Distribution and Characterisation of Tick-Borne Flavi-, Flavi-like, and Phenuiviruses in the Chelyabinsk Region of Russia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Processing of Ticks
2.2. Infection of Tick Cell Line
2.3. Infection of Mammalian Cell Line
2.4. Reverse-Transcriptase PCR (RT-PCR) and Sequencing of Amplified Products
2.5. Phylogenetic Analysis
2.5.1. Tick-Borne Encephalitis Virus
2.5.2. Flavi-like Viruses
2.5.3. Phenuiviruses
2.6. Data Analysis
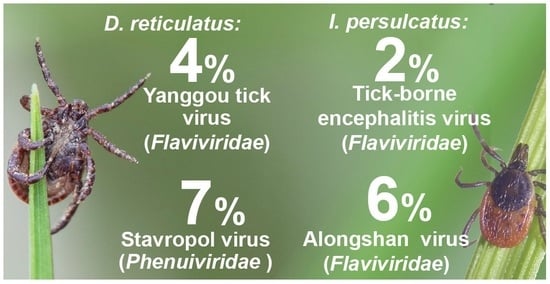
3. Results
3.1. Tick-Borne Encephalitis Virus
3.2. Segmented Flavi-like Viruses
3.2.1. Alongshan Virus
3.2.2. Yanggou Tick Virus
3.3. Phenuiviruses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shi, M.; Lin, X.-D.; Vasilakis, N.; Tian, J.-H.; Li, C.-X.; Chen, L.-J.; Eastwood, G.; Diao, X.-N.; Chen, M.-H.; Chen, X.; et al. Divergent Viruses Discovered in Arthropods and Vertebrates Revise the Evolutionary History of the Flaviviridae and Related Viruses. J. Virol. 2016, 90, 659–669. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; He, B.; Huang, S.Y.; Wei, F.; Zhu, X.Q. Severe fever with thrombocytopenia syndrome, an emerging tick-borne zoonosis. Lancet Infect. Dis. 2014, 14, 763–772. [Google Scholar] [CrossRef] [PubMed]
- López, Y.; Miranda, J.; Mattar, S.; Gonzalez, M.; Rovnak, J. First report of Lihan Tick virus (Phlebovirus, Phenuiviridae) in ticks, Colombia. Virol. J. 2020, 17, 63. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.-C.; Shi, M.; Tian, J.-H.; Lin, X.-D.; Gao, D.-Y.; He, J.-R.; Wang, J.-B.; Li, C.-X.; Kang, Y.-J.; Yu, B.; et al. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl. Acad. Sci. USA 2014, 111, 6744–6749. [Google Scholar] [CrossRef] [Green Version]
- Litov, A.G.; Belova, O.A.; Kholodilov, I.S.; Gadzhikurbanov, M.N.; Gmyl, L.V.; Oorzhak, N.D.; Saryglar, A.A.; Ishmukhametov, A.A.; Karganova, G.G. Possible Arbovirus Found in Virome of Melophagus ovinus. Viruses 2021, 13, 2375. [Google Scholar] [CrossRef]
- Jia, N.; Liu, H.B.; Ni, X.B.; Bell-Sakyi, L.; Zheng, Y.C.; Song, J.L.; Li, J.; Jiang, B.G.; Wang, Q.; Sun, Y.; et al. Emergence of human infection with Jingmen tick virus in China: A retrospective study. EBioMedicine 2019, 43, 317–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.-D.; Wang, B.; Wei, F.; Han, S.-Z.; Zhang, L.; Yang, Z.-T.; Yan, Y.; Lv, X.-L.; Li, L.; Wang, S.-C.; et al. A New Segmented Virus Associated with Human Febrile Illness in China. N. Engl. J. Med. 2019, 380, 2116–2125. [Google Scholar] [CrossRef]
- McMullan, L.K.; Folk, S.M.; Kelly, A.J.; MacNeil, A.; Goldsmith, C.S.; Metcalfe, M.G.; Batten, B.C.; Albariño, C.G.; Zaki, S.R.; Rollin, P.E.; et al. A New Phlebovirus Associated with Severe Febrile Illness in Missouri. N. Engl. J. Med. 2012, 367, 834–841. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Bukh, J.; Gould, E.A.; Meyers, G.; Monath, T.; Muerhoff, S.; Pletnev, A.; Rico-Hesse, R.; Smith, D.B.; et al. ICTV Virus Taxonomy Profile: Flaviviridae. J. Gen. Virol. 2017, 98, 2–3. [Google Scholar] [CrossRef]
