Challenges and Opportunities for Global Genomic Surveillance Strategies in the COVID-19 Era
Abstract
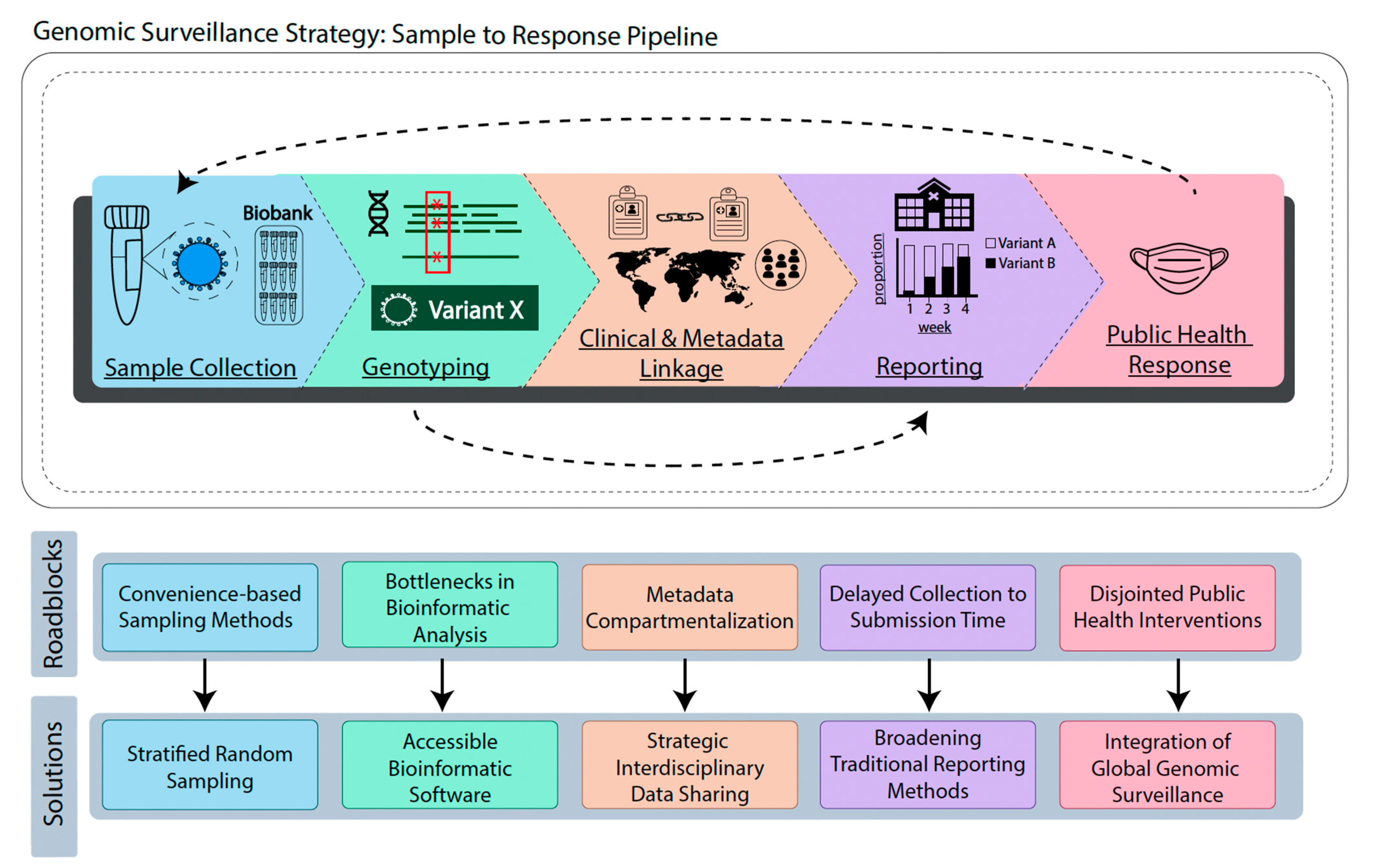
:1. Introduction
2. Sample Collection
3. Genotyping
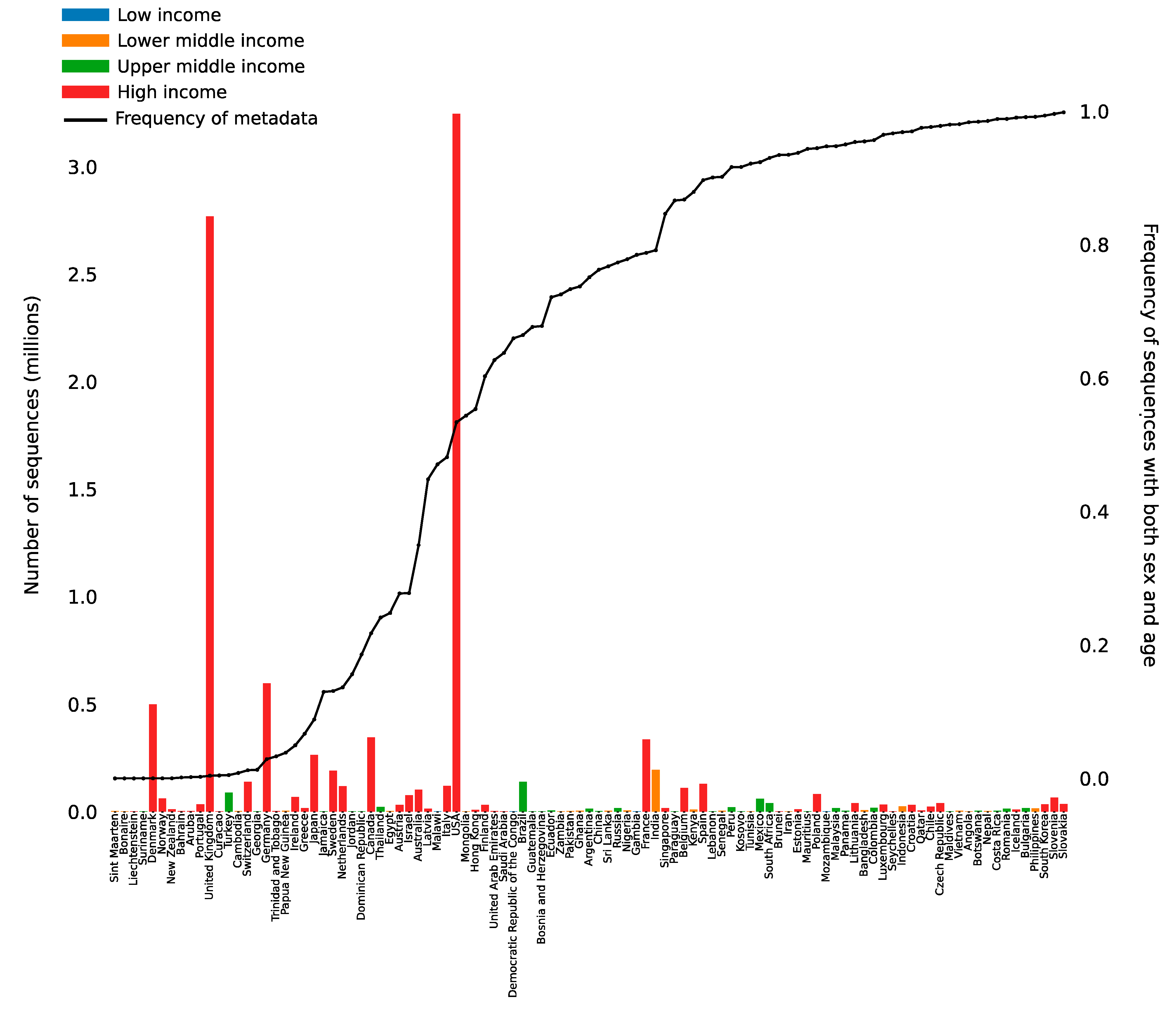
4. Clinical and Metadata Linkage
5. Reporting
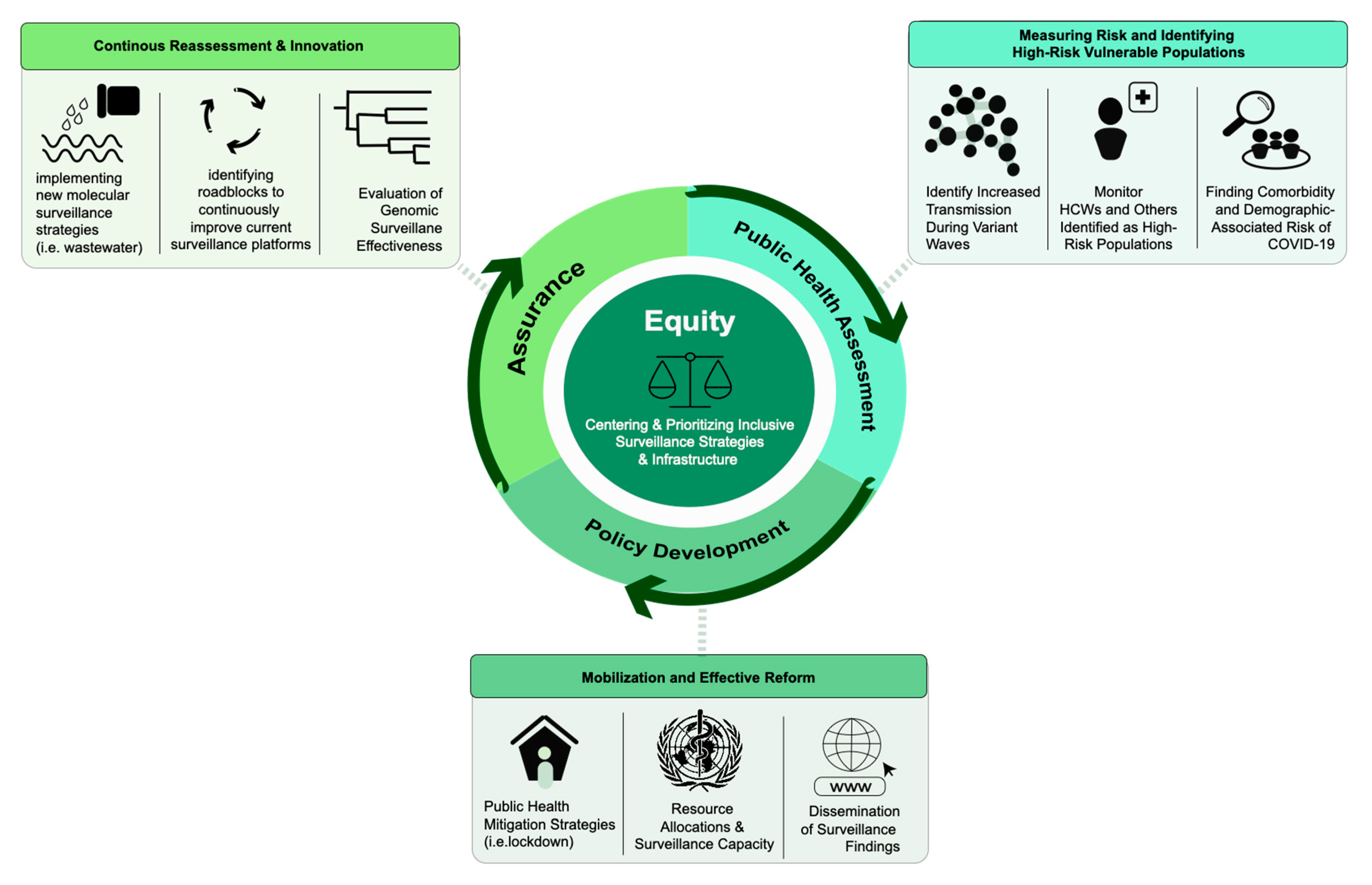
6. Public Health Response
6.1. Assessment
6.2. Policy Development
6.3. Assurance
6.4. Equity
7. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 17 August 2022).
- Hou, Y.J.; Chiba, S.; Halfmann, P.; Ehre, C.; Kuroda, M.; Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Nakajima, N.; Takahashi, K.; et al. SARS-CoV-2 D614G Variant Exhibits Efficient Replication Ex Vivo and Transmission in Vivo. Science 2020, 370, 1464–1468. [Google Scholar] [CrossRef] [PubMed]
- Lythgoe, K.A.; Hall, M.; Ferretti, L.; de Cesare, M.; MacIntyre-Cockett, G.; Trebes, A.; Andersson, M.; Otecko, N.; Wise, E.L.; Moore, N.; et al. SARS-CoV-2 within-Host Diversity and Transmission. Science 2021, 372, eabg0821. [Google Scholar] [CrossRef] [PubMed]
- Tracking SARS-CoV-2 Variants. Available online: https://www.who.int/health-topics/typhoid/tracking-SARS-CoV-2-variants (accessed on 16 February 2022).
- CDC. What Is Genomic Surveillance? Available online: https://www.cdc.gov/coronavirus/2019-ncov/variants/genomic-surveillance.html (accessed on 12 October 2022).
