Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soybean Materials and Growth Conditions
2.2. Virus Resource and Mechanical Inoculation
2.3. Enzyme-Linked Immunosorbent Assay (ELISA)
2.4. DNA Extraction, Library Preparation, and High-Throughput Sequencing
2.5. Bulk Segregation Analysis
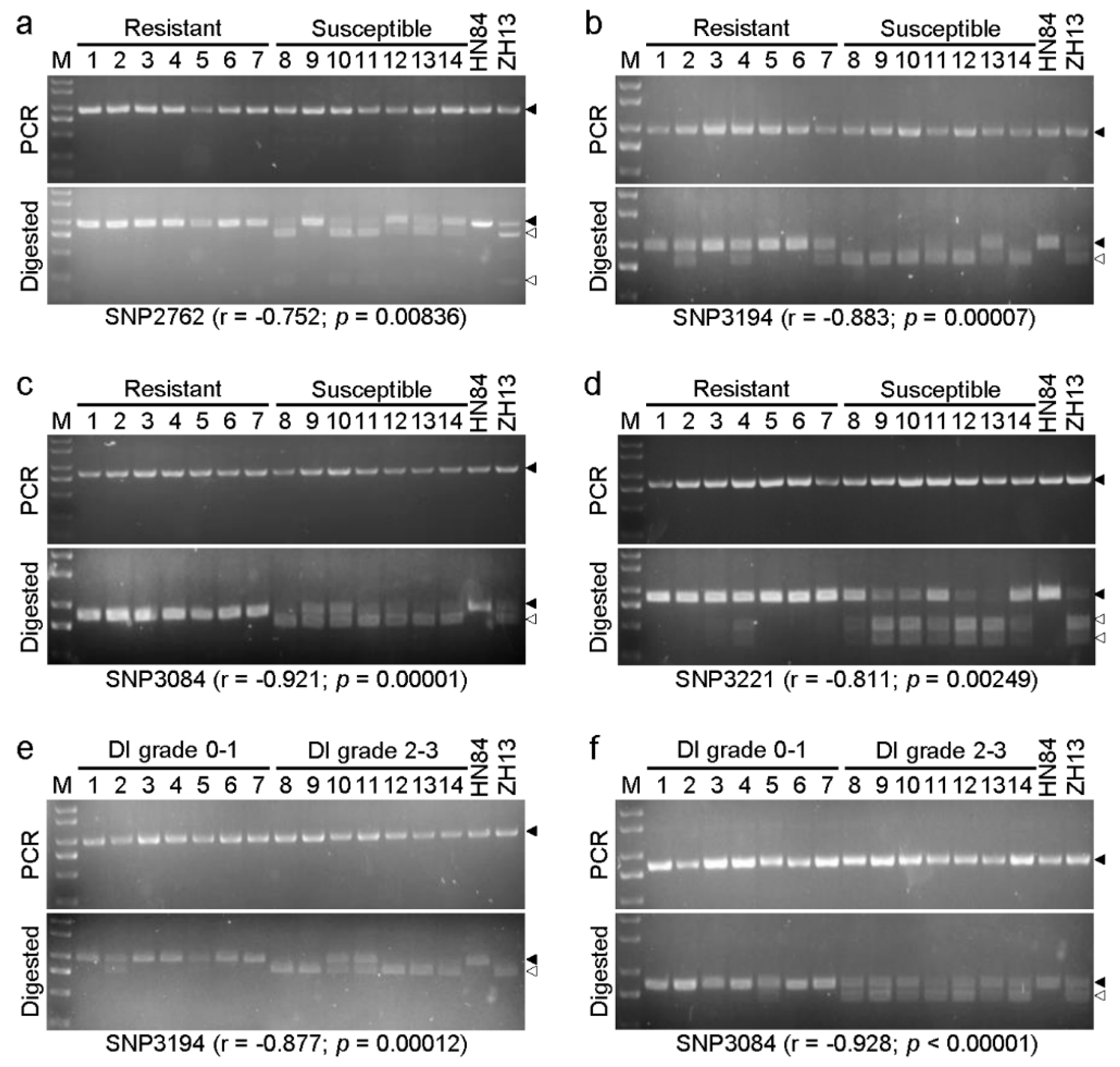
2.6. Cleaved Amplified Polymorphic Sequences (CAPS) Primer Design and Validation
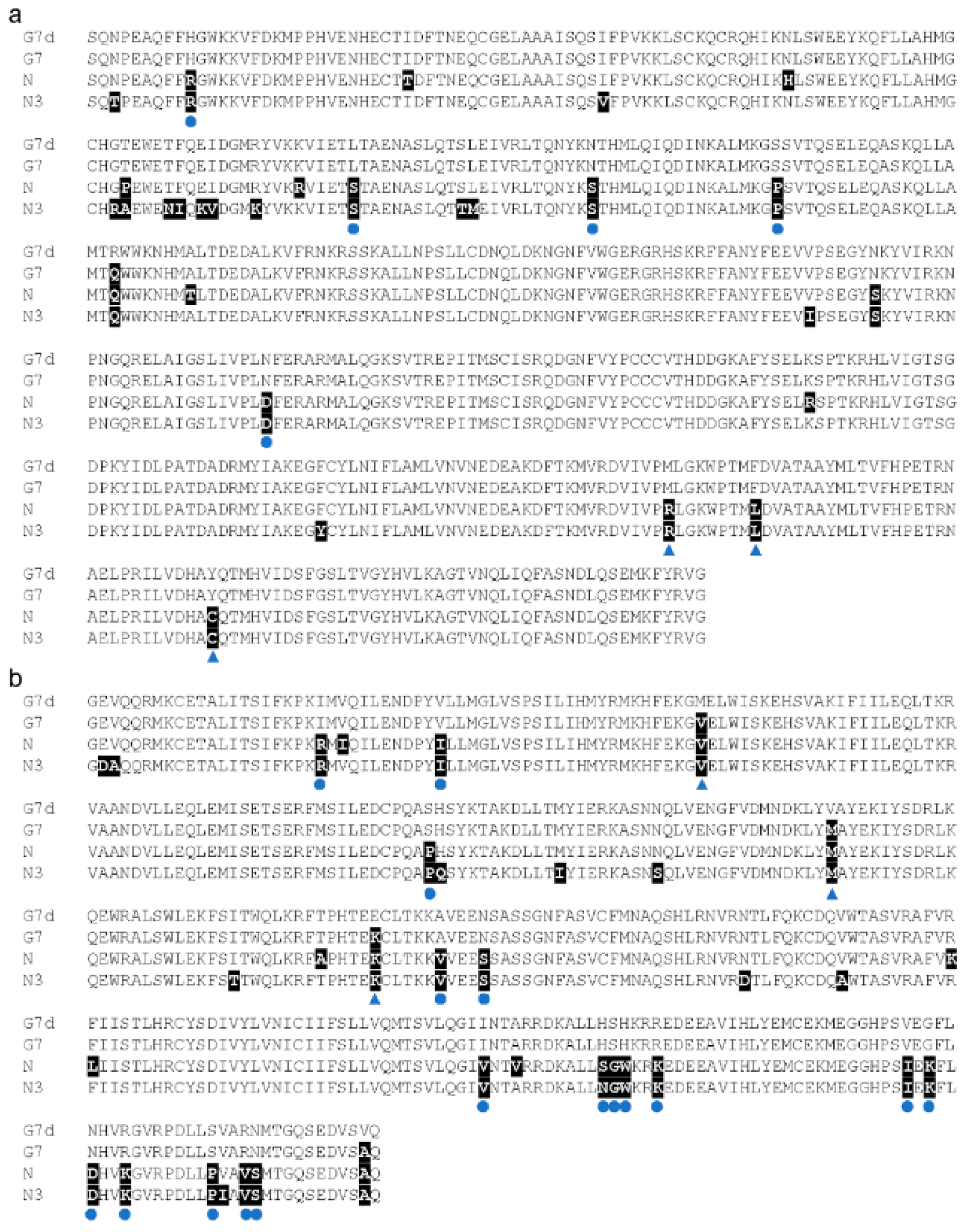
2.7. Sequence Clone, Alignment, and Comparison
3. Results
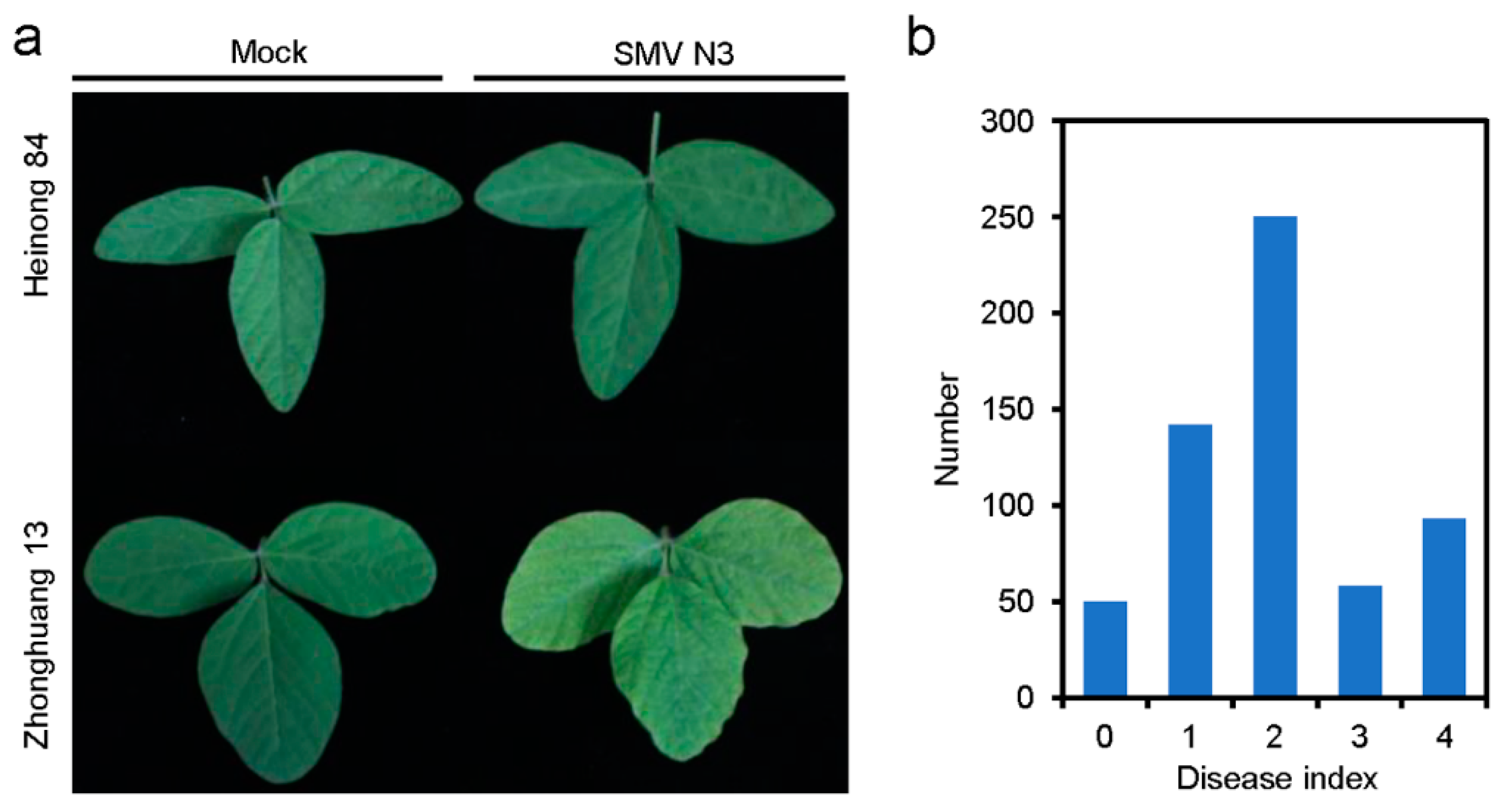
3.1. Genetic Analysis of the Resistance of Heinong 84 to SMV N3
3.2. BSA Pool Preparation and Sequencing
3.3. Mapping the Resistance Locus in Heinong 84
3.4. Resistant Gene Prediction
3.5. Development of CAPS Marker for Rapid Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Whitham, S.A.; Qi, M.; Innes, R.W.; Ma, W.; Lopes-Caitar, V.; Hewezi, T. Molecular Soybean-Pathogen Interactions. Annu. Rev. Phytopathol. 2016, 54, 443–468. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.H.; Whitham, S.A. Advances in Virus Research; Elsevier: Amsterdam, The Netherlands, 2014; pp. 355–390. [Google Scholar]
- Hajimorad, M.R.; Domier, L.L.; Tolin, S.A.; Whitham, S.A.; Maroof, M.A.S. Soybean mosaic virus: A successful potyvirus with a wide distribution but restricted natural host range. Mol. Plant Pathol. 2017, 19, 1563–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, B.Y.; Miller, W.A.; Atkins, J.F.; Firth, A.E. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 2008, 105, 5897–5902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; López-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef]
- Cho, E.K.; Goodman, R.M. Strains of soybean mosaic virus: Classification based on virulence in resistant soybean cultivars. Phytopathology 1979, 69, 467–470. [Google Scholar] [CrossRef]