- Růžek, D.; Yakimenko, V.V.; Karan, L.S.; Tkachev, S.E. Omsk haemorrhagic fever. Lancet 2010, 376, 2104–2113. [Google Scholar] [CrossRef]
- Ruzek, D.; Avšič Županc, T.; Borde, J.; Chrdle, A.; Eyer, L.; Karganova, G.; Kholodilov, I.; Knap, N.; Kozlovskaya, L.; Matveev, A.; et al. Tick-borne encephalitis in Europe and Russia: Review of pathogenesis, clinical features, therapy, and vaccines. Antiviral. Res. 2019, 164, 23–51. [Google Scholar] [CrossRef] [PubMed]
- Chancey, C.; Grinev, A.; Volkova, E.; Rios, M. The global ecology and epidemiology of West Nile virus. Biomed. Res. Int. 2015, 2015, 376230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, A.; Gandhi, S. Japanese encephalitis virus: A review on emerging diagnostic techniques. Front. Biosci. 2020, 25, 1875–1893. [Google Scholar] [CrossRef]
- Guzman, M.G.; Harris, E. Dengue. Lancet 2015, 385, 453–465. [Google Scholar] [CrossRef]
- Platonov, A.E. West Nile encephalitis in Russia 1999–2001: Were we ready? Are we ready? Ann. N. Y. Acad. Sci. 2001, 951, 102–116. [Google Scholar] [CrossRef]
- Shartova, N.; Mironova, V.; Zelikhina, S.; Korennoy, F.; Grishchenko, M. Spatial patterns of West Nile virus distribution in the Volgograd region of Russia, a territory with long-existing foci. PLoS Negl. Trop. Dis. 2022, 16, e0010145. [Google Scholar] [CrossRef]
- Demenev, V.A.; Zhang, F.; Makarova, T.E.; Huanyu, W.; Savin, S.Z.; Yan, L. Japanese encephalitis, as a problem of reemerging arboviral neuroinfections on border territory of Far East Russian and China. Zdr. Dal’Nego Vostoka 2016, 68, 52–57. [Google Scholar]
- Yastrebov, V.K.; Yakimenko, V.V. The Omsk hemorrhagic fever: Research results (1946–2013). Vopr. Virusol. 2014, 59, 5–11. (In Russian) [Google Scholar]
- Leonova, G.N.; Kondratov, I.G.; Ternovoi, V.; Romanova, E.V.; Protopopova, E.V.; Chausov, E.; Pavlenko, E.V.; Ryabchikova, E.I.; Belikov, S.I.; Loktev, V.B. Characterization of Powassan viruses from Far Eastern Russia. Arch. Virol. 2009, 154, 811–820. (In Russian) [Google Scholar] [CrossRef]
- Leonova, G.N.; Kondratov, I.G.; Maystrovskaya, O.S.; Takashima, I.; Belikov, S.I. Louping ill virus (LIV) in the Far East. Arch. Virol. 2015, 160, 663–673. (In Russian) [Google Scholar] [CrossRef]
- Dinçer, E.; Hacıoğlu, S.; Kar, S.; Emanet, N.; Brinkmann, A.; Nitsche, A.; Özkul, A.; Linton, Y.-M.; Ergünay, K. Survey and Characterization of Jingmen Tick Virus Variants. Viruses 2019, 11, 1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garry, C.E.; Garry, R.F. Proteomics Computational Analyses Suggest that the Envelope Glycoproteins of Segmented Jingmen Flavi-Like Viruses Are Class II Viral Fusion Proteins (β-Penetrenes) with Mucin-Like Domains. Viruses 2020, 12, 260. [Google Scholar] [CrossRef] [PubMed]
