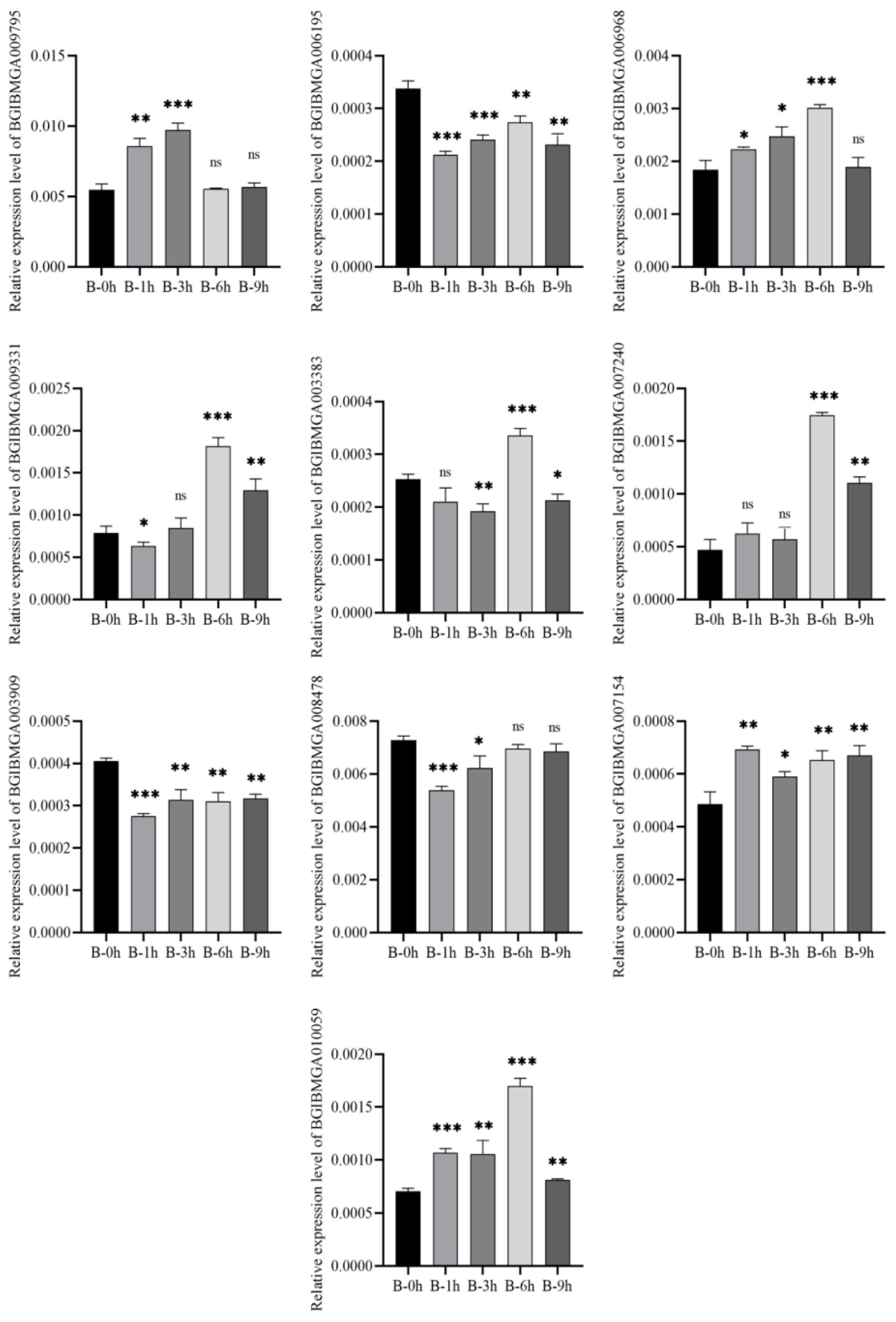
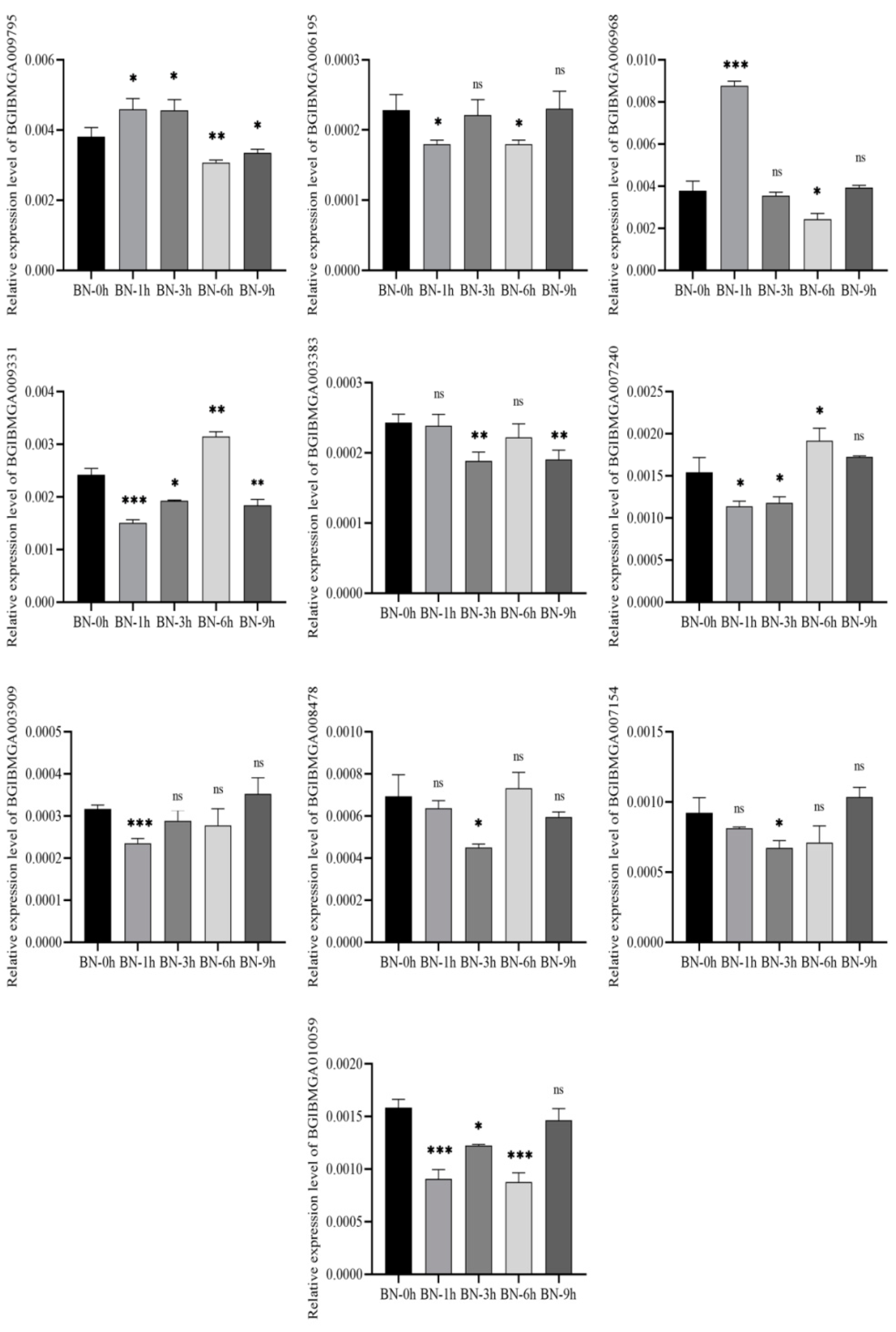
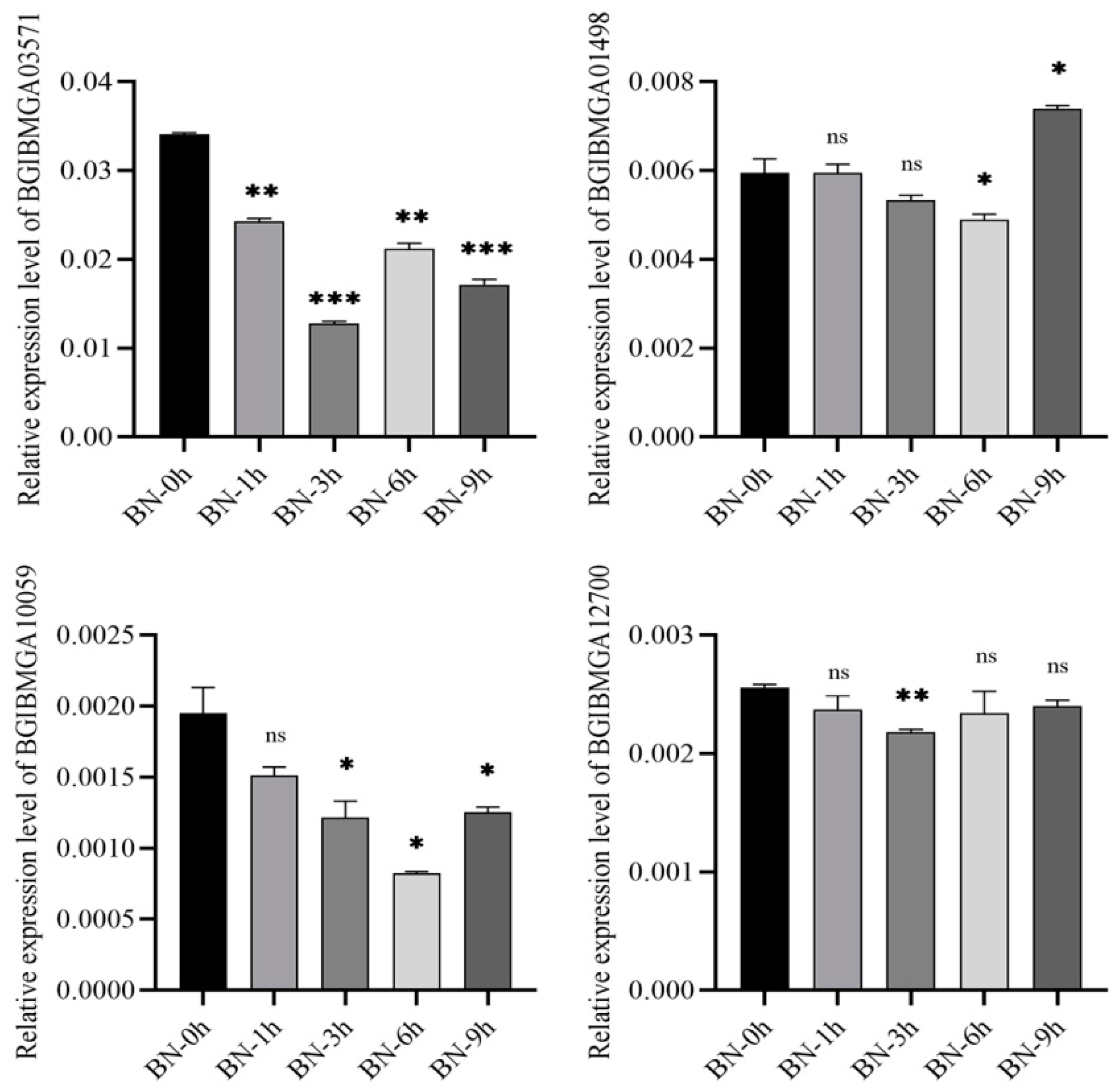
3.8. Validation of DEGs by qRT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Luan, J.B.; Li, J.M.; Varela, N.; Wang, Y.L.; Li, F.F.; Bao, Y.Y.; Zhang, C.X.; Liu, S.S.; Wang, X.W. Global analysis of the transcriptional response of whitefly to tomato yellow leaf curl China virus reveals the relationship of coevolved adaptations. J. Virol. 2011, 85, 3330–3340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.Q.; Yao, Q.; Bao, F.; Chen, K.P.; Liu, X.Y.; Li, J.; Wang, L. Comparative proteome analysis of silkworm in its susceptibility and resistance responses to Bombyx mori densonucleosis virus. Intervirology 2012, 55, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Li, F.; Ma, L.; Wang, B.; Zhang, H.; Ni, M.; Hong, F.; Shen, W.; Li, B. Mechanism of enhanced Bombyx mori nucleopolyhedrovirus-resistance by titanium dioxide nanoparticles in silkworm. PLoS ONE 2015, 10, e0118222. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Qian, H.; Luo, X.; Xu, P.; Yang, J.; Liu, M.; Xu, A. Transcriptomic analysis of resistant and susceptible Bombyx mori strains following BmNPV infection provides insights into the antiviral mechanisms. Int. J. Genom. 2016, 2016, 2086346. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.H.; Dong, X.L.; Pan, C.X.; Du, G.Y.; Wu, Y.F.; Yang, J.G.; Chen, P.; Lu, C.; Pan, M.H. A newly discovered member of the Atlastin family, BmAtlastin-n, has an antiviral effect against BmNPV in Bombyx mori. Sci. Rep. 2016, 6, 28946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.Y.; Tang, X.D.; Lv, Z.Y.; Wang, X.Y.; Tian, C.H.; Xu, Y.P.; Zhang, C.X. Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 2009, 94, 138–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.Y.; Yu, H.Z.; Xu, J.P.; Zhang, S.Z.; Yu, D.; Liu, M.H.; Wang, L.L. Comparative subcellular proteomics analysis of susceptible and near-isogenic resistant Bombyx mori (Lepidoptera) larval midgut response to BmNPV infection. Sci. Rep. 2017, 7, 45690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, A.Y.; Qian, H.Y.; Sun, J.P.; Liu, M.Z.; Lin, C.Q.; Li, G.; Li, L.; Zhang, Y.H.; Zhao, G.D. Breeding of a new silkworm variety Huakang 3 with resistance to Bombyx mori nucleopolyhedrosis. Sci. Seric. 2019, 45, 201–211. (In Chinese) [Google Scholar] [CrossRef]
- Guo, R.; Wang, S.; Xue, R.; Cao, G.; Hu, X.; Huang, M.; Zhang, Y.; Lu, Y.; Zhu, L.; Chen, F.; et al. The gene expression profile of resistant and susceptible Bombyx mori strains reveals cypovirus-associated variations in host gene transcript levels. Appl. Microbiol. Biotechnol. 2015, 99, 5175–5187. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.Y.; Yu, H.Z.; Geng, L.; Xu, J.P.; Yu, D.; Zhang, S.Z.; Ma, Y.; Fei, D.Q. Comparative transcriptome analysis of Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic resistant strains. PLoS ONE 2016, 11, e0155341. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Xue, B.; Li, Y.Y.; Li, F.C.; Ni, M.; Shen, W.D.; Gu, Z.Y.; Li, B.; Shen, W.D.; Gu, Z.Y.; et al. Construction of silkworm midgut cDNA library for screen and sequence analysis of peritrophic membrane protein genes. Arch. Insect Biochem. Physiol. 2016, 91, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Song, G.; Liu, B.; Li, Z.; Wu, H.; Wang, P.; Zhao, K.; Jiang, G.; Zhang, L.; Gao, C. E3 ubiquitin ligase RNF128 promotes innate antiviral immunity through K63-linked ubiquitination of TBK1. Nat. Immunol. 2016, 17, 1342–1351. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.Y.; Hu, H.; Killiny, N.; Xu, J.P. A hypothetical model of crossing Bombyx mori nucleopolyhedrovirus through its host midgut physical barrier. PLoS ONE 2014, 9, e115032. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Cheng, T.; Dang, Y.; Peng, Z.; Zhao, P.; Liu, S.; Jin, S.; Lin, P.; Sun, Q.; Xia, Q. Identification of a midgut-specific promoter in the silkworm Bombyx mori. Biochem. Biophys. Res. Commun. 2013, 433, 542–546. [Google Scholar] [CrossRef]
