Virus Prevalence in Egg Samples Collected from Naturally Selected and Traditionally Managed Honey Bee Colonies across Europe
Abstract
:1. Introduction
2. Materials and Methods
2.1. Virus Screening in Eggs
2.2. Egg Sample Collection across Europe
2.3. RNA Extraction and cDNA Synthesis
2.4. End-Point PCR
2.5. qPCR
2.6. Statistics
3. Results
3.1. SOV Phenotyping Method
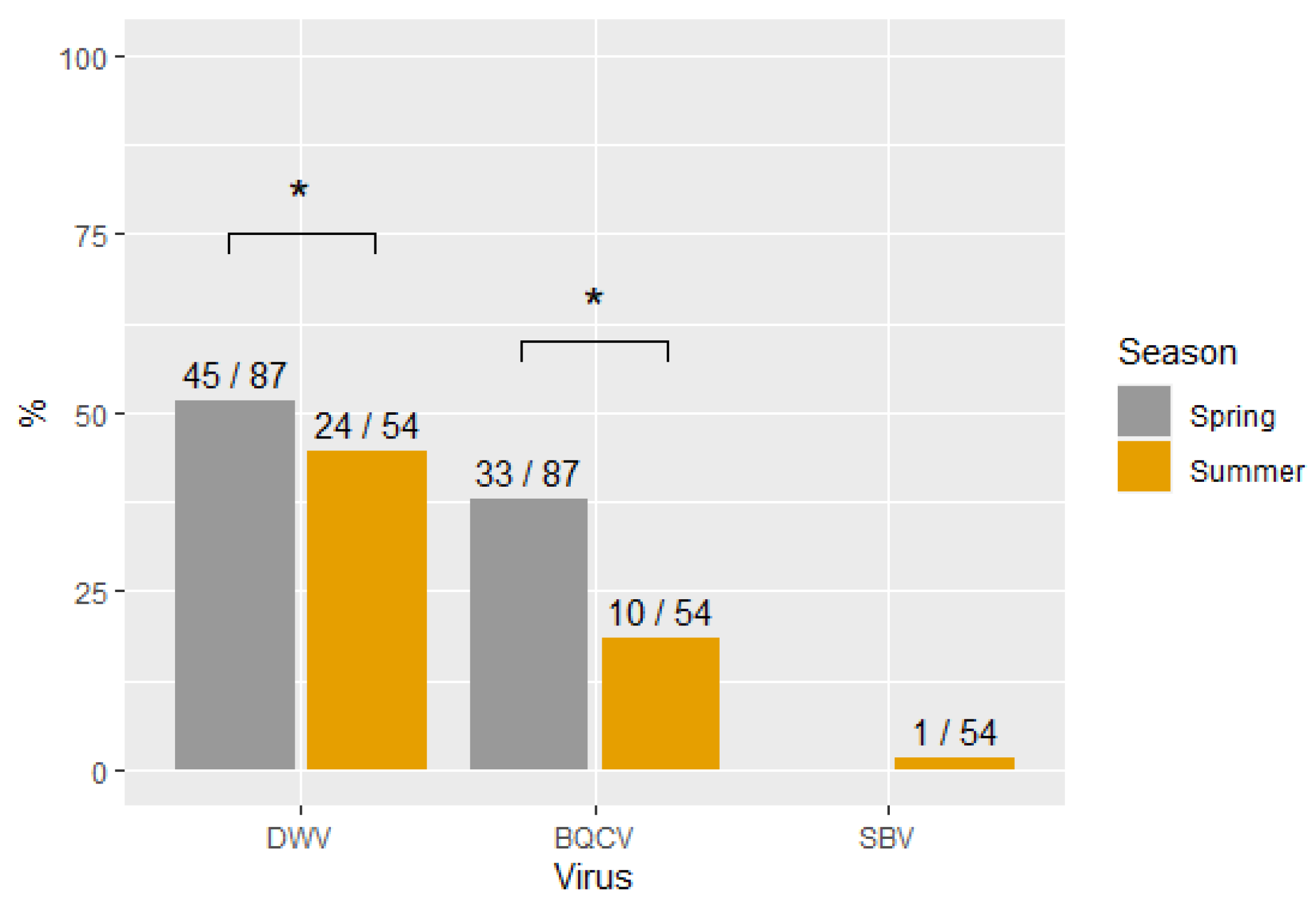
3.2. Virus Prevalence
3.3. Natural Survivors vs. Traditionally Managed Colonies
3.4. Queen Age
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nowak, M.A.; Tarnita, C.E.; Wilson, E.O. The evolution of eusociality. Nature 2010, 466, 1057–1062. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, X.Y.; Tang, Q.B.; Lei, C.L.; Huang, Q.Y. The Mechanisms of Social Immunity Against Fungal Infections in Eusocial Insects. Toxins 2019, 11, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Udiani, O.; Fefferman, N.H. How disease constrains the evolution of social systems. Proc. R. Soc. B 2020, 287, 20201284. [Google Scholar] [CrossRef] [PubMed]
- Sumpter, D.; Martin, S.J. The dynamics of virus epidemics in Varroa -infested honey bee colonies. J. Anim. Ecol. 2004, 73, 51–63. [Google Scholar] [CrossRef]
- Dainat, B.; Neumann, P. Clinical signs of deformed wing virus infection are predictive markers for honey bee colony losses. J. Invertebr. Pathol. 2013, 112, 278–280. [Google Scholar] [CrossRef]
- Wegener, J.; Ruhnke, H.; Scheller, K.; Mispagel, S.; Knollmann, U.; Kamp, G.; Bienefeld, K. Pathogenesis of varroosis at the level of the honey bee (Apis mellifera) colony. Insect Physiol. 2016, 91–92, 1–9. [Google Scholar] [CrossRef]
- Gisder, S.; Möckel, N.; Eisenhardt, D.; Genersch, E. In vivo evolution of viral virulence: Switching of deformed wing virus between hosts results in virulence changes and sequence shifts. Environ. Microbiol. 2018, 20, 4612–4628. [Google Scholar] [CrossRef] [Green Version]
- Ryabov, E.V.; Childers, A.K.; Lopez, D.; Grubbs, K.; Posada-Florez, F.; Weaver, D.; Girten, W.; VanEngelsdorp, D.; Chen, Y.; Evans, J.D. Dynamic evolution in the key honey bee pathogen deformed wing virus: Novel insights into virulence and competition using reverse genetics. PLoS Biol. 2019, 17, e3000502. [Google Scholar] [CrossRef] [Green Version]
- Norton, A.M.; Remnant, E.J.; Tom, J.; Buchmann, G.; Blacquiere, T.; Beekman, M. Adaptation to vector-based transmission in a honeybee virus. J. Anim. Ecol. 2021, 90, 2254–2267. [Google Scholar] [CrossRef]
- Neumann, P.; Yañez, O.; Fries, I.; De Miranda, J.R.; de Miranda, J. Varroa invasion and virus adaptation. Trends Parasitol. 2012, 28, 353–354. [Google Scholar] [CrossRef]
- Wilfert, L.; Long, G.; Leggett, H.; Schmid-Hempel, P.; Butlin, R.; Martin, S.J.; Boots, M. Honeybee disease: Deformed wing virus is a recent global epidemic in honeybees driven by Varroa mites. Science 2016, 351, 594–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traynor, K.S.; Mondet, F.; de Miranda, J.R.; Techer, M.; Kowallik, V.; Oddie, M.A.Y.; Chantawannakul, P.; McAfee, A. Varroa destructor: A Complex Parasite, Crippling Honey Bees Worldwide. Trends Parasitol. 2020, 36, 592–606. [Google Scholar] [CrossRef] [PubMed]
- Mondet, F.; Beaurepaire, A.; McAfee, A.; Locke, B.; Alaux, C.; Blanchard, S.; Danka, B.; Le Conte, Y. Honey bee survival mechanisms against the parasite Varroa destructor: A systematic review of phenotypic and genomic research efforts. Int. J. Parasitol. 2020, 50, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Highfield, A.C.; Nagar, A.E.; Mackinder, L.; Noël, L.; Hall, M.J.; Martin, S.J.; El Nagar, A.; Mackinder, L. Deformed wing virus implicated in overwintering honeybee colony losses. Appl. Environ. Microbiol. 2009, 75, 7212–7220. [Google Scholar] [CrossRef] [Green Version]