- Kholodilov, I.S.; Belova, O.A.; Morozkin, E.S.; Litov, A.G.; Ivannikova, A.Y.; Makenov, M.T.; Shchetinin, A.M.; Aibulatov, S.V.; Bazarova, G.K.; Bell-Sakyi, L.; et al. Geographical and Tick-Dependent Distribution of Flavi-like Alongshan and Yanggou Tick Viruses in Russia. Viruses 2021, 13, 458. [Google Scholar] [CrossRef] [PubMed]
- Temmam, S.; Bigot, T.; Chrétien, D.; Gondard, M.; Pérot, P.; Pommelet, V.; Dufou, E.; Petres, S.; Devillers, E.; Hoem, T.; et al. Insights into the Host Range, Genetic Diversity, and Geographical Distribution of Jingmenviruses. mSphere 2019, 4, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kuivanen, S.; Levanov, L.; Kareinen, L.; Sironen, T.; Jääskeläinen, A.J.; Plyusnin, I.; Zakham, F. Detection of novel tick-borne pathogen, Alongshan virus, in Ixodes ricinus ticks, south-eastern Finland. Eurosurveillance 2019, 24, 1900394. [Google Scholar] [CrossRef] [Green Version]
- Villa, E.C.; Maruyama, S.R.; de Miranda-Santos, I.K.F.; Palacios, G.; Ladner, J.T. Complete Coding Genome Sequence for Mogiana Tick Virus, a Jingmenvirus Isolated from Ticks in Brazil. Genome Announc. 2017, 5, 17–18. [Google Scholar] [CrossRef] [Green Version]
- Ladner, J.T.; Wiley, M.R.; Beitzel, B.; Auguste, A.J.; Dupuis, A.P.; Lindquist, M.E.; Sibley, S.D.; Kota, K.P.; Fetterer, D.; Eastwood, G.; et al. A Multicomponent Animal Virus Isolated from Mosquitoes. Cell Host. Microbe. 2016, 20, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Ogola, E.O.; Kopp, A.; Bastos, A.D.S.; Slothouwer, I.; Marklewitz, M.; Omoga, D.; Rotich, G.; Getugi, C.; Sang, R.; Torto, B.; et al. Jingmen Tick Virus in Ticks from Kenya. Viruses 2022, 14, 1041. [Google Scholar] [CrossRef]
- De Souza, W.M.; Fumagalli, M.J.; de Torres Carrasco, A.O.; Romeiro, M.F.; Modha, S.; Seki, M.C.; Gheller, J.M.; Daffre, S.; Nunes, M.R.T.; Murcia, P.R.; et al. Viral diversity of Rhipicephalus microplus parasitizing cattle in southern Brazil. Sci. Rep. 2018, 8, 16315. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-D.; Wang, W.; Wang, N.N.; Qiu, K.; Zhang, X.; Tana, G.; Liu, Q. Prevalence of the emerging novel Alongshan virus infection in sheep and cattle in Inner Mongolia, northeastern China. Parasit. Vectors 2019, 12, 450. [Google Scholar] [CrossRef] [Green Version]
- Maruyama, S.R.; Castro-Jorge, L.A.; Ribeiro, J.M.C.; Gardinassi, L.G.; Garcia, G.R.; Brandão, L.G.; Rodrigues, A.R.; Okada, M.I.; Abrão, E.P.; Ferreira, B.R.; et al. Characterisation of divergent flavivirus NS3 and NS5 protein sequences detected in Rhipicephalus microplus ticks from Brazil. Mem. Inst. Oswaldo Cruz. 2014, 109, 38–50. [Google Scholar] [CrossRef]