- Saruta, M.; Kikuchi, A.; Okabe, A.; Sasaya, T. Molecular characterization of A2 and D strains of Soybean mosaic virus, which caused a recent virus outbreak in soybean cultivar Sachiyutaka in Chugoku and Shikoku regions of Japan. J. Gen. Plant Pathol. 2005, 71, 431–435. [Google Scholar] [CrossRef]
- Gibbs, A.J.; Trueman, J.W.; Gibbs, M.J. The bean common mosaic virus lineage of potyviruses: Where did it arise and when? Arch. Virol. 2008, 153, 2177–2187. [Google Scholar] [CrossRef]
- Zhou, G.-C.; Shao, Z.-Q.; Ma, F.-F.; Wu, P.; Wu, X.-Y.; Xie, Z.-Y.; Yu, D.-Y.; Cheng, H.; Liu, Z.-H.; Jiang, Z.-F.; et al. The evolution of soybean mosaic virus: An updated analysis by obtaining 18 new genomic sequences of Chinese strains/isolates. Virus Res. 2015, 208, 189–198. [Google Scholar] [CrossRef]
- Li, K. Strain Identification of Soybean Mosaic Virus and Inheritance and Gene Mapping of Its Resistance in Soybeans in Southern China; Nanjing Agricultural University: Nanjing, China, 2009. [Google Scholar]
- Lv, W.; Zhang, M.; Wei, P.; Xie, S.; Guo, J.; Jiang, Y.; Geng, Y. Classification and distribution of strains of soybean mosaic virus in Northeast China. Acta Phytopathol. Sin. 1985, 15, 225–229. [Google Scholar]
- Gao, L.; Wu, Y.; An, J.; Huang, W.; Liu, X.; Xue, Y.; Luan, X.; Lin, F.; Sun, L. Pathogenicity and genome-wide sequence analysis reveals relationships between soybean mosaic virus strains. Arch. Virol. 2022, 167, 517–529. [Google Scholar] [CrossRef]