- Berthoud, H.; Imdorf, A.; Haueter, M.; Radloff, S.; Neumann, P. Virus infections and winter losses of honey bee colonies (Apis mellifera). J. Apic. Res. 2010, 49, 60–65. [Google Scholar] [CrossRef]
- Iqbal, J.; Mueller, U. Virus infection causes specific learning deficits in honeybee foragers. Proc. R. Soc. B Biol. Sci. 2007, 274, 1517–1521. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.J. The role of varroa and viral pathogens in the collapse of honeybee colonies: A modelling approach. J. Appl. Ecol. 2001, 38, 1082–1093. [Google Scholar] [CrossRef] [Green Version]
- Peck, D.T.; Seeley, T.D. Mite bombs or robber lures? The roles of drifting and robbing in Varroa destructor transmission from collapsing honey bee colonies to their neighbors. PLoS ONE 2019, 14, e0218392. [Google Scholar] [CrossRef]
- Barroso-Arévalo, S.; Fernández-Carrión, E.; Goyache, J.; Molero, F.; Puerta, F.; Sánchez-Vizcaíno, J.M. High load of deformed wing virus and Varroa destructor infestation are related to weakness of honey bee colonies in Southern Spain. Front. Microbiol. 2019, 10, 1331. [Google Scholar] [CrossRef]
- Dainat, B.; Evans, J.D.; Chen, Y.P.; Gauthier, L.; Neumann, P. Predictive markers of honey bee colony collapse. PLoS ONE 2012, 7, e32151. [Google Scholar] [CrossRef] [Green Version]
- McMenamin, A.J.; Genersch, E. Honey bee colony losses and associated viruses. Curr. Opin. Insect Sci. 2015, 8, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Beaurepaire, A.; Piot, N.; Doublet, V.; Antunez, K.; Campbell, E.; Chantawannakul, P.; Chejanovsky, N.; Gajda, A.; Heerman, M.; Panziera, D.; et al. Diversity and global distribution of viruses of the western honey bee, Apis mellifera. Insects 2020, 11, 239. [Google Scholar] [CrossRef] [PubMed]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E. ICTV Virus Taxonomy Profile: Dicistroviridae. J. Gen. Virol. 2017, 98, 355. [Google Scholar] [CrossRef] [PubMed]
- McMahon, D.P.; Wilfert, L.; Paxton, R.J.; Brown, M.J.F. Emerging Viruses in Bees: From Molecules to Ecology. Adv. Virus Res. 2018, 101, 251–291. [Google Scholar] [CrossRef] [PubMed]
- Yañez, O.; Piot, N.; Dalmon, A.; de Miranda, J.R.; Chantawannakul, P.; Panziera, D.; Amiri, E.; Smagghe, G.; Schroeder, D.; Chejanovsky, N. Bee Viruses: Routes of Infection in Hymenoptera. Front. Microbiol. 2020, 11, 943. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Brettell, L.E.; Chejanovsky, N.; Childers, A.K.; Dalmon, A.; Deboutte, W.; de Graaf, D.C.; Doublet, V.; Gebremedhn, H.; Genersch, E.; et al. Cold case: The disappearance of Egypt bee virus, a fourth distinct master strain of deformed wing virus linked to honeybee mortality in 1970’s Egypt. Virol. J. 2022, 19, 12. [Google Scholar] [CrossRef]
- Mordecai, G.J.; Wilfert, L.; Martin, S.J.; Jones, I.M.; Schroeder, D.C. Diversity in a honey bee pathogen: First report of a third master variant of the Deformed Wing Virus quasispecies. ISME J. 2016, 10, 1264–1273. [Google Scholar] [CrossRef] [Green Version]
- Ryabov, E.V.; Wood, G.R.; Fannon, J.M.; Moore, J.D.; Bull, J.C.; Chandler, D.; Mead, A.; Burroughs, N.; Evans, D.J. A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees (Apis mellifera) Prevails after Varroa destructor-Mediated, or In Vitro, Transmission. PLoS Pathog. 2014, 10, e1004230. [Google Scholar] [CrossRef] [Green Version]