- Emmerich, P.; Jakupi, X.; von Possel, R.; Berisha, L.; Halili, B.; Günther, S.; Cadar, D.; Ahmeti, S.; Schmidt-Chanasit, J. Viral metagenomics, genetic and evolutionary characteristics of Crimean-Congo hemorrhagic fever orthonairovirus in humans, Kosovo. Infect. Genet. Evol. 2018, 65, 6–11. [Google Scholar] [CrossRef]
- Kholodilov, I.S.; Litov, A.G.; Klimentov, A.S.; Belova, O.A.; Polienko, A.E.; Nikitin, N.A.; Shchetinin, A.M.; Ivannikova, A.Y.; Bell-Sakyi, L.; Yakovlev, A.S.; et al. Isolation and characterisation of Alongshan virus in Russia. Viruses 2020, 12, 362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ternovoi, V.A.; Gladysheva, A.V.; Sementsova, A.O.; Zaykovskaya, A.V.; Volynkina, A.S.; Kotenev, E.S.; Agafonov, A.P.; Loktev, V.B. Detection of the RNA for New Multicomponent Virus in Patients with Crimean-Congo Hemorrhagic Fever in Southern Russia. Vestn. Ross. Akad. Meditsinskikh Nauk 2020, 75, 129–134. [Google Scholar] [CrossRef]
- Kuhn, J.H.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Amarasinghe, G.K.; Anthony, S.J.; Avšič-Županc, T.; Ayllón, M.A.; Bahl, J.; Balkema-Buschmann, A.; et al. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch. Virol. 2020, 165, 3023–3072. [Google Scholar] [CrossRef] [PubMed]
- Wright, D.; Kortekaas, J.; Bowden, T.A.; Warimwe, G.M. Rift Valley fever: Biology and epidemiology. J. Gen. Virol. 2019, 100, 1187–1199. [Google Scholar] [CrossRef]
- Klimentov, A.S.; Belova, O.A.; Kholodilov, I.S.; Butenko, A.M.; Bespyatova, L.A.; Bugmyrin, S.V.; Chernetsov, N.; Ivannikova, A.Y.; Kovalchuk, I.V.; Nafeev, A.A.; et al. Phlebovirus sequences detected in ticks collected in the Russian Federation: Novel species, distinguish criteria, high tick specificity. Infect. Genet. Evol. 2020, 85, 104524. [Google Scholar] [CrossRef]
- Bugmyrin, S.V.; Romanova, L.Y.; Belova, O.A.; Kholodilov, I.S.; Bespyatova, L.A.; Chernokhaeva, L.L.; Gmyl, L.V.; Klimentov, A.S.; Ivannikova, A.Y.; Polienko, A.E.; et al. Ticks and Tick-borne Diseases Pathogens in Ixodes persulcatus and Ixodes ricinus ticks (Acari, Ixodidae) in Karelia. Ticks Tick. Borne. Dis. 2022, 13, 102045. [Google Scholar] [CrossRef]
- Cutler, S.J.; Vayssier-Taussat, M.; Estrada-Peña, A.; Potkonjak, A.; Mihalca, A.D.; Zeller, H. Tick-borne diseases and co-infection: Current considerations. Ticks Tick. Borne. Dis. 2021, 12, 101607. [Google Scholar] [CrossRef]
- Capligina, V.; Seleznova, M.; Akopjana, S.; Freimane, L.; Lazovska, M.; Krumins, R.; Kivrane, A.; Namina, A.; Aleinikova, D.; Kimsis, J.; et al. Large-scale countrywide screening for tick-borne pathogens in field-collected ticks in Latvia during 2017–2019. Parasites Vectors 2020, 13, 351. [Google Scholar] [CrossRef]
- General Characteristics of the Chelyabinsk Region. Available online: https://chelreglib.ru/ru/pages/kray/chelregion/general/ (accessed on 2 June 2022).