- Widyasari, K.; Alazem, M.; Kim, K.-H. Soybean Resistance to Soybean Mosaic Virus. Plants 2020, 9, 219. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, K.; Saruta, M.; Shimizu, T.; Shu, M.; Anai, T.; Komatsu, K.; Yamada, N.; Katayose, Y.; Ishikawa, M.; Ishimoto, M.; et al. Soybean antiviral immunity conferred by dsRNase targets the viral replication complex. Nat. Commun. 2019, 10, 4033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, J.; Wang, L.; Jin, T.; Nie, Y.; Liu, H.; Qiu, Y.; Yang, Y.; Li, B.; Zhang, J.; Wang, D.; et al. A cell wall-localized NLR confers resistance to Soybean mosaic virus by recognizing viral-encoded cylindrical inclusion protein. Mol. Plant 2021, 14, 1881–1900. [Google Scholar] [CrossRef]
- Zhao, X.; Jing, Y.; Luo, Z.; Gao, S.; Teng, W.; Zhan, Y.; Qiu, L.; Zheng, H.; Li, W.; Han, Y. GmST1, which encodes a sulfotransferase, confers resistance to soybean mosaic virus strains G2 and G3. Plant Cell Environ. 2021, 44, 2777–2792. [Google Scholar] [CrossRef] [PubMed]
- Luan, X.; Liu, X.; Xue, Y.; Ma, Y.; Wang, J.; Zhang, B.; Yu, B.; Wang, G. Innovating soybean Heinong 84 with high quality and multi-resisantce by molecular marker gene polymerization. Soybean Sci. 2018, 37, 839–842. [Google Scholar] [CrossRef]
- Luan, X. Inhereitance of soybean plant resistant to No III (173) strain of soybean mosaic virus. Soybean Sci. 1997, 16, 223–226. [Google Scholar]
- Magwene, P.M.; Willis, J.H.; Kelly, J.K. The Statistics of Bulk Segregant Analysis Using Next Generation Sequencing. PLoS Comput. Biol. 2011, 7, e1002255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Xu, Y. Bulk segregation analysis in the NGS era: A review of its teenage years. Plant J. 2021, 109, 1355–1374. [Google Scholar] [CrossRef]
- Shen, Y.; Liu, J.; Geng, H.; Zhang, J.; Liu, Y.; Zhang, H.; Xing, S.; Du, J.; Ma, S.; Tian, Z. De novo assembly of a Chinese soybean genome. Sci. China Life Sci. 2018, 61, 871–884. [Google Scholar] [CrossRef]