- Smith, G.J.D.; Vijaykrishna, D.; Bahl, J.; Lycett, S.J.; Worobey, M.; Pybus, O.G.; Ma, S.K.; Cheung, C.L.; Raghwani, J.; Bhatt, S.; et al. Origins and Evolutionary Genomics of the 2009 Swine-Origin H1N1 Influenza A Epidemic. Nature 2009, 459, 1122–1125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gire, S.K.; Goba, A.; Andersen, K.G.; Sealfon, R.S.G.; Park, D.J.; Kanneh, L.; Jalloh, S.; Momoh, M.; Fullah, M.; Dudas, G.; et al. Genomic Surveillance Elucidates Ebola Virus Origin and Transmission during the 2014 Outbreak. Science 2014, 345, 1369–1372. [Google Scholar] [CrossRef] [Green Version]
- Coll, F.; Harrison, E.M.; Toleman, M.S.; Reuter, S.; Raven, K.E.; Blane, B.; Palmer, B.; Kappeler, A.R.M.; Brown, N.M.; Török, M.E.; et al. Longitudinal Genomic Surveillance of MRSA in the UK Reveals Transmission Patterns in Hospitals and the Community. Sci. Transl. Med. 2017, 9, eaak9745. [Google Scholar] [CrossRef] [Green Version]
- Faria, N.R.; Azevedo, R.; Kraemer, M.U.G.; Souza, R.; Cunha, M.S.; Hill, S.C.; Thézé, J.; Bonsall, M.B.; Bowden, T.A.; Rissanen, I.; et al. Zika Virus in the Americas: Early Epidemiological and Genetic Findings. Science 2016, 352, 345–349. [Google Scholar] [CrossRef] [Green Version]
- Hemelaar, J.; Elangovan, R.; Yun, J.; Dickson-Tetteh, L.; Fleminger, I.; Kirtley, S.; Williams, B.; Gouws-Williams, E.; Ghys, P.D.; Abimiku, A.G.; et al. Global and Regional Molecular Epidemiology of HIV-1, 1990–2015: A Systematic Review, Global Survey, and Trend Analysis. Lancet Infect. Dis. 2019, 19, 143–155. [Google Scholar] [CrossRef]
- CDC. Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (accessed on 12 October 2022).