- Dalmon, A.; Desbiez, C.; Coulon, M.; Thomasson, M.; Le Conte, Y.; Alaux, C.; Vallon, J.; Moury, B. Evidence for positive selection and recombination hotspots in Deformed wing virus (DWV). Sci. Rep. 2017, 7, srep41045. [Google Scholar] [CrossRef] [Green Version]
- Moore, J.; Jironkin, A.; Chandler, D.; Burroughs, N.; Evans, D.J.; Ryabov, E.V. Recombinants between Deformed wing virus and Varroa destructor virus-1 may prevail in Varroa destructor-infested honeybee colonies. J. Gen. Virol. 2011, 92, 156–161. [Google Scholar] [CrossRef]
- Wang, H.; Xie, J.; Shreeve, T.G.; Ma, J.; Pallett, D.W.; King, L.A.; Possee, R.D. Sequence Recombination and Conservation of Varroa destructor Virus-1 and Deformed Wing Virus in Field Collected Honey Bees (Apis mellifera). PLoS ONE 2013, 8, e74508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zioni, N.; Soroker, V.; Chejanovsky, N. Replication of varroa destructor virus 1 (VDV-1) and a varroa destructor virus 1-deformed wing virus recombinant (VDV-1-DWV) in the head of the honey bee. Virology 2011, 417, 106–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- di Prisco, G.; Zhang, X.; Pennacchio, F.; Caprio, E.; Li, J.; Evans, J.D.; DeGrandi-Hoffman, G.; Hamilton, M.; Chen, Y.; Prisco, G.; et al. Dynamics of Persistent and Acute Deformed Wing Virus Infections in Honey Bees, Apis mellifera. Viruses 2011, 3, 2425–2441. [Google Scholar] [CrossRef] [Green Version]
- Glenny, W.; Cavigli, I.; Daughenbaugh, K.F.; Radford, R.; Kegley, S.E.; Flenniken, M.L. Honey bee (Apis mellifera) colony health and pathogen composition in migratory beekeeping operations involved in California almond pollination. PLoS ONE 2017, 12, e0128214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thaduri, S.; Marupakula, S.; Terenius, O.; Onorati, P.; Tellgren-Roth, C.; Locke, B.; de Miranda, J.R. Global similarity, and some key differences, in the metagenomes of Swedish varroa-surviving and varroa-susceptible honeybees. Sci. Rep. 2021, 11, 23214. [Google Scholar] [CrossRef] [PubMed]
- Moro, A.; Blacquière, T.; Panziera, D.; Dietemann, V.; Neumann, P. Host-Parasite Co-Evolution in Real-Time: Changes in Honey Bee Resistance Mechanisms and Mite Reproductive Strategies. Insects 2021, 12, 120. [Google Scholar] [CrossRef]
- Moro, A.; Blacquière, T.; Dahle, B.; Dietemann, V.; Le Conte, Y.; Locke, B.; Neumann, P.; Beaurepaire, A. Adaptive population structure shifts in invasive parasitic mites, Varroa destructor. Ecol. Evol. 2021, 11, 5937–5949. [Google Scholar] [CrossRef]
- Thaduri, S.; Stephan, J.G.; de Miranda, J.R.; Locke, B. Disentangling host-parasite-pathogen interactions in a varroa-resistant honeybee population reveals virus tolerance as an independent, naturally adapted survival mechanism. Sci. Rep. 2019, 9, 6221. [Google Scholar] [CrossRef] [Green Version]
- Locke, B. Natural Varroa mite-surviving Apis mellifera honeybee populations. Apidologie 2016, 47, 467–482. [Google Scholar] [CrossRef] [Green Version]
- Le Conte, Y.; Meixner, M.D.; Brandt, A.; Carreck, N.L.; Costa, C.; Mondet, F.; Büchler, R. Geographical distribution and selection of european honey bees resistant to varroa destructor. Insects 2020, 11, 873. [Google Scholar] [CrossRef]
- Thompson, J.N. The Geographic Mosaic of Coevolution; University of Chicago Press: Chicago, IL, USA, 2005. [Google Scholar]