- Jiang, L.; Peng, Z.; Guo, Y.; Cheng, T.; Guo, H.; Sun, Q.; Huang, C.; Zhao, P.; Xia, Q. Transcriptome analysis of interactions between silkworm and cytoplasmic polyhedrosis virus. Sci. Rep. 2016, 6, 24894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, L.; Xia, Q. The progress and future of enhancing antiviral capacity by transgenic technology in the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2014, 48, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callea, M.; Vinciguerra, A.; Willoughby, C.E.; Deroma, L.; Clarich, G. Infantile bilateral glaucoma in a child with ectodermal dysplasia. Ophthalmic Genet. 2013, 34, 58–60. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Florea, L.; Song, L.; Salzberg, S.L. Thousands of exon skipping events differentiate among splicing patterns in sixteen human tissues. F1000Res 2013, 2, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method A Companion Methods Enzymol. 2013, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Li, G.; Zhao, G.; Liu, M.; Xu, A. Metabolic characterisation of the midgut of Bombyx mori varieties after BmNPV infection using GC-MS-Based metabolite profiling. Int. J. Mol. Sci. 2020, 21, 4707. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Wang, X.; Xu, J.; Ma, Y.; Zhang, S.; Yu, D.; Fei, D.; Muhammad, A. iTRAQ-based quantitative proteomics analysis of molecular mechanisms associated with Bombyx mori (Lepidoptera) larval midgut response to BmNPV in susceptible and near-isogenic strains. J. Proteom. 2017, 165, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Haas-Stapleton, E.J.; Washburn, J.O.; Volkman, L.E. Spodoptera frugiperda resistance to oral infection by autographa californica multiple nucleopolyhedrovirus linked to aberrant occlusion-derived virus binding in the midgut. J. Gen. Virol. 2005, 86, 1349–1355. [Google Scholar] [CrossRef]