- Zerbinati, R.M.; Palmieri, M.; Schwab, G.; Felix, A.C.; Martinho, H.; Giannecchini, S.; To, K.K.-W.; Lindoso, J.A.L.; Romano, C.M.; Braz-Silva, P.H. Use of Saliva and RT-PCR Screening for SARS-CoV-2 Variants of Concern: Surveillance and Monitoring. J. Med. Virol. 2022, 94, 4518–4521. [Google Scholar] [CrossRef]
- Wang, Y.; Upadhyay, A.; Pillai, S.; Khayambashi, P.; Tran, S.D. Saliva as a Diagnostic Specimen for SARS-CoV-2 Detection: A Scoping Review. Oral Dis. 2022, 28, 2362–2390. [Google Scholar] [CrossRef]
- Stratton, S.J. Population Research: Convenience Sampling Strategies. Prehospital Disaster Med. 2021, 36, 373–374. [Google Scholar] [CrossRef]
- Goswami, C.; Sheldon, M.; Bixby, C.; Keddache, M.; Bogdanowicz, A.; Wang, Y.; Schultz, J.; McDevitt, J.; LaPorta, J.; Kwon, E.; et al. Identification of SARS-CoV-2 Variants Using Viral Sequencing for the Centers for Disease Control and Prevention Genomic Surveillance Program. BMC Infect. Dis. 2022, 22, 404. [Google Scholar] [CrossRef] [PubMed]
- Hill, V.; Ruis, C.; Bajaj, S.; Pybus, O.G.; Kraemer, M.U.G. Progress and Challenges in Virus Genomic Epidemiology. Trends Parasitol. 2021, 37, 1038–1049. [Google Scholar] [CrossRef] [PubMed]
- Griffith, G.J.; Morris, T.T.; Tudball, M.J.; Herbert, A.; Mancano, G.; Pike, L.; Sharp, G.C.; Sterne, J.; Palmer, T.M.; Davey Smith, G.; et al. Collider Bias Undermines Our Understanding of COVID-19 Disease Risk and Severity. Nat. Commun. 2020, 11, 5749. [Google Scholar] [CrossRef] [PubMed]
- Zejda, J.E.; Brożek, G.M.; Kowalska, M.; Barański, K.; Kaleta-Pilarska, A.; Nowakowski, A.; Xia, Y.; Buszman, P. Seroprevalence of Anti-SARS-CoV-2 Antibodies in a Random Sample of Inhabitants of the Katowice Region, Poland. Int. J. Environ. Res. Public Health 2021, 18, 3188. [Google Scholar] [CrossRef]
- Pollán, M.; Pérez-Gómez, B.; Pastor-Barriuso, R.; Oteo, J.; Hernán, M.A.; Pérez-Olmeda, M.; Sanmartín, J.L.; Fernández-García, A.; Cruz, I.; Fernández de Larrea, N.; et al. Prevalence of SARS-CoV-2 in Spain (ENE-COVID): A Nationwide, Population-Based Seroepidemiological Study. Lancet Lond. Engl. 2020, 396, 535–544. [Google Scholar] [CrossRef]
- Suhail, Y.; Afzal, J. Incorporating and Addressing Testing Bias within Estimates of Epidemic Dynamics for SARS-CoV-2. BMC Med. Res. Methodol. 2021, 21, 11. [Google Scholar] [CrossRef]
- CDC. Guidance for Antigen Testing for SARS-CoV-2 for Healthcare Providers Testing Individuals in the Community. Available online: https://www.cdc.gov/coronavirus/2019-ncov/lab/resources/antigen-tests-guidelines.html (accessed on 12 October 2022).
- McLaughlin, K. Growing Use of Home COVID-19 Tests Leaves Health Agencies in the Dark about Unreported Cases. 2021. Available online: https://www.statnews.com/2021/12/07/growing-use-of-home-covid19-tests-leaves-health-agencies-in-the-dark/ (accessed on 12 October 2022).
- Anthes, E. Why Didn’t the U.S. Detect Omicron Cases Sooner? The New York Times, 2 December 2021. Available online: https://www.nytimes.com/2021/12/02/health/omicron-variant-genetic-surveillance.html (accessed on 12 October 2022).
- Andrews, N.; Tessier, E.; Stowe, J.; Gower, C.; Kirsebom, F.; Simmons, R.; Gallagher, E.; Thelwall, S.; Groves, N.; Dabrera, G.; et al. Duration of Protection against Mild and Severe Disease by COVID-19 Vaccines. N. Engl. J. Med. 2022, 386, 340–350. [Google Scholar] [CrossRef]
- Cohn, B.A.; Cirillo, P.M.; Murphy, C.C.; Krigbaum, N.Y.; Wallace, A.W. SARS-CoV-2 Vaccine Protection and Deaths among US Veterans during 2021. Science 2022, 375, 331–336. [Google Scholar] [CrossRef]
- Tenforde, M.W.; Self, W.H.; Adams, K.; Gaglani, M.; Ginde, A.A.; McNeal, T.; Ghamande, S.; Douin, D.J.; Talbot, H.K.; Casey, J.D.; et al. Association Between MRNA Vaccination and COVID-19 Hospitalization and Disease Severity. JAMA 2021, 326, 2043–2054. [Google Scholar] [CrossRef]
- CDC. National Wastewater Surveillance System. Available online: https://www.cdc.gov/healthywater/surveillance/wastewater-surveillance/wastewater-surveillance.html (accessed on 12 October 2022).
- Vo, V.; Tillett, R.L.; Papp, K.; Shen, S.; Gu, R.; Gorzalski, A.; Siao, D.; Markland, R.; Chang, C.-L.; Baker, H.; et al. Use of Wastewater Surveillance for Early Detection of Alpha and Epsilon SARS-CoV-2 Variants of Concern and Estimation of Overall COVID-19 Infection Burden. Sci. Total Environ. 2022, 835, 155410. [Google Scholar] [CrossRef]
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516. [Google Scholar] [CrossRef]
- Simeon-Dubach, D.; Henderson, M.K. Opportunities and Risks for Research Biobanks in the COVID-19 Era and Beyond. Biopreserv. Biobanking 2020, 18, 503–510. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Boeras, D.; Wilder-Smith, A.; Sall, A.; Nkengasong, J. Need for Sustainable Biobanking Networks for COVID-19 and Other Diseases of Epidemic Potential. Lancet Infect. Dis. 2020, 20, e268–e273. [Google Scholar] [CrossRef]
- Abdaljaleel, M.; Singer, E.J.; Yong, W.H. Sustainability in Biobanking. Methods Mol. Biol. Clifton NJ 2019, 1897, 1–6. [Google Scholar] [CrossRef]
- Abdelhafiz, A.S.; Ahram, M.; Ibrahim, M.E.; Elgamri, A.; Gamel, E.; Labib, R.; Silverman, H. Biobanks in the Low- and Middle-Income Countries of the Arab Middle East Region: Challenges, Ethical Issues, and Governance Arrangements-a Qualitative Study Involving Biobank Managers. BMC Med. Ethics 2022, 23, 83. [Google Scholar] [CrossRef]
- Matharoo-Ball, B.; Diop, M.; Kozlakidis, Z. Harmonizing the COVID-19 Sample Biobanks: Barriers and Opportunities for Standards, Best Practices and Networks. Biosaf. Health 2022, 4, 280–282. [Google Scholar] [CrossRef]
- Final Report Summary—ERINHA (European Research Infrastructure on Highly Pathogenic Agents)|FP7|CORDIS|European Commission. Available online: https://cordis.europa.eu/project/id/262042/reporting (accessed on 12 October 2022).