- Rubel, F.; Brugger, K.; Belova, O.A.; Kholodilov, I.S.; Didyk, Y.M.; Kurzrock, L.; García-Pérez, A.L.; Kahl, O. Vectors of disease at the northern distribution limit of the genus Dermacentor in Eurasia: D. reticulatus and D. silvarum. Exp. Appl. Acarol. 2020, 82, 95–123. [Google Scholar] [CrossRef] [PubMed]
- Luchinina, S.V.; Feldblyum, I.V.; Usoltseva, N.M. The role of ticks of the genus Dermacentor in transmission of tick-borne encephalitis in the Southern Urals. In Results and Prospects of Studying the Problems of Infectious and Parasitic Diseases; University of Tyumen: Tyumen, Russia, 2015; pp. 202–205. (In Russian) [Google Scholar]
- Luchinina, S.V.; Stepanova, O.N.; Pogodina, V.V.; Sten’ko, E.A.; Chirkova, G.G.; Gerasimov, S.G.; Kolesnikova, L.I. Modern Epidemiological Situation of Tick-Borne Encephalitis in Chelyabinsk Region, Russia. Epidemiol. Vaccinal Prev. 2014, 75, 32–37. (In Russian) [Google Scholar]
- Leonovich, S.A. Types of Parasitism of Hard Ticks (Ixodidae). Entomol. Rev. 2020, 100, 1201–1204. [Google Scholar] [CrossRef]
- Luchinina, S.V.; Semenov, A.I.; Stepanova, O.N.; Pogodina, V.V.; Gerasimov, S.G.; Shcherbinina, M.S.; Kolesnikova, L.I.; Suslova, T.A. Vaccinal Prevention of Tick-Borne Encephalitis in Chelyabinsk Region: Dynamics of Vaccination, Population Immunity, Analysis of TBE Cases in Vaccinated Persons. Epidemiol. Vaccinal Prev. 2016, 86, 67–76. (In Russian) [Google Scholar] [CrossRef] [Green Version]
- Morozova, O.V.; Grishechkin, A.E.; Konkova-Reidman, A.B. Quantitations of Borrelia and Bartonella DNA and tick-borne encephalitis virus RNA in Ixodes persulcatus ticks collected in Chelyabinsk region. Mol. Genet. Microbiol. Virol. 2011, 26, 41–45. [Google Scholar] [CrossRef]
- Filippova, N.A. Ixodid Ticks of the Subfamily Ixodinae. Fauna of the USSR: Arachnoides; Nauka: Leningrad, Russia, 1977; Volume 4. (In Russian) [Google Scholar]
- Filippova, N.A. Fauna of Russia and Neighbouring Countries. Ixodid Ticks of Subfamily Amblyomminae. Arachnoidea; Nauka: Saint Petersburg, Russia, 1997; Volume 4. (In Russian) [Google Scholar]
- Bell-Sakyi, L. Continuous cell lines from the tick Hyalomma anatolicum anatolicum. J. Parasitol. 1991, 77, 1006–1008. [Google Scholar] [CrossRef]
- Kholodilov, I.; Belova, O.; Burenkova, L.; Korotkov, Y.; Romanova, L.; Morozova, L.; Kudriavtsev, V.; Gmyl, L.; Belyaletdinova, I.; Chumakov, A.; et al. Ixodid ticks and tick-borne encephalitis virus prevalence in the South Asian part of Russia (Republic of Tuva). Ticks Tick. Borne. Dis. 2019, 10, 959–969. [Google Scholar] [CrossRef]
- Scaramozzino, N.; Crance, J.-M.; Jouan, A.; DeBriel, D.A.; Stoll, F.; Garin, D. Comparison of Flavivirus universal primer pairs and development of a rapid, highly sensitive heminested reverse transcription-PCR assay for detection of flaviviruses targeted to a conserved region of the NS5 gene sequences. J. Clin. Microbiol. 2001, 39, 1922–1927. [Google Scholar] [CrossRef] [Green Version]
- Klimentov, A.S.; Butenko, A.M.; Khutoretskaya, N.V.; Shustova, E.Y.; Larichev, V.F.; Isaeva, O.V.; Karganova, G.G.; Lukashev, A.N.; Gmyl, A.P. Development of pan-phlebovirus RT-PCR assay. J. Virol. Methods 2016, 232, 29–32. [Google Scholar] [CrossRef]
- Romanova, L.I.; Kozlovskaya, L.I.; Shevtsova, A.S.; Karganova, G.G. Evidence for the absence of tick-borne encephalitis virus RNA in bioassays. Vopr. Virusol. 2006, 51, 38–41. (In Russian) [Google Scholar]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Williams, C.J.; Moffitt, C.M. A Critique of Methods of Sampling and Reporting Pathogens in Populations of Fish. J. Aquat. Anim. Health 2001, 13, 300–309. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 2 June 2022).
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. Available online: http://qgis.osgeo.org (accessed on 24 May 2022).