- Ikeda, M.; Kobayashi, M. Cell-Cycle Perturbation in Sf9 cells infected with Autographa californica nucleopolyhedrovirus. Virology 1999, 258, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Kannan, R.P.; Hensley, L.L.; Evers, L.E.; Lemon, S.M.; McGivern, D.R. Hepatitis C virus infection causes cell cycle arrest at the level of initiation of mitosis. J. Virol. 2011, 85, 7989–8001. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhou, W.; Zhou, Y.; Wu, J.; Zhou, X. Transcriptome and comparative gene expression analysis of Sogatella furcifera (Horvath) in response to southern rice black-streaked dwarf virus. PLoS ONE 2012, 7, e36238. [Google Scholar] [CrossRef]
- Pullen, S.; Friesen, P. Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5’ noncoding leader region. Virology 1995, 69, 176–188. [Google Scholar] [CrossRef] [Green Version]
- Crouch, E.A.; Morales, K.G.; Passarelli, A.L.; Cox, L.T. Inter-subunit interactions of the Autographa californica M nucleopolyhedrovirus RNA polymerase. Virology 2007, 367, 265–274. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Gao, L.; Shi, H.; Xia, H.; Gao, L.; Lian, C.; Chen, L.; Yao, Q.; Chen, K.; Liu, X. Microarray analysis of gene expression profile in resistant and susceptible Bombyx mori strains reveals resistance-related genes to nucleopolyhedrovirus. Genomics 2013, 101, 256–262. [Google Scholar] [CrossRef] [Green Version]
- Govindaraj, L.; Gupta, T.; Esvaran, V.G.; Awasthi, A.K.; Ponnuvel, K.M. Genome-wide identification, characterization of sugar transporter genes in the silkworm Bombyx mori and role in Bombyx mori nucleopolyhedrovirus (BmNPV) infection. Gene 2016, 579, 162–171. [Google Scholar] [CrossRef]
- Yao, Q.; Gao, L.; Chen, K.P.; Hu, Z.G. Detection of proliferation of Bombyx mori nucleopolyhedrovirus in its host by fluorescence quantitative PCR. Acta Entomol. Sin. 2005, 48, 5. [Google Scholar] [CrossRef]