- Coutard, B.; Romette, J.-L.; Miyauchi, K.; Charrel, R.; Prat, C.M.A.; EVA Zika Workgroup; EVA COVID-19 Workgroup. The Importance of Biobanking for Response to Pandemics Caused by Emerging Viruses: The European Virus Archive As an Observatory of the Global Response to the Zika Virus and COVID-19 Crisis. Biopreserv. Biobanking 2020, 18, 561–569. [Google Scholar] [CrossRef]
- Romette, J.L.; Prat, C.M.; Gould, E.A.; de Lamballerie, X.; Charrel, R.; Coutard, B.; Fooks, A.R.; Bardsley, M.; Carroll, M.; Drosten, C.; et al. The European Virus Archive Goes Global: A Growing Resource for Research. Antivir. Res. 2018, 158, 127–134. [Google Scholar] [CrossRef]
- Harris, J.R.; Burton, P.; Knoppers, B.M.; Lindpaintner, K.; Bledsoe, M.; Brookes, A.J.; Budin-Ljøsne, I.; Chisholm, R.; Cox, D.; Deschênes, M.; et al. Toward a Roadmap in Global Biobanking for Health. Eur. J. Hum. Genet. 2012, 20, 1105–1111. [Google Scholar] [CrossRef]
- Rambaut, A.; Holmes, E.C.; O’Toole, Á.; Hill, V.; McCrone, J.T.; Ruis, C.; du Plessis, L.; Pybus, O.G. A Dynamic Nomenclature Proposal for SARS-CoV-2 Lineages to Assist Genomic Epidemiology. Nat. Microbiol. 2020, 5, 1403–1407. [Google Scholar] [CrossRef]
- O’Toole, Á.; Scher, E.; Underwood, A.; Jackson, B.; Hill, V.; McCrone, J.T.; Colquhoun, R.; Ruis, C.; Abu-Dahab, K.; Taylor, B.; et al. Assignment of Epidemiological Lineages in an Emerging Pandemic Using the Pangolin Tool. Virus Evol. 2021, 7, veab064. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, E.; Giovanetti, M.; Tegally, H.; San, J.E.; Lessells, R.; Cuadros, D.; Martin, D.P.; Rasmussen, D.A.; Zekri, A.-R.N.; Sangare, A.K.; et al. A Year of Genomic Surveillance Reveals How the SARS-CoV-2 Pandemic Unfolded in Africa. Science 2021, 374, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Campbell, F.; Archer, B.; Laurenson-Schafer, H.; Jinnai, Y.; Konings, F.; Batra, N.; Pavlin, B.; Vandemaele, K.; Van Kerkhove, M.D.; Jombart, T.; et al. Increased Transmissibility and Global Spread of SARS-CoV-2 Variants of Concern as at June 2021. Eurosurveillance 2021, 26, 2100509. [Google Scholar] [CrossRef]
- Yu, H.; Chen, Z.; Azman, A.; Chen, X.; Zou, J.; Tian, Y.; Sun, R.; Xu, X.; Wu, Y.; Lu, W.; et al. Global Landscape of SARS-CoV-2 Genomic Surveillance, Public Availability Extent of Genomic Data, and Epidemic Shaped by Variants. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Esper, F.P.; Cheng, Y.-W.; Adhikari, T.M.; Tu, Z.J.; Li, D.; Li, E.A.; Farkas, D.H.; Procop, G.W.; Ko, J.S.; Chan, T.A.; et al. Genomic Epidemiology of SARS-CoV-2 Infection During the Initial Pandemic Wave and Association With Disease Severity. JAMA Netw. Open 2021, 4, e217746. [Google Scholar] [CrossRef] [PubMed]
- Bull, R.A.; Adikari, T.N.; Ferguson, J.M.; Hammond, J.M.; Stevanovski, I.; Beukers, A.G.; Naing, Z.; Yeang, M.; Verich, A.; Gamaarachchi, H.; et al. Analytical Validity of Nanopore Sequencing for Rapid SARS-CoV-2 Genome Analysis. Nat. Commun. 2020, 11, 6272. [Google Scholar] [CrossRef]
- Kames, J.; Holcomb, D.D.; Kimchi, O.; DiCuccio, M.; Hamasaki-Katagiri, N.; Wang, T.; Komar, A.A.; Alexaki, A.; Kimchi-Sarfaty, C. Sequence Analysis of SARS-CoV-2 Genome Reveals Features Important for Vaccine Design. Sci. Rep. 2020, 10, 15643. [Google Scholar] [CrossRef]
- Gonzalez-Reiche, A.S.; Hernandez, M.M.; Sullivan, M.J.; Ciferri, B.; Alshammary, H.; Obla, A.; Fabre, S.; Kleiner, G.; Polanco, J.; Khan, Z.; et al. Introductions and Early Spread of SARS-CoV-2 in the New York City Area. Science 2020, 369, 297–301. [Google Scholar] [CrossRef]
- Liu, T.; Chen, Z.; Chen, W.; Chen, X.; Hosseini, M.; Yang, Z.; Li, J.; Ho, D.; Turay, D.; Gheorghe, C.P.; et al. A Benchmarking Study of SARS-CoV-2 Whole-Genome Sequencing Protocols Using COVID-19 Patient Samples. iScience 2021, 24, 102892. [Google Scholar] [CrossRef]
- Lam, C.; Gray, K.; Gall, M.; Sadsad, R.; Arnott, A.; Johnson-Mackinnon, J.; Fong, W.; Basile, K.; Kok, J.; Dwyer, D.E.; et al. SARS-CoV-2 Genome Sequencing Methods Differ in Their Abilities To Detect Variants from Low-Viral-Load Samples. J. Clin. Microbiol. 2021, 59, e0104621. [Google Scholar] [CrossRef]
- Lambisia, A.W.; Mohammed, K.S.; Makori, T.O.; Ndwiga, L.; Mburu, M.W.; Morobe, J.M.; Moraa, E.O.; Musyoki, J.; Murunga, N.; Mwangi, J.N.; et al. Optimization of the SARS-CoV-2 ARTIC Network V4 Primers and Whole Genome Sequencing Protocol. Front. Med. 2022, 9, 836728. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence That D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Gupta, A.K.; Kumar, M. Benchmarking and Assessment of Eight De Novo Genome Assemblers on Viral Next-Generation Sequencing Data, Including the SARS-CoV-2. OMICS J. Integr. Biol. 2022, 26, 372–381. [Google Scholar] [CrossRef]
- Jacot, D.; Pillonel, T.; Greub, G.; Bertelli, C. Assessment of SARS-CoV-2 Genome Sequencing: Quality Criteria and Low-Frequency Variants. J. Clin. Microbiol. 2021, 59, e00944-21. [Google Scholar] [CrossRef] [PubMed]
- Popa, A.; Genger, J.-W.; Nicholson, M.D.; Penz, T.; Schmid, D.; Aberle, S.W.; Agerer, B.; Lercher, A.; Endler, L.; Colaço, H.; et al. Genomic Epidemiology of Superspreading Events in Austria Reveals Mutational Dynamics and Transmission Properties of SARS-CoV-2. Sci. Transl. Med. 2020, 12, eabe2555. [Google Scholar] [CrossRef] [PubMed]
- Kubik, S.; Marques, A.C.; Xing, X.; Silvery, J.; Bertelli, C.; De Maio, F.; Pournaras, S.; Burr, T.; Duffourd, Y.; Siemens, H.; et al. Recommendations for Accurate Genotyping of SARS-CoV-2 Using Amplicon-Based Sequencing of Clinical Samples. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 27, 1036.e1–1036.e8. [Google Scholar] [CrossRef]
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef] [Green Version]