- Shen, Y.; Du, H.; Liu, Y.; Ni, L.; Wang, Z.; Liang, C.; Tian, Z. Update soybean Zhonghuang 13 genome to a golden reference. Sci. China Life Sci. 2019, 62, 1257–1260. [Google Scholar] [CrossRef]
- Che, X.; Jiang, X.; Liu, X.; Luan, X.; Liu, Q.; Cheng, X.; Wu, X. First Report of Alfalfa Mosaic Virus on Soybean in Heilongjiang, China. Plant Dis. 2020, 104, 3085. [Google Scholar] [CrossRef]
- Lin, J.; Lan, Z.; Hou, W.; Yang, C.; Wang, D.; Zhang, M.; Zhi, H. Identification and fine-mapping of a genetic locus underlying soybean tolerance to SMV infections. Plant Sci. 2019, 292, 110367. [Google Scholar] [CrossRef] [PubMed]
- Usovsky, M.; Chen, P.; Li, D.; Wang, A.; Shi, A.; Zheng, C.; Shakiba, E.; Lee, D.; Vieira, C.C.; Lee, Y.C.; et al. Decades of Genetic Research on Soybean mosaic virus Resistance in Soybean. Viruses 2022, 14, 1122. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; O Pollard, M.; Whitwham, A.; Keane, T.; A McCarthy, S.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Grumet, R. QTLseqr: An R Package for Bulk Segregant Analysis with Next-Generation Sequencing. Plant Genome 2018, 11, 180006. [Google Scholar] [CrossRef] [Green Version]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11121–111234. [Google Scholar] [CrossRef] [Green Version]
- Thiel, T.; Kota, R.; Grosse, I.; Stein, N.; Graner, A. SNP2CAPS: A SNP and INDEL analysis tool for CAPS marker development. Nucleic Acids Res. 2004, 32, e5. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2017, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Hajimorad, M.R.; Hill, J.H. Rsv1-mediated resistance against soybean mosaic virus-N is hypersensitive response-independent at inoculation site, but has the potential to initiate a hypersensitive response-like mechanism. Mol. Plant-Microbe Interact. 