- Gao, K.; Deng, X.Y.; Qian, H.Y.; Qin, G.; Guo, X.J. Digital gene expression analysis in the midgut of 4008 silkworm strain infected with cytoplasmic polyhedrosis virus. J. Invertebr. Pathol. 2014, 115, 8–13. [Google Scholar] [CrossRef]
- Gao, K.; Deng, X.Y.; Qian, H.Y.; Qin, G.X.; Hou, C.X.; Guo, X.J. Cytoplasmic polyhedrosis virus-induced differential gene expression in two silkworm strains of different susceptibility. Gene 2014, 539, 230–237. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Lee, B.L.; Soderhall, K. Cell-mediated immunity in arthropods: Hematopoiesis, coagulation, melanization and opsonization. Immunobiology 2006, 211, 213–236. [Google Scholar] [CrossRef]
- Waterhouse, R.M.; Kriventseva, E.V.; Meister, S.; Xi, Z.; Alvarez, K.S.; Bartholomay, L.C.; Barillas-Mury, C.; Bian, G.; Blandin, S.; Christensen, B.M.; et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 2007, 316, 1738–1743. [Google Scholar] [CrossRef] [Green Version]
- Etebari, K.; Palfreyman, R.W.; Schlipalius, D.; Nielsen, L.K.; Glatz, R.V.; Asgari, S. Deep sequencing-based transcriptome analysis of Plutella xylostella larvae parasitized by Diadegma semiclausum. BMC Genom. 2011, 12, 446. [Google Scholar] [CrossRef] [PubMed]
- Qian, H.; Guo, H.; Zhang, X.; Liu, M.; Zhao, G.; Xu, A.; Li, G. Metabolic characterization of hemolymph in Bombyx mori varieties after Bombyx mori nucleopolyhedrovirus infection by GC-MS-based metabolite profiling. Arch. Virol. 2022, 167, 1637–1648. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Afek, A.; Gilburd, B.; Shoenfeld, Y.; Harats, D. Cellular and humoral immune responses to heat shock protein 65 are both involved in promoting fatty-streak formation in LDL-receptor deficient mice. J. Am. Coll. Cardiol. 2001, 38, 900–905. [Google Scholar] [CrossRef] [Green Version]
- Tezel, G.; Yang, J.; Wax, M.B. Heat shock proteins, immunity and glaucoma. Brain Res. Bull. 2004, 62, 473–480. [Google Scholar] [CrossRef]