- Lim, H.G.-M.; Hsiao, S.-H.; Fann, Y.C.; Lee, Y.-C.G. Robust Mutation Profiling of SARS-CoV-2 Variants from Multiple Raw Illumina Sequencing Data with Cloud Workflow. Genes 2022, 13, 686. [Google Scholar] [CrossRef]
- Brown, K.A.; Gubbay, J.; Hopkins, J.; Patel, S.; Buchan, S.A.; Daneman, N.; Goneau, L.W. S-Gene Target Failure as a Marker of Variant B.1.1.7 Among SARS-CoV-2 Isolates in the Greater Toronto Area, December 2020 to March 2021. JAMA 2021, 325, 2115–2116. [Google Scholar] [CrossRef]
- Wolter, N.; Jassat, W.; Walaza, S.; Welch, R.; Moultrie, H.; Groome, M.; Amoako, D.G.; Everatt, J.; Bhiman, J.N.; Scheepers, C.; et al. Early Assessment of the Clinical Severity of the SARS-CoV-2 Omicron Variant in South Africa: A Data Linkage Study. Lancet Lond. Engl. 2022, 399, 437–446. [Google Scholar] [CrossRef]
- Stefan, C.P.; Hall, A.T.; Graham, A.S.; Minogue, T.D. Comparison of Illumina and Oxford Nanopore Sequencing Technologies for Pathogen Detection from Clinical Matrices Using Molecular Inversion Probes. J. Mol. Diagn. JMD 2022, 24, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, Z.; Zeeshan, S.; Foran, D.J.; Kleinman, L.C.; Wondisford, F.E.; Dong, X. Integrative Clinical, Genomics and Metabolomics Data Analysis for Mainstream Precision Medicine to Investigate COVID-19. BMJ Innov. 2021, 7, 6–10. [Google Scholar] [CrossRef]
- Oude Munnink, B.B.; Worp, N.; Nieuwenhuijse, D.F.; Sikkema, R.S.; Haagmans, B.; Fouchier, R.A.M.; Koopmans, M. The next Phase of SARS-CoV-2 Surveillance: Real-Time Molecular Epidemiology. Nat. Med. 2021, 27, 1518–1524. [Google Scholar] [CrossRef]
- The COVID-19 Genomics UK (COG-UK) consortium An Integrated National Scale SARS-CoV-2 Genomic Surveillance Network. Lancet Microbe 2020, 1, e99–e100. [CrossRef]
- Petros, B.A.; Paull, J.S.; Tomkins-Tinch, C.H.; Loftness, B.C.; DeRuff, K.C.; Nair, P.; Gionet, G.L.; Benz, A.; Brock-Fisher, T.; Hughes, M.; et al. Multimodal Surveillance of SARS-CoV-2 at a University Enables Development of a Robust Outbreak Response Framework. Med 2022, in press. [Google Scholar] [CrossRef]
- As SARS-CoV-2 Virus Evolves, Genomic Data Should Be Collected Alongside Patient and Public Health Data, Says New Report|National Academies. Available online: https://www.nationalacademies.org/news/2020/07/as-sars-cov-2-virus-evolves-genomic-data-should-be-collected-alongside-patient-and-public-health-data-says-new-report (accessed on 13 October 2022).
- Colijn, C.; Earn, D.J.; Dushoff, J.; Ogden, N.H.; Li, M.; Knox, N.; Van Domselaar, G.; Franklin, K.; Jolly, G.; Otto, S.P. The Need for Linked Genomic Surveillance of SARS-CoV-2. Can. Commun. Dis. Rep. Releve Mal. Transm. Au Can. 2022, 48, 131–139. [Google Scholar] [CrossRef]
- Gozashti, L.; Corbett-Detig, R. Shortcomings of SARS-CoV-2 Genomic Metadata. BMC Res. Notes 2021, 14, 189. [Google Scholar] [CrossRef]
- Becker, S.J.; Taylor, J.; Sharfstein, J.M. Identifying and Tracking SARS-CoV-2 Variants—A Challenge and an Opportunity. N. Engl. J. Med. 2021, 385, 389–391. [Google Scholar] [CrossRef]
- Schriml, L.M.; Chuvochina, M.; Davies, N.; Eloe-Fadrosh, E.A.; Finn, R.D.; Hugenholtz, P.; Hunter, C.I.; Hurwitz, B.L.; Kyrpides, N.C.; Meyer, F.; et al. COVID-19 Pandemic Reveals the Peril of Ignoring Metadata Standards. Sci. Data 2020, 7, 188. [Google Scholar] [CrossRef]
- Yilmaz, P.; Kottmann, R.; Field, D.; Knight, R.; Cole, J.R.; Amaral-Zettler, L.; Gilbert, J.A.; Karsch-Mizrachi, I.; Johnston, A.; Cochrane, G.; et al. Minimum Information about a Marker Gene Sequence (MIMARKS) and Minimum Information about Any (x) Sequence (MIxS) Specifications. Nat. Biotechnol. 2011, 29, 415–420. [Google Scholar] [CrossRef]
- Bauer, D.C.; Metke-Jimenez, A.; Maurer-Stroh, S.; Tiruvayipati, S.; Wilson, L.O.W.; Jain, Y.; Perrin, A.; Ebrill, K.; Hansen, D.P.; Vasan, S.S. Interoperable Medical Data: The Missing Link for Understanding COVID-19. Transbound. Emerg. Dis. 2021, 68, 1753–1760. [Google Scholar] [CrossRef] [PubMed]
- Bernasconi, A.; Ceri, S. Interoperability of COVID-19 Clinical Phenotype Data with Host and Viral Genetics Data. BioMed 2022, 2, 69–81. [Google Scholar] [CrossRef]
- Austin, C.C.; Bernier, A.; Bezuidenhout, L.; Bicarregui, J.; Biro, T.; Cambon-Thomsen, A.; Carroll, S.R.; Cournia, Z.; Dabrowski, P.W.; Diallo, G.; et al. Fostering Global Data Sharing: Highlighting the Recommendations of the Research Data Alliance COVID-19 Working Group. Wellcome Open Res. 2020, 5, 267. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.-W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef] [Green Version]
- Dagliati, A.; Malovini, A.; Tibollo, V.; Bellazzi, R. Health Informatics and EHR to Support Clinical Research in the COVID-19 Pandemic: An Overview. Brief. Bioinform. 2021, 22, 812–822. [Google Scholar] [CrossRef]
- Madhavan, S.; Bastarache, L.; Brown, J.S.; Butte, A.J.; Dorr, D.A.; Embi, P.J.; Friedman, C.P.; Johnson, K.B.; Moore, J.H.; Kohane, I.S.; et al. Use of Electronic Health Records to Support a Public Health Response to the COVID-19 Pandemic in the United States: A Perspective from 15 Academic Medical Centers. J. Am. Med. Inform. Assoc. JAMIA 2021, 28, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Kadirvelu, B.; Burcea, G.; Quint, J.K.; Costelloe, C.E.; Faisal, A.A. Variation in Global COVID-19 Symptoms by Geography and by Chronic Disease: A Global Survey Using the COVID-19 Symptom Mapper. EClinicalMedicine 2022, 45, 101317. [Google Scholar] [CrossRef]
- Suthar, A.B.; Schubert, S.; Garon, J.; Couture, A.; Brown, A.M.; Charania, S. Coronavirus Disease Case Definitions, Diagnostic Testing Criteria, and Surveillance in 25 Countries with Highest Reported Case Counts. Emerg. Infect. Dis. 2022, 28, 148–156. [Google Scholar] [CrossRef]
- SNOMED Home Page. Available online: https://www.snomed.org/ (accessed on 13 October 2022).