- Locke, B.; Fries, I. Characteristics of honey bee colonies (Apis mellifera) in Sweden surviving Varroa destructor infestation. Apidologie 2011, 42, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Blacquière, T.; Boot, W.; Calis, J.; Moro, A.; Neumann, P.; Panziera, D. Darwinian black box selection for resistance to settled invasive Varroa destructor parasites in honey bees. Biol. Invasions 2019, 21, 2519–2528. [Google Scholar] [CrossRef] [Green Version]
- Locke, B.; Thaduri, S.; Stephan, J.G.; Low, M.; Blacquière, T.; Dahle, B.; Le Conte, Y.; Neumann, P.; de Miranda, J.R. Adapted tolerance to virus infections in four geographically distinct Varroa destructor-resistant honeybee populations. Sci. Rep. 2021, 11, 12359. [Google Scholar] [CrossRef] [PubMed]
- Locke, B.; Forsgren, E.; De Miranda, J.R. Increased tolerance and resistance to virus infections: A possible factor in the survival of Varroa destructor-resistant honey bees (Apis mellifera). PLoS ONE 2014, 9, e99998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amiri, E.; Strand, M.K.; Rueppell, O.; Tarpy, D.R. Queen quality and the impact of honey bee diseases on queen health: Potential for interactions between two major threats to colony health. Insects 2017, 8, 48. [Google Scholar] [CrossRef]
- Gauthier, L.; Ravallec, M.; Tournaire, M.; Cousserans, F.F.; Bergoin, M.; Dainat, B.; de Miranda, J.R. Viruses Associated with Ovarian Degeneration in Apis mellifera L. Queens. PLoS ONE 2011, 6, e16217. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Highfield, A.; Brettell, L.; Villalobos, E.; Budge, G.E.; Powell, M.; Nikaido, S.; Schroeder, D.C. Global honey bee viral landscape altered by a parasitic mite. Science 2012, 336, 1304–1306. [Google Scholar] [CrossRef]
- de Graaf, D.C.; Laget, D.; De Smet, L.; Claeys Boúúaert, D.; Brunain, M.; Veerkamp, R.F.; Brascamp, E.W. Heritability estimates of the novel trait ‘suppressed in ovo virus infection’ in honey bees (Apis mellifera). Sci. Rep. 2020, 10, 14310. [Google Scholar] [CrossRef]
- Claeys Bouuaert, D.; De Smet, L.; de Graaf, D.C. Breeding for virus resistance and its effects on deformed wing virus infection patterns in honey bee queens. Viruses 2021, 13, 1074. [Google Scholar] [CrossRef]
- Woyke, J. Drone Larvae from Fertilized Eggs of the Honeybee. J. Apic. Res. 2015, 2, 19–24. [Google Scholar] [CrossRef]
- Chen, Y.; Pettis, J.S.; Corona, M.; Chen, W.P.; Li, C.J.; Spivak, M.; Visscher, P.K.; DeGrandi-Hoffman, G.; Boncristiani, H.; Zhao, Y.; et al. Israeli Acute Paralysis Virus: Epidemiology, Pathogenesis and Implications for Honey Bee Health. PLoS Pathog. 2014, 10, e1004261. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Cui, L.; Ostiguy, N.; Cox-Foster, D. Intricate transmission routes and interactions between picorna-like viruses (Kashmir bee virus and sacbrood virus) with the honeybee host and the parasitic varroa mite. J. Gen. Virol. 2005, 86, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Amiri, E.; Kryger, P.; Meixner, M.D.; Strand, M.K.; Tarpy, D.R.; Rueppell, O. Quantitative patterns of vertical transmission of deformed wing virus in honey bees. PLoS ONE 2018, 13, e0195283. [Google Scholar] [CrossRef]
- Bradford, E.L.; Christie, C.R.; Campbell, E.M.; Bowman, A.S. A real-time PCR method for quantification of the total and major variant strains of the deformed wing virus. PLoS ONE 2017, 12, e0190017. [Google Scholar] [CrossRef] [Green Version]
- De Miranda, J.R.; Bailey, L.; Ball, B.V.; Blanchard, P.; Budge, G.E.; Chejanovsky, N.; Chen, Y.; Gauthier, L.; Genersch, E.; De Graaf, D.C.; et al. Standard methods for virus research in Apis mellifera. J. Apic. Res. 2013, 52, 1–56. [Google Scholar] [CrossRef] [Green Version]