- Hvidsten, D.; Stuen, S.; Jenkins, A.; Dienus, O.; Olsen, R.S.; Kristiansen, B.E.; Mehl, R.; Matussek, A. Ixodes ricinus and Borrelia prevalence at the Arctic Circle in Norway. Ticks Tick. Borne. Dis. 2014, 5, 107–112. [Google Scholar] [CrossRef]
- Paulsen, K.M.; Pedersen, B.N.; Soleng, A.; Okbaldet, Y.B.; Pettersson, J.H.O.; Dudman, S.G.; Ottesen, P.; Vik, I.S.S.; Vainio, K.; Andreassen, Å. Prevalence of tick-borne encephalitis virus in Ixodes ricinus ticks from three islands in north-western Norway. Apmis 2015, 123, 759–764. [Google Scholar] [CrossRef]
- Soleng, A.; Edgar, K.S.; Paulsen, K.M.; Pedersen, B.N.; Okbaldet, Y.B.; Skjetne, I.E.B.; Gurung, D.; Vikse, R.; Andreassen, Å.K. Distribution of Ixodes ricinus ticks and prevalence of tick-borne encephalitis virus among questing ticks in the Arctic Circle region of northern Norway. Ticks Tick. Borne. Dis. 2018, 9, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Kreusch, T.M.; Holding, M.; Hewson, R.; Harder, T.; Medlock, J.M.; Hansford, K.M.; Dowall, S.; Semper, A.; Brooks, T.; Walsh, A.; et al. A probable case of tick-borne encephalitis (TBE) acquired in England, July 2019. Eurosurveillance 2019, 24, 1900679. [Google Scholar] [CrossRef] [Green Version]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; Pullan, S.T.; Lewis, J.; Vipond, R.; Rocchi, M.S.; Baylis, M.; Hewson, R. Tick-borne encephalitis virus, United Kingdom. Emerg. Infect. Dis. 2020, 26, 90–96. [Google Scholar] [CrossRef]
- Holding, M.; Dowall, S.D.; Medlock, J.M.; Carter, D.P.; McGinley, L.; Curran-French, M.; Pullan, S.T.; Chamberlain, J.; Hansford, K.M.; Baylis, M.; et al. Detection of new endemic focus of tick-borne encephalitis virus (TBEV), Hampshire/Dorset border, England, September 2019. Eurosurveillance 2019, 24, 1900658. [Google Scholar] [CrossRef] [Green Version]
- Jahfari, S.; De Vries, A.; Rijks, J.M.; Van Gucht, S.; Vennema, H.; Sprong, H.; Rockx, B. Tick-borne encephalitis virus in ticks and roe deer, the Netherlands. Emerg. Infect. Dis. 2017, 23, 1028–1030. [Google Scholar] [CrossRef]
- Dekker, M.; Laverman, G.D.; de Vries, A.; Reimerink, J.; Geeraedts, F. Emergence of tick-borne encephalitis (TBE) in the Netherlands. Ticks Tick. Borne. Dis. 2019, 10, 176–179. [Google Scholar] [CrossRef] [PubMed]
- Makenov, M.; Karan, L.; Shashina, N.; Akhmetshina, M.; Zhurenkova, O.; Kholodilov, I.; Karganova, G.; Smirnova, N.; Grigoreva, Y.; Yankovskaya, Y.; et al. First detection of tick-borne encephalitis virus in Ixodes ricinus ticks and their rodent hosts in Moscow, Russia. Ticks Tick. Borne. Dis. 2019, 10, 101265. [Google Scholar] [CrossRef]
- Khamassi Khbou, M.; Romdhane, R.; Foughali, A.A.; Sassi, L.; Suin, V.; Rekik, M.; Benzarti, M. Presence of antibodies against tick-borne encephalitis virus in sheep in Tunisia, North Africa. BMC Vet. Res. 2020, 16, 441. [Google Scholar] [CrossRef] [PubMed]
- Fares, W.; Dachraoui, K.; Cherni, S.; Barhoumi, W.; Ben Slimane, T.; Younsi, H.; Zhioua, E. Tick-borne encephalitis virus in Ixodes ricinus (Acari: Ixodidae) ticks, Tunisia. Ticks Tick. Borne. Dis. 2021, 12, 101606. [Google Scholar] [CrossRef]