- Duarte, J.; Castro, S.; Santos, M.; Abelha, A.; Machado, J. Improving Quality of Electronic Health Records with SNOMED. Procedia Technol. 2014, 16, 1342–1350. [Google Scholar] [CrossRef] [Green Version]
- General Data Protection Regulation (GDPR)—Official Legal Text. Available online: https://gdpr-info.eu/ (accessed on 13 October 2022).
- Bentzen, H.B.; Castro, R.; Fears, R.; Griffin, G.; ter Meulen, V.; Ursin, G. Remove Obstacles to Sharing Health Data with Researchers Outside of the European Union. Nat. Med. 2021, 27, 1329–1333. [Google Scholar] [CrossRef]
- DePuccio, M.J.; Di Tosto, G.; Walker, D.M.; McAlearney, A.S. Patients’ Perceptions About Medical Record Privacy and Security: Implications for Withholding of Information During the COVID-19 Pandemic. J. Gen. Intern. Med. 2020, 35, 3122–3125. [Google Scholar] [CrossRef] [PubMed]
- Mukaigawara, M.; Hassan, I.; Fernandes, G.; King, L.; Patel, J.; Sridhar, D. An Equitable Roadmap for Ending the COVID-19 Pandemic. Nat. Med. 2022, 28, 893–896. [Google Scholar] [CrossRef] [PubMed]
- Maxmen, A. Why Some Researchers Oppose Unrestricted Sharing of Coronavirus Genome Data. Nature 2021, 593, 176–177. [Google Scholar] [CrossRef]
- Rieke, N.; Hancox, J.; Li, W.; Milletarì, F.; Roth, H.R.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.A.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. NPJ Digit. Med. 2020, 3, 119. [Google Scholar] [CrossRef]
- Hardhantyo, M.; Djasri, H.; Nursetyo, A.A.; Yulianti, A.; Adipradipta, B.R.; Hawley, W.; Mika, J.; Praptiningsih, C.Y.; Mangiri, A.; Prasetyowati, E.B.; et al. Quality of National Disease Surveillance Reporting before and during COVID-19: A Mixed-Method Study in Indonesia. Int. J. Environ. Res. Public Health 2022, 19, 2728. [Google Scholar] [CrossRef] [PubMed]
- Umeozuru, C.M.; Usman, A.B.; Olorukooba, A.A.; Abdullahi, I.N.; John, D.J.; Lawal, L.A.; Uwazie, C.C.; Balogun, M.S. Performance of COVID-19 Case-Based Surveillance System in FCT, Nigeria, March 2020–January 2021. PLoS ONE 2022, 17, e0264839. [Google Scholar] [CrossRef]
- Clare, T.; Twohig, K.A.; O’Connell, A.-M.; Dabrera, G. Timeliness and Completeness of Laboratory-Based Surveillance of COVID-19 Cases in England. Public Health 2021, 194, 163–166. [Google Scholar] [CrossRef]
- Hart, O.E.; Halden, R.U. Computational Analysis of SARS-CoV-2/COVID-19 Surveillance by Wastewater-Based Epidemiology Locally and Globally: Feasibility, Economy, Opportunities and Challenges. Sci. Total Environ. 2020, 730, 138875. [Google Scholar] [CrossRef]
- Jajosky, R.A.; Groseclose, S.L. Evaluation of Reporting Timeliness of Public Health Surveillance Systems for Infectious Diseases. BMC Public Health 2004, 4, 29. [Google Scholar] [CrossRef] [Green Version]
- Mackey, T.; Purushothaman, V.; Li, J.; Shah, N.; Nali, M.; Bardier, C.; Liang, B.; Cai, M.; Cuomo, R. Machine Learning to Detect Self-Reporting of Symptoms, Testing Access, and Recovery Associated with COVID-19 on Twitter: Retrospective Big Data Infoveillance Study. JMIR Public Health Surveill. 2020, 6, e19509. [Google Scholar] [CrossRef]
- COVID-19 Contact Tracing Self-Reporting System to Be Launched on May 1 to Simplify Contact Tracing Procedures of Health Departments. Available online: https://www.cdc.gov.tw/En/Bulletin/Detail/qcdA-5yI0rjAIwj5RwI9dA?typeid=158 (accessed on 13 October 2022).
- Kalinich, C.C.; Jensen, C.G.; Neugebauer, P.; Petrone, M.E.; Peña-Hernández, M.; Ott, I.M.; Wyllie, A.L.; Alpert, T.; Vogels, C.B.F.; Fauver, J.R.; et al. Real-Time Public Health Communication of Local SARS-CoV-2 Genomic Epidemiology. PLoS Biol. 2020, 18, e3000869. [Google Scholar] [CrossRef] [PubMed]
- Gough, A.; Hunter, R.F.; Ajao, O.; Jurek, A.; McKeown, G.; Hong, J.; Barrett, E.; Ferguson, M.; McElwee, G.; McCarthy, M.; et al. Tweet for Behavior Change: Using Social Media for the Dissemination of Public Health Messages. JMIR Public Health Surveill. 2017, 3, e14. [Google Scholar] [CrossRef] [PubMed]
- Alipanah, N.; Jarlsberg, L.; Miller, C.; Linh, N.N.; Falzon, D.; Jaramillo, E.; Nahid, P. Adherence Interventions and Outcomes of Tuberculosis Treatment: A Systematic Review and Meta-Analysis of Trials and Observational Studies. PLoS Med. 2018, 15, e1002595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhagen, L.M.; de Groot, R.; Lawrence, C.A.; Taljaard, J.; Cotton, M.F.; Rabie, H. COVID-19 Response in Low- and Middle-Income Countries: Don’t Overlook the Role of Mobile Phone Communication. Int. J. Infect. Dis. IJID Off. Publ. Int. Soc. Infect. Dis. 2020, 99, 334–337. [Google Scholar] [CrossRef]
- World Health Organization; United Nations Foundation. UNDP/UNFPA/WHO/World Bank Special Programme of Research, Development. and Research Training in Human Reproduction & Johns Hopkins University; The MAPS Toolkit: MHealth Assessment and Planning for Scale; World Health Organization: Geneva, Switzerland, 2015; ISBN 978-92-4-150951-0. [Google Scholar]
- Castrucci, B.C. The “10 Essential Public Health Services” Is the Common Framework Needed to Communicate About Public Health. Am. J. Public Health 2021, 111, 598–599. [Google Scholar] [CrossRef]
- Smith, D.R.M.; Duval, A.; Pouwels, K.B.; Guillemot, D.; Fernandes, J.; Huynh, B.-T.; Temime, L.; Opatowski, L. AP-HP/Universities/Inserm COVID-19 research collaboration Optimizing COVID-19 Surveillance in Long-Term Care Facilities: A Modelling Study. BMC Med. 2020, 18, 386. [Google Scholar] [CrossRef]
- Davies, N.G.; Kucharski, A.J.; Eggo, R.M.; Gimma, A.; Edmunds, W.J.; Jombart, T.; O’Reilly, K.; Endo, A.; Hellewell, J.; Nightingale, E.S.; et al. Effects of Non-Pharmaceutical Interventions on COVID-19 Cases, Deaths, and Demand for Hospital Services in the UK: A Modelling Study. Lancet Public Health 2020, 5, e375–e385. [Google Scholar] [CrossRef]
- World Health Organization. WHO Mass Gathering COVID-19 Risk Assessment Tool—Generic Events; World Health Organization: Geneva, Switzerland, 2020. [Google Scholar]
- Zhang, M.; Xiao, J.; Deng, A.; Zhang, Y.; Zhuang, Y.; Hu, T.; Li, J.; Tu, H.; Li, B.; Zhou, Y.; et al. Transmission Dynamics of an Outbreak of the COVID-19 Delta Variant B.1.617.2—Guangdong Province, China, May-June 2021. China CDC Wkly. 2021, 3, 584–586. [Google Scholar] [CrossRef]
- Shi, Q.; Gao, X.; Hu, B. Research Progress on Characteristics, Epidemiology and Control Measure of SARS-CoV-2 Delta VOC. Chin. J. Nosocomiol. 2021, 31, 3703–3707. [Google Scholar]
- Mackie, D. Global Vulnerabilities to the COVID-19 Variant B.1.617.2, SUERF Policy Brief SUERF—The European Money and Finance Forum. Available online: https://www.suerf.org/suer-policy-brief/26931/global-vulnerabilities-to-the-covid-19-variant-b-1-617-2 (accessed on 13 October 2022).