- Yañez, O.; Chávez-Galarza, J.; Tellgren-Roth, C.; Pinto, M.A.; Neumann, P.; de Miranda, J.R. The honeybee (Apis mellifera) developmental state shapes the genetic composition of the deformed wing virus-A quasispecies during serial transmission. Sci. Rep. 2020, 10, 5956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, B.; Forsgren, E.; Fries, I.; De Miranda, J.R. Acaricide Treatment Affects Viral Dynamics in Varroa destructor-Infested Honey Bee Colonies via both Host Physiology and Mite Control. Appl. Env. Microbiol. 2012, 78, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Thaduri, S.; Locke, B.; Granberg, F.; De Miranda, J.R. Temporal changes in the viromes of swedish varroa-resistant and varroa-susceptible honeybee populations. PLoS ONE 2018, 13, e0206938. [Google Scholar] [CrossRef]
- Locke, B.; Semberg, E.; Forsgren, E.; De Miranda, J.R. Persistence of subclinical deformed wing virus infections in honeybees following Varroa mite removal and a bee population turnover. PLoS ONE 2017, 12, e0180910. [Google Scholar] [CrossRef] [Green Version]
- Bhatia, S.; Baral, S.; Vega Melendez, C.; Amiri, E.; Rueppell, O. Comparing survival of israeli acute paralysis virus infection among stocks of U.S. honey bees. Insects 2021, 12, 60. [Google Scholar] [CrossRef]
- Faurot-Daniels, C.; Glenny, W.; Daughenbaugh, K.F.; McMenamin, A.J.; Burkle, L.A.; Flenniken, M.L. Longitudinal monitoring of honey bee colonies reveals dynamic nature of virus abundance and indicates a negative impact of Lake Sinai virus 2 on colony health. PLoS ONE 2020, 15, e0237544. [Google Scholar] [CrossRef] [PubMed]
- Harizanis, P. Infestation of queen cells by the mite Varroa-Jacobsoni. Apidologie 1991, 22, 533–538. [Google Scholar] [CrossRef]
- Anderson, D.; Gibbs, A.J. Inapparent Virus Infections and their Interactions in Pupae of the Honey Bee (Apis mellifera Linnaeus) in Australia. J. Gen. Virol. 1988, 69, 1617–1625. [Google Scholar] [CrossRef]
- Chen, Y.; Pettis, J.S.; Collins, A.; Feldlaufer, M.F. Prevalence and transmission of honeybee viruses. Appl. Environ. Microbiol. 2006, 72, 606–611. [Google Scholar] [CrossRef] [Green Version]
- Gray, A.; Adjlane, N.; Arab, A.; Ballis, A.; Brusbardis, V.; Charrière, J.D.; Chlebo, R.; Coffey, M.F.; Cornelissen, B.; Amaro da Costa, C.; et al. Honey bee colony winter loss rates for 35 countries participating in the COLOSS survey for winter 2018–2019, and the effects of a new queen on the risk of colony winter loss. Apidologie 2020, 59, 744–751. [Google Scholar] [CrossRef]
- Ravoet, J.; Maharramov, J.; Meeus, I.; De Smet, L.; Wenseleers, T.; Smagghe, G.; de Graaf, D.C. Comprehensive Bee Pathogen Screening in Belgium Reveals Crithidia mellificae as a New Contributory Factor to Winter Mortality. PLoS ONE 2013, 8, e72443. [Google Scholar] [CrossRef] [Green Version]
- Francis, R.M.; Nielsen, S.L.; Kryger, P. Patterns of viral infection in honey bee queens. J. Gen. Virol. 2013, 94, 668–676. [Google Scholar] [CrossRef]
- de Miranda, J.R.; Genersch, E. Deformed wing virus. J. Invertebr. Pathol. 2010, 103, S48–S61. [Google Scholar] [CrossRef]
- Genersch, E.; Aubert, M. Emerging and re-emerging viruses of the honey bee ( Apis mellifera L.). Vet. Res. 2010, 41, 54. [Google Scholar] [CrossRef] [Green Version]
- de Miranda, J.R.; Fries, I. Venereal and vertical transmission of deformed wing virus in honeybees (Apis mellifera L.). J. Invertebr. Pathol. 2008, 98, 184–189. [Google Scholar] [CrossRef]