2001, 14, 587–598. [Google Scholar] [CrossRef] [Green Version]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajimorad, M.R.; Eggenberger, A.L.; Hill, J.H. Loss and Gain of Elicitor Function of Soybean Mosaic Virus G7 Provoking Rsv1 -Mediated Lethal Systemic Hypersensitive Response Maps to P3. J. Virol. 2005, 79, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Eggenberger, A.L.; Hajimorad, M.R.; Hill, J.H. Gain of Virulence on Rsv1-Genotype Soybean by an Avirulent Soybean mosaic virus Requires Concurrent Mutations in Both P3 and HC-Pro. Mol. Plant-Microbe Interact. 2008, 21, 931–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajimorad, M.; Eggenberger, A.; Hill, J. Evolution of Soybean mosaic virus-G7 molecularly cloned genome in Rsv1-genotype soybean results in emergence of a mutant capable of evading Rsv1-mediated recognition. Virology 2003, 314, 497–509. [Google Scholar] [CrossRef] [Green Version]
- Luan, X.; Li, Z.; Man, W.; Bai, Y.; Ma, S.; Liu, X.; Zheng, B.; Du, W. Identification of molecular markers linked to resistance for SMV3 in soybean. Mol. Plant Breed. 2006, 4, 841–845. [Google Scholar]
- Yu, Y.G.; Buss, G.R.; A Maroof, M. Isolation of a superfamily of candidate disease-resistance genes in soybean based on a conserved nucleotide-binding site. Proc. Natl. Acad. Sci. USA 1996, 93, 11751–11756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, R.-H.; Khatabi, B.; Ashfield, T.; Maroof, M.A.S.; Hajimorad, M.R. The HC-Pro and P3 Cistrons of an Avirulent Soybean mosaic virus Are Recognized by Different Resistance Genes at the Complex Rsv1 Locus. Mol. Plant-Microbe Interact. 2013, 26, 203–215. [Google Scholar] [CrossRef]


| Primer Names | Sequences (5′–3′) | Usage |
|---|---|---|