- Pogodina, V.; Luchinina, S.V.; Stepanova, O.N.; Stenko, E.A.; Gorfinke, A.N.; Karmysheva, V.Y.; Gerasimov, S.G.; Levina, L.S.; Chirkova, G.G.; Karan, L.S.; et al. Unusual case of lethal tick-borne encephalitis in patient vaccinated with vaccines produced from different viruses strains (the Chelyabinsk Region). Epidemiohgiya Indectsionnye Bolezn. 2015, 20, 56–64. [Google Scholar] [CrossRef]
- Shchuchinova, L.D.; Kozlova, I.V.; Zlobin, V.I. Influence of altitude on tick-borne encephalitis infection risk in the natural foci of the Altai Republic, Southern Siberia. Ticks Tick. Borne. Dis. 2015, 6, 322–329. [Google Scholar] [CrossRef]
- Wójcik-Fatla, A.; Cisak, E.; Zajac, V.; Zwoliński, J.; Dutkiewicz, J. Prevalence of tick-borne encephalitis virus in Ixodes ricinus and Dermacentor reticulatus ticks collected from the Lublin region (eastern Poland). Ticks Tick. Borne. Dis. 2011, 2, 16–19. [Google Scholar] [CrossRef]
- Belova, O.A.; Kholodilov, I.S.; Litov, A.G.; Karganova, G.G. The Ability of Ixodid Ticks (Acari: Ixodidae) to Support Reproduction of the Tick-Borne Encephalitis Virus. Entomol. Rev. 2018, 98, 1369–1378. [Google Scholar] [CrossRef]
- Belova, O.A.; Litov, A.G.; Kholodilov, I.S.; Kozlovskaya, L.I.; Bell-Sakyi, L.; Romanova, L.I.; Karganova, G.G. Properties of the tick-borne encephalitis virus population during persistent infection of ixodid ticks and tick cell lines. Ticks Tick. Borne. Dis. 2017, 8, 895–906. [Google Scholar] [CrossRef]
- Valitskaya, A.V.; Katin, A.A.; Bragina, E.A.; Shuman, V.A. Comparative study of the role of the ixodid ticks Ixodes persulcatus and Dermacentor reticulatus in the epidemiological process of tick-borne encephalitis in the western part of Western Siberia. In Proceedings of the VIII International Scientifc Conference on ‘Reading in Memory of Professor I. I. Barabash-Nikiforov’, Voronezh, Russian, 10 March 2016; pp. 21–26. (In Russian). [Google Scholar]
- Rubel, F.; Brugger, K.; Pfeffer, M.; Chitimia-Dobler, L.; Didyk, Y.M.; Leverenz, S.; Dautel, H.; Kahl, O. Geographical distribution of Dermacentor marginatus and Dermacentor reticulatus in Europe. Ticks Tick. Borne. Dis. 2016, 7, 224–233. [Google Scholar] [CrossRef] [Green Version]
- Li, L.F.; Zhang, M.Z.; Zhu, J.G.; Cui, X.M.; Zhang, C.F.; Niu, T.Y.; Li, J.; Sun, Y.; Wei, W.; Liu, H.B.; et al. Dermacentor silvarum, a Medically Important Tick, May Not Be a Competent Vector to Transmit Jingmen Tick Virus. Vector-Borne Zoonotic Dis. 2022, 22, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-J.; Lin, X.-D.; Chen, Y.-M.; Hao, Z.-Y.; Wang, Z.-X.; Yu, Z.-M.; Lu, M.; Li, K.; Qin, X.-C.; Wang, W.; et al. Diversity and circulation of Jingmen tick virus in ticks and mammals. Virus Evol. 2020, 6, veaa051. [Google Scholar] [CrossRef] [PubMed]