- Li, J.; Wang, T.; Evans, J.D.; Rose, R.; Zhao, Y.; Li, Z.; Li, J.; Huang, S.; Heerman, M.; Rodríguez-García, C.; et al. The phylogeny and pathogenesis of sacbrood virus (SBV) infection in European honey bees, Apis mellifera. Viruses 2019, 11, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandrock, C.; Tanadini, M.; Tanadini, L.G.; Fauser-Misslin, A.; Potts, S.G.; Neumann, P. Impact of chronic neonicotinoid exposure on honeybee colony performance and queen supersedure. PLoS ONE 2014, 9, e103592. [Google Scholar] [CrossRef] [Green Version]
- Di Prisco, G.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; Evans, J.D.; Chen, Y.; Prisco, G.D.; Pennacchio, F.; Caprio, E.; Boncristiani, H.F.; et al. Varroa destructor is an effective vector of Israeli acute paralysis virus in the honeybee, Apis mellifera. J. Gen. Virol. 2011, 92, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Traynor, K.S.; Rennich, K.; Forsgren, E.; Rose, R.; Pettis, J.; Kunkel, G.; Madella, S.; Evans, J.; Lopez, D.; VanEngelsdorp, D. Multiyear survey targeting disease incidence in US honey bees. Apidologie 2016, 47, 325–347. [Google Scholar] [CrossRef] [Green Version]
- Allsopp, M.H. Analysis of Varroa Destructor Infestation of Southern African Honeybee Populations. Ph.D. Thesis, University of Pretoria, Pretoria, South Africa, 2006. [Google Scholar]
- Tentcheva, D.; Gauthier, L.; Zappulla, N.; Dainat, B.; Cousserans, F.; Colin, M.E.; Bergoin, M. Prevalence and seasonal variations of six bee viruses in Apis mellifera L. and Varroa destructor mite populations in France. Appl. Environ. Microbiol. 2004, 70, 7185–7191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, C.; Rivkin, H.; Slabezki, Y.; Chejanovsky, N. Dynamics of the presence of israeli acute paralysis virus in honey bee colonies with colony collapse disorder. Viruses 2014, 6, 2012–2027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blažytė-Čereškienė, L.; Skrodenytė-Arbačiauskienė, V.; Radžiutė, S.; Čepulytė-Rakauskienė, R.; Apšegaitė, V.; Būda, V. A three-year survey of honey bee viruses in Lithuania. J. Apic. Res. 2016, 55, 176–184. [Google Scholar] [CrossRef]
- Porrini, C.; Mutinelli, F.; Bortolotti, L.; Granato, A.; Laurenson, L.; Roberts, K.; Gallina, A.; Silvester, N.; Medrzycki, P.; Renzi, T.; et al. The status of honey bee health in Italy: Results from the nationwide bee monitoring network. PLoS ONE 2016, 11, e0155411. [Google Scholar] [CrossRef] [Green Version]
- Walton, A.; Toth, A.L.; Dolezal, A.G. Developmental environment shapes honeybee worker response to virus infection. Sci. Rep. 2021, 11, 13961. [Google Scholar] [CrossRef]
- Dolezal, A.G.; Carrillo-Tripp, J.; Judd, T.M.; Allen Miller, W.; Bonning, B.C. Interacting stressors matter: Diet quality and virus infection in honeybee health. R. Soc. OPEN Sci. 2019, 6, 181803. [Google Scholar] [CrossRef]
- Seeley, T.D.; Smith, M.L. Crowding honeybee colonies in apiaries can increase their vulnerability to the deadly ectoparasite Varroa destructor. Apidologie 2015, 46, 716–727. [Google Scholar] [CrossRef] [Green Version]
- Brosi, B.J.; Delaplane, K.S.; Boots, M.; De Roode, J.C. Ecological and evolutionary approaches to managing honeybee disease. Nat. Ecol. Evol. 2017, 1, 1250–1262. [Google Scholar] [CrossRef]
- Lloyd-Smith, J.O.; Cross, P.C.; Briggs, C.J.; Daugherty, M.; Getz, W.M.; Latto, J.; Sanchez, M.S.; Smith, A.B.; Swei, A. Should we expect population thresholds for wildlife disease? Trends Ecol. Evol. 2005, 20, 511–519. [Google Scholar] [CrossRef] [PubMed]