- Khachatoorian, R.; Riahi, R.; Ganapathy, E.; Shao, H.; Wheatley, N.M.; Sundberg, C.; Jung, C.L.; Ruchala, P.; Dasgupta, A.; Arumugaswami, V.; et al. Allosteric heat shock protein 70 inhibitors block hepatitis C virus assembly. Int. J. Antimicrob. Agents 2016, 47, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Wallin, R.P.; Lundqvist, A.; Moré, S.H.; von Bonin, A.; Kiessling, R.; Ljunggren, H.-G. Heat-shock proteins as activators of the innate immune system. Trends Immunol. 2002, 23, 130–135. [Google Scholar] [CrossRef]
- Fallouh, H.; Mahana, W. Antibody to heat shock protein 70 (HSP70) inhibits human T-cell lymphoptropic virus type I (HTLV-I) production by transformed rabbit T-cell lines. Toxins 2012, 4, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Li, G.; Zhang, J.; Tong, X.; Liu, W.; Ye, X. Heat shock protein 70 inhibits the activity of Influenza A virus ribonucleoprotein and blocks the replication of virus in vitro and in vivo. PLoS ONE 2011, 6, e16546. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Bao, X.Y.; Dong, Z.Q.; Kang, T.T.; Zhu, Y.; Pan, M.H.; LU, C. Gene Hsc70-4 promotes the replication and proliferation of Bombyx mori nucleopolyhedrovirus. Microbiology 2019, 59, 10. [Google Scholar] [CrossRef]
- Shang, Q.; Wu, P.; Huang, H.L.; Zhang, S.L.; Tang, X.D.; Guo, X.J. Inhibition of heat shock protein 90 suppresses Bombyx mori nucleopolyhedrovirus replication in B. mori. Insect Mol. Biol. 2020, 29, 205–213. [Google Scholar] [CrossRef]

| Variety Name | Mortality Rate of the 2nd Instar after BmNPV Infection with 1.0 × 108 mL−1 Solution | Regression Equation | LC50/mL−1 |
|---|---|---|---|
| Baiyu N | 0% | Y = −7.252 + 0.510 X | 2.52 × 109 |
| Baiyu | 100% | Y = −10.019 + 1.603 X | 2.13 × 105 |
| Qiufeng N × Baiyu N | 10% | Y = −8.361 + 0.610 X | 2.16 × 109 |
| Baiyu N × Qiufeng N | 0% | Y = −8.233 + 0.702 X | 1.99 × 109 |