- SPI-M-O: Summary of Further Modelling of Easing Restrictions—Roadmap Step 4 on 19 July 2021, 7 July 2021. Available online: https://www.gov.uk/government/publications/spi-m-o-summary-of-further-modelling-of-easing-restrictions-roadmap-step-4-on-19-july-2021-7-july-2021/spi-m-o-summary-of-further-modelling-of-easing-restrictions-roadmap-step-4-on-19-july-2021-7-july-2021 (accessed on 13 October 2022).
- De Vogli, R.; Gimeno, D.; Kivimaki, M. Socioeconomic Inequalities in Health in 22 European Countries. N. Engl. J. Med. 2008, 359, 1290; author reply 1290–1291. [Google Scholar] [CrossRef] [Green Version]
- Lieberman-Cribbin, W.; Tuminello, S.; Flores, R.M.; Taioli, E. Disparities in COVID-19 Testing and Positivity in New York City. Am. J. Prev. Med. 2020, 59, 326–332. [Google Scholar] [CrossRef] [PubMed]
- Abrams, E.M.; Szefler, S.J. COVID-19 and the Impact of Social Determinants of Health. Lancet Respir. Med. 2020, 8, 659–661. [Google Scholar] [CrossRef]
- Mena, G.E.; Martinez, P.P.; Mahmud, A.S.; Marquet, P.A.; Buckee, C.O.; Santillana, M. Socioeconomic Status Determines COVID-19 Incidence and Related Mortality in Santiago, Chile. Science 2021, 372, eabg5298. [Google Scholar] [CrossRef]
- Aggarwal, D.; Myers, R.; Hamilton, W.L.; Bharucha, T.; Tumelty, N.M.; Brown, C.S.; Meader, E.J.; Connor, T.; Smith, D.L.; Bradley, D.T.; et al. The Role of Viral Genomics in Understanding COVID-19 Outbreaks in Long-Term Care Facilities. Lancet Microbe 2022, 3, e151–e158. [Google Scholar] [CrossRef]
- Ricoca Peixoto, V.; Nunes, C.; Abrantes, A. Epidemic Surveillance of COVID-19: Considering Uncertainty and Under-Ascertainment. Port. J. Public Health 2020, 38, 23–29. [Google Scholar] [CrossRef]
- Costa-Santos, C.; Neves, A.L.; Correia, R.; Santos, P.; Monteiro-Soares, M.; Freitas, A.; Ribeiro-Vaz, I.; Henriques, T.S.; Pereira Rodrigues, P.; Costa-Pereira, A.; et al. COVID-19 Surveillance Data Quality Issues: A National Consecutive Case Series. BMJ Open 2021, 11, e047623. [Google Scholar] [CrossRef]
- German, R.R.; Lee, L.M.; Horan, J.M.; Milstein, R.L.; Pertowski, C.A.; Waller, M.N. Guidelines Working Group Centers for Disease Control and Prevention (CDC) Updated Guidelines for Evaluating Public Health Surveillance Systems: Recommendations from the Guidelines Working Group. MMWR Recomm. Rep. Morb. Mortal. Wkly. Rep. Recomm. Rep. 2001, 50, 1–35, quiz CE1-7. [Google Scholar]
- Austin, E.W.; Austin, B.W.; Willoughby, J.F.; Amram, O.; Domgaard, S. How Media Literacy and Science Media Literacy Predicted the Adoption of Protective Behaviors Amidst the COVID-19 Pandemic. J. Health Commun. 2021, 26, 239–252. [Google Scholar] [CrossRef]
- Kondilis, E.; Papamichail, D.; McCann, S.; Carruthers, E.; Veizis, A.; Orcutt, M.; Hargreaves, S. The Impact of the COVID-19 Pandemic on Refugees and Asylum Seekers in Greece: A Retrospective Analysis of National Surveillance Data from 2020. EClinicalMedicine 2021, 37, 100958. [Google Scholar] [CrossRef]
- Shragai, T.; Summers, A.; Olushayo, O.; Rumunu, J.; Mize, V.; Laku, R.; Bunga, S. Impact of Policy and Funding Decisions on COVID-19 Surveillance Operations and Case Reports—South Sudan, April 2020-February 2021. MMWR Morb. Mortal. Wkly. Rep. 2021, 70, 811–817. [Google Scholar] [CrossRef]
- DeSalvo, K.; Hughes, B.; Bassett, M.; Benjamin, G.; Fraser, M.; Galea, S.; Gracia, J.N. Public Health COVID-19 Impact Assessment: Lessons Learned and Compelling Needs. NAM Perspect. 2021, 2021. [Google Scholar] [CrossRef] [PubMed]
- Burstein, P. The Impact of Public Opinion on Public Policy: A Review and an Agenda. Polit. Res. Q. 2003, 56, 29–40. [Google Scholar] [CrossRef]
- Gollust, S.E.; Nagler, R.H.; Fowler, E.F. The Emergence of COVID-19 in the US: A Public Health and Political Communication Crisis. J. Health Polit. Policy Law 2020, 45, 967–981. [Google Scholar] [CrossRef]
- Qazi, A.; Qazi, J.; Naseer, K.; Zeeshan, M.; Hardaker, G.; Maitama, J.Z.; Haruna, K. Analyzing Situational Awareness through Public Opinion to Predict Adoption of Social Distancing amid Pandemic COVID-19. J. Med. Virol. 2020, 92, 849–855. [Google Scholar] [CrossRef] [PubMed]