- Keesing, F.; Belden, L.K.; Daszak, P.; Dobson, A.; Harvell, C.D.; Holt, R.D.; Hudson, P.; Jolles, A.; Jones, K.E.; Mitchell, C.E.; et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 2010, 468, 647–652. [Google Scholar] [CrossRef] [PubMed]
- Civitello, D.J.; Cohen, J.; Fatima, H.; Halstead, N.T.; Liriano, J.; McMahon, T.A.; Ortega, C.N.; Sauer, E.L.; Sehgal, T.; Young, S.; et al. Biodiversity inhibits parasites: Broad evidence for the dilution effect. Proc. Natl. Acad. Sci. USA 2015, 112, 8667–8671. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Chen, H.; Fan, J.X.; Wang, Y.; Li, Y.; Chen, J.; Fan, J.X.; Yang, S.; Hu, L.; Leung, H.; et al. Genetic diversity and disease control in rice. Nature 2000, 406, 718–722. [Google Scholar] [CrossRef]
- Mundt, C.C. Use of multiline cultivars and cultivar mixtures for disease management. Annu. Rev. Entomol. 2002, 40, 381–410. [Google Scholar] [CrossRef] [Green Version]
- Beaurepaire, A.; Sann, C.; Arredondo, D.; Mondet, F.; Le Conte, Y. Behavioral Genetics of the Interactions between Apis mellifera and Varroa destructor. Insects 2019, 10, 299. [Google Scholar] [CrossRef] [Green Version]
- Bartlett, L.J.; Boots, M.; Brosi, B.J.; De Roode, J.C.; Delaplane, K.S.; Hernandez, C.A.; Wilfert, L. Persistent effects of management history on honeybee colony virus abundances. J. Invertebr. Pathol. 2021, 179, 107520. [Google Scholar] [CrossRef]
- Francis, R.M.; Amiri, E.; Meixner, M.D.; Kryger, P.; Gajda, A.; Andonov, S.; Uzunov, A.; Topolska, G.; Charistos, L.; Costa, C.; et al. Effect of genotype and environment on parasite and pathogen levels in one apiary—A case study. J. Apic. Res. 2015, 53, 230–232. [Google Scholar] [CrossRef]
- Brutscher, L.M.; Daughenbaugh, K.F.; Flenniken, M.L. Antiviral defense mechanisms in honey bees. Curr. Opin. Insect Sci. 2015, 10, 71–82. [Google Scholar] [CrossRef] [Green Version]
- Di Prisco, G.; Cavaliere, V.; Annoscia, D.; Varricchio, P.; Caprio, E.; Nazzi, F.; Gargiulo, G.; Pennacchio, F. Neonicotinoid clothianidin adversely affects insect immunity and promotes replication of a viral pathogen in honey bees. Proc. Natl. Acad. Sci. USA 2013, 110, 18466–18471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coulon, M.; Dalmon, A.; Di Prisco, G.; Prado, A.; Arban, F.; Dubois, E.; Ribière-Chabert, M.; Alaux, C.; Thiéry, R.; Le Conte, Y. Interactions Between Thiamethoxam and Deformed Wing Virus Can Drastically Impair Flight Behavior of Honey Bees. Front. Microbiol. 2020, 11, 766. [Google Scholar] [CrossRef] [PubMed]
- Kurze, C.; Routtu, J.; Moritz, R.F.A. Parasite resistance and tolerance in honeybees at the individual and social level. Zoology 2016, 119, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Gregorc, A.; Bakonyi, T. Viral infections in queen bees (Apis mellifera carnica) from rearing apiaries. Acta Vet. Brno 2012, 81, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Žvokelj, L.; Bakonyi, T.; Korošec, T.; Gregorc, A. Appearance of acute bee paralysis virus, black queen cell virus and deformed wing virus in Carnolian honey bee (Apis mellifera carnica) queen rearing. J. Apic. Res. 2020, 59, 53–58. [Google Scholar] [CrossRef]
- Amiri, E.; Meixner, M.D.; Kryger, P. Deformed wing virus can be transmitted during natural mating in honey bees and infect the queens. Sci. Rep. 2016, 6, 33065. [Google Scholar] [CrossRef] [Green Version]
- Gregorc, A.; Smodiš Škerl, M.I. Characteristics of honey bee (Apis mellifera carnica, Pollman 1879) queens reared in Slovenian commercial breeding stations. J. Apic. Sci. 2015, 59, 5–12. [Google Scholar] [CrossRef] [Green Version]