| N3-HcPro_F | TCCCAAACTCCTGAAGCTCAA | Cloning |
| N3-HcPro_R | ACCAACTCTATAAAATTTCATCTC | Cloning |
| N3-P3_F | GGTGATGCGCAACAAAGGATG | Cloning |
| N3-P3_R | CTGTGCGGAAACATCTTCTGATTG | Cloning |
| SNP2762_F | TAGTGGTGGATGGTTATGC | CAPS assay |
| SNP2762_R | TTTCCTGGCTGTTCCTATT | CAPS assay |
| SNP2805_F | TGAAAGTGGCTATGCTAT | CAPS assay |
| SNP2805_R | AATCAACCCTCCAAATCG | CAPS assay |
| SNP3084_F | CAACTGTATGGTTTAGGGATT | CAPS assay |
| SNP3084_R | AATTAGAGTGACCTGCAAGAT | CAPS assay |
| SNP3194_F | TTCTCCTACGGTCATTGTT | CAPS assay |
| SNP3194_R | TTTCTTATGTATGCTGGTG | CAPS assay |
| SNP3221_F | TCAATGACCCTTTGTGAG | CAPS assay |
| SNP3221_R | GGGAGGCTTGTCTACTGC | CAPS assay |
| Parents and Offspring | Resistant | Susceptible | Total | Theoretical Separation Ratio | χ2 | p |
|---|---|---|---|---|---|---|
| Heinong 84 | 14 | 0 | 14 | |||
| Zhonghuang 13 | 0 | 14 | 14 | |||
| F1 | 12 | 0 | 12 | |||
| F2 | 442 | 151 | 593 | 3:1 | 0.667 | 0.414 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Liu, X.; Deng, W.; Liu, J.; Fang, Y.; Liu, Y.; Ma, T.; Zhang, Y.; Xue, Y.; Tang, X.; et al. Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification. Viruses 2022, 14, 2533. https://doi.org/10.3390/v14112533
Li Y, Liu X, Deng W, Liu J, Fang Y, Liu Y, Ma T, Zhang Y, Xue Y, Tang X, et al. Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification. Viruses. 2022; 14(11):2533. https://doi.org/10.3390/v14112533
Chicago/Turabian StyleLi, Yong, Xinlei Liu, Wenjia Deng, Jiahui Liu, Yue Fang, Ye Liu, Tingshuai Ma, Ying Zhang, Yongguo Xue, Xiaofei Tang, and et al. 2022. "Fine Mapping the Soybean Mosaic Virus Resistance Gene in Soybean Cultivar Heinong 84 and Development of CAPS Markers for Rapid Identification" Viruses 14, no. 11: 2533. https://doi.org/10.3390/v14112533