| Qiufeng × Baiyu | 100% | Y = −10.240 + 1.692 X | 1.13 × 105 |
| Baiyu × Qiufeng | 100% | Y = −12.080 + 1.904 X | 2.21 × 105 |
| Sample | Raw Reads | Raw Bases | Clean Reads | Clean Bases | Valid Ratio (Base) | Q30 (%) | GC Content (%) | Ratio of Reads Mapped to Genome |
|---|---|---|---|---|---|---|---|---|
| B-0h-1 | 28562592 | 3570324000 | 27765390 | 3467734429 | 97.12% | 95.25% | 47.00% | 79.89% |
| B-0h-2 | 28876928 | 3609616000 | 28096334 | 3509155838 | 97.21% | 95.34% | 46.00% | 81.33% |
| B-0h-3 | 28794488 | 3599311000 | 27820182 | 3474441859 | 96.53% | 94.78% | 45.00% | 80.55% |
| B-1h-1 | 28088422 | 3511052750 | 27287032 | 3407989919 | 97.06% | 95.20% | 46.50% | 80.47% |
| B-1h-2 | 28960774 | 3620096750 | 28142744 | 3514894097 | 97.09% | 95.26% | 46.00% | 80.98% |
| B-1h-3 | 28012158 | 3501519750 | 27141482 | 3389744667 | 96.80% | 95.03% | 46.50% | 81.02% |
| B-3h-1 | 28319422 | 3539927750 | 27575244 | 3444110971 | 97.29% | 95.46% | 47.00% | 80.58% |
| B-3h-2 | 28880206 | 3610025750 | 28152056 | 3516148247 | 97.39% | 95.49% | 45.50% | 79.56% |
| B-3h-3 | 28080402 | 3510050250 | 27354780 | 3416555313 | 97.33% | 95.44% | 46.00% | 80.66% |
| B-6h-1 | 28368774 | 3546096750 | 27573602 | 3443838179 | 97.11% | 95.26% | 46.50% | 81.40% |
| B-6h-2 | 28045628 | 3505703500 | 27025310 | 3375100921 | 96.27% | 94.60% | 46.00% | 79.09% |
| B-6h-3 | 28399900 | 3549987500 | 27571816 | 3443565824 | 97.00% | 95.17% | 46.00% | 80.72% |
| B-9h-1 | 28317000 | 3539625000 | 27475118 | 3431461235 | 96.94% | 95.13% | 46.50% | 79.94% |
| B-9h-2 | 28119310 | 3514913750 | 27307226 | 3410564463 | 97.03% | 95.24% | 45.50% | 80.96% |
| B-9h-3 | 28648046 | 3581005750 | 27760426 | 3467084956 | 96.81% | 95.04% | 46.00% | 81.28% |
| BN-0h-1 | 28666864 | 3583358000 | 27841768 | 3477331144 | 97.04% | 95.17% | 47.50% | 81.01% |
| BN-0h-2 | 28298442 | 3537305250 | 27464334 | 3430189736 | 96.97% | 95.16% | 47.00% | 80.85% |