- Chaimanee, V.; Chantawannakul, P.; Chen, Y.; Evans, J.D.; Pettis, J.S. Effects of host age on susceptibility to infection and immune gene expression in honey bee queens (Apis mellifera) inoculated with Nosema ceranae. Apidologie 2014, 45, 451–463. [Google Scholar] [CrossRef] [Green Version]
- Van Der Zee, R.; Gray, A.; Holzmann, C.; Pisa, L.; Brodschneider, R.; Chlebo, R.; Coffey, M.F.; Kence, A.; Kristiansen, P.; Mutinelli, F.; et al. Standard survey methods for estimating colony losses and explanatory risk factors in Apis mellifera. J. Apic. Res. 2013, 52, 1–36. [Google Scholar] [CrossRef]


| Country | Sampling Season | NSC/TMC | No. of Sampled Colonies | No. of Virus-Free Samples | No. of Samples Positive For: | Mean Infection Load (Log10/Egg) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DWV | BQCV | SBV | ABPV | DWV | BQCV | SBV | ABPV | |||||
| Belgium | Spring | NSC | 10 | 1 | 9 (90%) | 0 | 0 | 0 | 4.3 | |||
| TMC | 11 | 4 | 7 (64%) | 2 (18%) | 0 | 0 | 6.1 | 4.2 | ||||
| Croatia | Summer | TMC | 10 | 3 | 4 (40%) | 3 (30%) | 1 (10%) | 0 | 4.7 | 5.3 | 3.3 | |
| France | Spring | NSC | 13 | 2 | 7 (54%) | 11 (85%) | 0 | 0 | 5.6 | 5.5 | ||
| TMC | 10 | 2 | 1 (10%) | 8 (80%) | 0 | 0 | 5.8 | 6.4 | ||||
| the Netherlands | Partly in spring and summer | NSC | 10 | 2 | 8 (80%) | 3 (30%) | 0 | 0 | 6.2 | 5.4 | ||
| TMC | 6 | 1 | 5 (83%) | 3 (50%) | 0 | 0 | 5.4 | 4.9 | ||||
| Norway | Summer | NSC | 10 | 1 | 9 (90%) | 1 (10%) | 0 | 0 | 5.1 | 4.9 | ||
| TMC | 10 | 5 | 4 (40%) | 3 (30%) | 0 | 0 | 4.3 | 4.7 | ||||
| Portugal | Spring | TMC | 10 | 1 | 8 (80%) | 1 (10%) | 0 | 0 | 5.1 | 3.3 | ||
| Romania | Spring | NSC | 4 | 1 | 0 | 3 (75%) | 0 | 0 | 4.5 | |||
| TMC | 9 | 4 | 2 (22%) | 4 (44%) | 0 | 0 | 4.0 | 4.0 | ||||
| Slovenia | Spring | TMC | 72 | 27 | 38 (53%) | 11 (15%) | 2 (2%) | 2 (2%) | 5.3 | 5.2 | 3.2 | 3.8 |
| Spain | Spring | TMC | 10 | 4 | 5 (50%) | 1 (10%) | 0 | 0 | 5.5 | 4.8 | ||
| Sweden | Summer | NSC | 6 | 5 | 1 (16%) | 0 | 0 | 0 | 5.0 | |||
| TMC | 12 | 11 | 1 (8%) | 0 | 0 | 0 | 4.3 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Claeys Bouuaert, D.; De Smet, L.; Brunain, M.; Dahle, B.; Blacquière, T.; Dalmon, A.; Dezmirean, D.; Elen, D.; Filipi, J.; Giurgiu, A.; et al. Virus Prevalence in Egg Samples Collected from Naturally Selected and Traditionally Managed Honey Bee Colonies across Europe. Viruses 2022, 14, 2442. https://doi.org/10.3390/v14112442
Claeys Bouuaert D, De Smet L, Brunain M, Dahle B, Blacquière T, Dalmon A, Dezmirean D, Elen D, Filipi J, Giurgiu A, et al. Virus Prevalence in Egg Samples Collected from Naturally Selected and Traditionally Managed Honey Bee Colonies across Europe. Viruses. 2022; 14(11):2442. https://doi.org/10.3390/v14112442
Chicago/Turabian StyleClaeys Bouuaert, David, Lina De Smet, Marleen Brunain, Bjørn Dahle, Tjeerd Blacquière, Anne Dalmon, Daniel Dezmirean, Dylan Elen, Janja Filipi, Alexandru Giurgiu, and et al. 2022. "Virus Prevalence in Egg Samples Collected from Naturally Selected and Traditionally Managed Honey Bee Colonies across Europe" Viruses 14, no. 11: 2442. https://doi.org/10.3390/v14112442
APA StyleClaeys Bouuaert, D., De Smet, L., Brunain, M., Dahle, B., Blacquière, T., Dalmon, A., Dezmirean, D., Elen, D., Filipi, J., Giurgiu, A., Gregorc, A., Kefuss, J., Locke, B., de Miranda, J. R., Oddie, M., Panziera, D., Parejo, M., Pinto, M. A., & de Graaf, D. C. (2022). Virus Prevalence in Egg Samples Collected from Naturally Selected and Traditionally Managed Honey Bee Colonies across Europe. Viruses, 14(11), 2442. https://doi.org/10.3390/v14112442