- Mheidly, N.; Fares, J. Leveraging Media and Health Communication Strategies to Overcome the COVID-19 Infodemic. J. Public Health Policy 2020, 41, 410–420. [Google Scholar] [CrossRef] [PubMed]
- Hurie, A.H. School Choice, Exclusion, and Race Taming in Milwaukee: A Meta-Ethnography. Urban Rev. 2021, 53, 785–813. [Google Scholar] [CrossRef] [PubMed]
- Siddiqi, S.; Masud, T.I.; Nishtar, S.; Peters, D.H.; Sabri, B.; Bile, K.M.; Jama, M.A. Framework for Assessing Governance of the Health System in Developing Countries: Gateway to Good Governance. Health Policy 2009, 90, 13–25. [Google Scholar] [CrossRef]
- Groseclose, S.L.; Buckeridge, D.L. Public Health Surveillance Systems: Recent Advances in Their Use and Evaluation. Annu. Rev. Public Health 2017, 38, 57–79. [Google Scholar] [CrossRef] [Green Version]
- Hosch, S.; Mpina, M.; Nyakurungu, E.; Borico, N.S.; Obama, T.M.A.; Ovona, M.C.; Wagner, P.; Rubin, S.E.; Vickos, U.; Milang, D.V.N.; et al. Genomic Surveillance Enables the Identification of Co-Infections with Multiple SARS-CoV-2 Lineages in Equatorial Guinea. Front. Public Health 2022, 9, 818401. [Google Scholar] [CrossRef]
- Ozer, E.A.; Simons, L.M.; Adewumi, O.M.; Fowotade, A.A.; Omoruyi, E.C.; Adeniji, J.A.; Olayinka, O.A.; Dean, T.J.; Zayas, J.; Bhimalli, P.P.; et al. Multiple Expansions of Globally Uncommon SARS-CoV-2 Lineages in Nigeria. Nat. Commun. 2022, 13, 688. [Google Scholar] [CrossRef]
- Hall, K.L.; Vogel, A.L.; Crowston, K. Comprehensive Collaboration Plans: Practical Considerations Spanning Across Individual Collaborators to Institutional Supports. In Strategies for Team Science Success: Handbook of Evidence-Based Principles for Cross-Disciplinary Science and Practical Lessons Learned from Health Researchers; Hall, K.L., Vogel, A.L., Croyle, R.T., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 587–612. ISBN 978-3-030-20992-6. [Google Scholar]
- Wrigley, A.; Dawson, A. Vulnerability and Marginalized Populations. In Public Health Ethics: Cases Spanning the Globe; Barrett, D.H., Ortmann, L.W., Dawson, A., Saenz, C., Reis, A., Bolan, G., Eds.; Public Health Ethics Analysis; Springer International Publishing: Cham, Switzerland, 2016; pp. 203–240. ISBN 978-3-319-23847-0. [Google Scholar]
- Xu, S.; Li, Y. COVID-19 Clinical Research Coalition Global Coalition to Accelerate COVID-19 Clinical Research in Resource-Limited Settings. Lancet 2020, 395, 1322–1325. [Google Scholar] [CrossRef]
- Starr, T.N.; Greaney, A.J.; Hilton, S.K.; Ellis, D.; Crawford, K.H.D.; Dingens, A.S.; Navarro, M.J.; Bowen, J.E.; Tortorici, M.A.; Walls, A.C.; et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 2020, 182, 1295–1310.e20. [Google Scholar] [CrossRef] [PubMed]
- Obermeyer, F.; Jankowiak, M.; Barkas, N.; Schaffner, S.F.; Pyle, J.D.; Yurkovetskiy, L.; Bosso, M.; Park, D.J.; Babadi, M.; MacInnis, B.L.; et al. Analysis of 6.4 Million SARS-CoV-2 Genomes Identifies Mutations Associated with Fitness. Science 2022, 376, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Maher, M.C.; Bartha, I.; Weaver, S.; di Iulio, J.; Ferri, E.; Soriaga, L.; Lempp, F.A.; Hie, B.L.; Bryson, B.; Berger, B.; et al. Predicting the Mutational Drivers of Future SARS-CoV-2 Variants of Concern. Sci. Transl. Med. 2022, 14, eabk3445. [Google Scholar] [CrossRef] [PubMed]
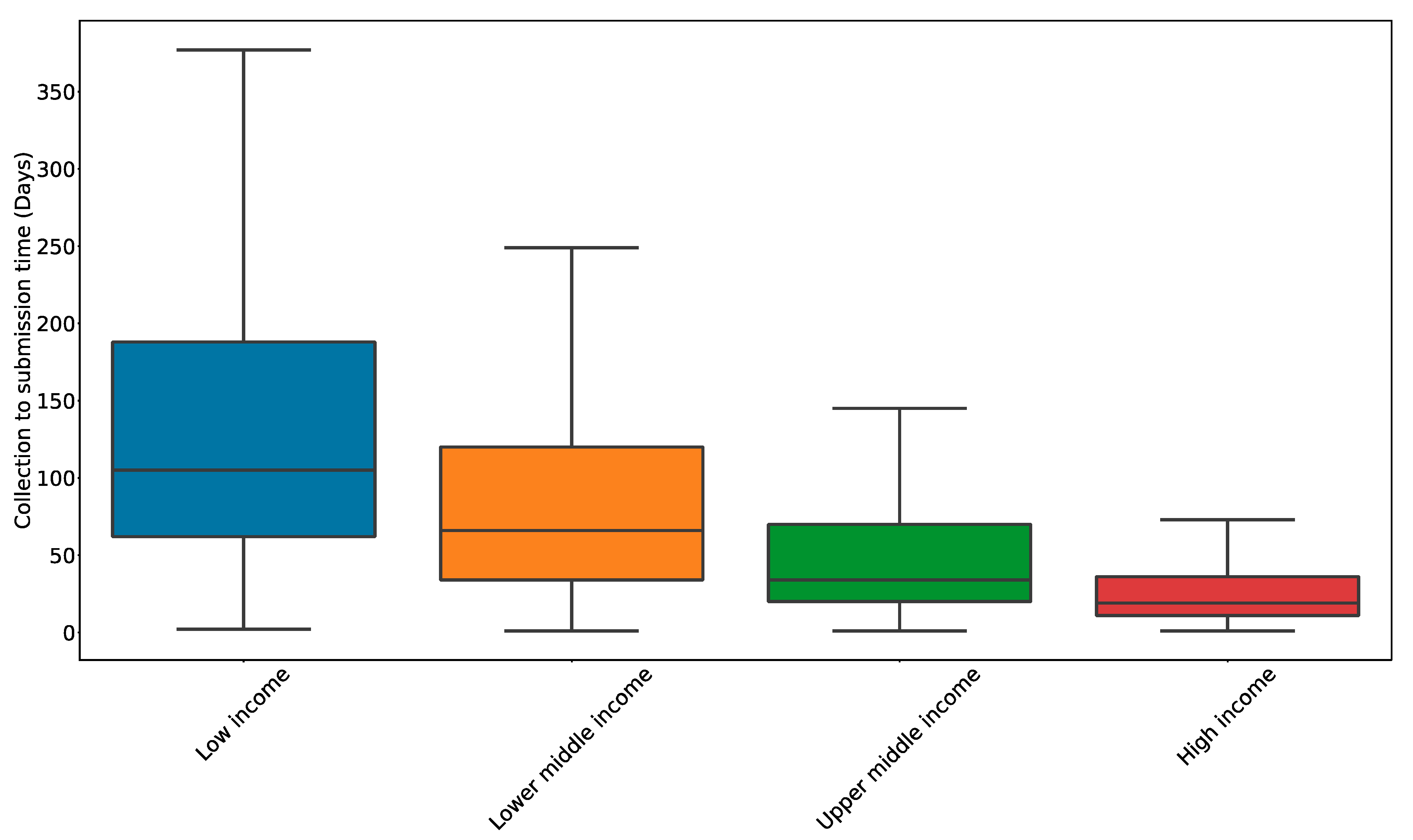
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling-Hu, T.; Rios-Guzman, E.; Lorenzo-Redondo, R.; Ozer, E.A.; Hultquist, J.F. Challenges and Opportunities for Global Genomic Surveillance Strategies in the COVID-19 Era. Viruses 2022, 14, 2532. https://doi.org/10.3390/v14112532
Ling-Hu T, Rios-Guzman E, Lorenzo-Redondo R, Ozer EA, Hultquist JF. Challenges and Opportunities for Global Genomic Surveillance Strategies in the COVID-19 Era. Viruses. 2022; 14(11):2532. https://doi.org/10.3390/v14112532
Chicago/Turabian StyleLing-Hu, Ted, Estefany Rios-Guzman, Ramon Lorenzo-Redondo, Egon A. Ozer, and Judd F. Hultquist. 2022. "Challenges and Opportunities for Global Genomic Surveillance Strategies in the COVID-19 Era" Viruses 14, no. 11: 2532. https://doi.org/10.3390/v14112532