| BN-0h-3 | 28012628 | 3501578500 | 27274002 | 3406499188 | 97.28% | 95.42% | 47.00% | 81.28% |
| BN-1h-1 | 28205818 | 3525727250 | 27406492 | 3422936978 | 97.08% | 95.21% | 46.50% | 79.69% |
| BN-1h-2 | 28279168 | 3534896000 | 27416632 | 3424133438 | 96.86% | 95.02% | 47.50% | 78.98% |
| BN-1h-3 | 28512350 | 3564043750 | 27675112 | 3456531187 | 96.98% | 95.20% | 46.50% | 80.65% |
| BN-3h-1 | 28011992 | 3501499000 | 27204406 | 3397702496 | 97.03% | 95.23% | 46.50% | 78.92% |
| BN-3h-2 | 28291120 | 3536390000 | 27537810 | 3439446648 | 97.25% | 95.45% | 47.00% | 81.19% |
| BN-3h-3 | 28764132 | 3595516500 | 27990216 | 3495896532 | 97.22% | 95.35% | 48.00% | 80.88% |
| BN-6h-1 | 28219152 | 3527394000 | 27113062 | 3385619587 | 95.98% | 94.55% | 46.50% | 77.41% |
| BN-6h-2 | 28662614 | 3582826750 | 27623744 | 3449506275 | 96.27% | 94.82% | 47.00% | 80.39% |
| BN-6h-3 | 28650550 | 3581318750 | 27580236 | 3444099370 | 96.16% | 94.79% | 46.00% | 81.16% |
| BN-9h-1 | 28010608 | 3501326000 | 27051472 | 3378073060 | 96.47% | 94.95% | 46.00% | 80.31% |
| BN-9h-2 | 28861830 | 3607728750 | 27704832 | 3459568321 | 95.89% | 94.56% | 46.50% | 80.07% |
| BN-9h-3 | 28443426 | 3555428250 | 27444454 | 3427214063 | 96.39% | 94.89% | 47.50% | 80.19% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, L.; Cao, Y.; Ge, S.; Xu, A.; Qian, H.; Li, G. Identification of Key Genes Involved in Resistance to Early Stage of BmNPV Infection in Silkworms. Viruses 2022, 14, 2405. https://doi.org/10.3390/v14112405
Yu L, Cao Y, Ge S, Xu A, Qian H, Li G. Identification of Key Genes Involved in Resistance to Early Stage of BmNPV Infection in Silkworms. Viruses. 2022; 14(11):2405. https://doi.org/10.3390/v14112405
Chicago/Turabian StyleYu, Linyuan, Yeqing Cao, Sicheng Ge, Anying Xu, Heying Qian, and Gang Li. 2022. "Identification of Key Genes Involved in Resistance to Early Stage of BmNPV Infection in Silkworms" Viruses 14, no. 11: 2405. https://doi.org/10.3390/v14112405