- Morozkin, E.S.; Makenov, M.T.; Zhurenkova, O.B.; Kholodilov, I.S.; Belova, O.A.; Radyuk, E.V.; Fyodorova, M.V.; Grigoreva, Y.E.; Litov, A.G.; Valdokhina, A.V.; et al. Integrated Jingmenvirus Polymerase Gene in Ixodes ricinus Genome. Viruses 2022, 14, 1908. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.S.; Kahl, O.; Lane, R.S.; Levin, M.L.; Tsao, J.I. Diapause in ticks of the medically important Ixodes ricinus species complex. Ticks Tick. Borne. Dis. 2016, 7, 992–1003. [Google Scholar] [CrossRef]
- Gray, J.S. The ecology of ticks transmitting Lyme borreliosis. Exp. Appl. Acarol. 1998, 22, 249–258. [Google Scholar] [CrossRef]
- Kahl, O.; Dautel, H. Seasonal Life Cycle Organisation of the Ixodid Tick Dermacentor reticulatus in Central Europe Implications on Its Vector Role and Distribution. Med. Kuzbass 2013, 12, 84–87. [Google Scholar]
- Földvári, G.; Široký, P.; Szekeres, S.; Majoros, G.; Sprong, H. Dermacentor reticulatus: A vector on the rise. Parasites and Vectors 2016, 9, 314. [Google Scholar] [CrossRef] [Green Version]
- Torii, S.; Matsuno, K.; Qiu, Y.; Mori-Kajihara, A.; Kajihara, M.; Nakao, R.; Nao, N.; Okazaki, K.; Sashika, M.; Hiono, T.; et al. Infection of newly identified phleboviruses in ticks and wild animals in Hokkaido, Japan indicating tick-borne life cycles. Ticks Tick. Borne. Dis. 2019, 10, 328–335. [Google Scholar] [CrossRef]






| Latitude, Longitude | Tick Species | Number of Analysed Ticks (Total) | Number of Analysed Pools | Number of Virus-Positive Pools | Prevalence a, % (CI 95%) | Virus, Strains |
|---|---|---|---|---|---|---|
| 53.881103, 59.163164 | D. reticulatus | 65 | 15 | 4 | 6.95 (2.21–15.50) | Stavropol, STA-Che-T19070 b |
| Stavropol, STA-Che-T19075 b | ||||||
| Stavropol, STA-Che-T19086 b | ||||||
| Stavropol, STA-Che-T19091 b | ||||||
| D. marginatus | 3 | 1 | 0 | 0 | - | |
| 53.382567, 59.9295 | D. reticulatus | 11 | 4 | 2 | 21.74 (3.93–54.60) c | YGTV, Fershampenuaz14-T19014 |
| YGTV, Paris14-T19197 | ||||||
| D. marginatus | 39 | 9 | 0 | 0 | - | |
| I. persulcatus | 1 | 1 | 0 | 0 | - | |
| 52.885633, 60.051833 | D. reticulatus | 125 | 26 | 3 | 2.52 (0.63–6.42) | YGTV, Kartaly14-T19309 |
| YGTV, Kartaly14-T19314 | ||||||
| YGTV, Kartaly14-T19551 | ||||||
| D. marginatus | 112 | 23 | 2 | 1.85 (0.31–5.61) | YGTV, Kartaly14-T19623 | |
| YGTV, Kartaly14-T19658 | ||||||
| I. persulcatus | 2 | 1 | 0 | 0 | - | |
| 52.940933, 59.935533 | D. reticulatus | 7 | 2 | 1 | 17.02 (1.05–57.59) c | YGTV, Kartaly14-T19346 |
| 52.053183, 59.957667 | D. reticulatus | 6 | 2 | 0 | 0 | - |
| D. marginatus | 130 | 27 | 1 | 0.78 (0.04–3.39) | YGTV, Bredy14-T19463 | |
| 52.497667, 60.00035 | D. reticulatus | 21 | 5 | 2 | 11.70 (2.02–32.62) c | YGTV, Bredy14-T19736 |
| YGTV, Bredy14-T19741 | ||||||
| D. marginatus | 30 | 6 | 0 | 0 | - | |
| 52.47595, 59.871383 | D. reticulatus | 14 | 3 | 2 | 20.47 (3.60–54.09) c | YGTV, Bredy14-T19763 |
| YGTV, Bredy14-T19767 | ||||||
| D. marginatus | 6 | 2 | 0 | 0 | - | |
| 52.459233, 60.249483 | D. reticulatus | 3 | 1 | 0 | 0 | - |
| D. marginatus | 53 | 11 | 1 | 1.96 (0.11–8.36) | YGTV, Bredy14-T19813 | |
| 55.02145, 60.168283 | I. persulcatus | 169 | 33 | 9 | 6.00 (2.93–10.59) | ALSV, Miass501 b |
| ALSV, Miass502 | ||||||
| ALSV, Miass506 | ||||||
| ALSV, Miass508 b | ||||||
| ALSV, Miass510 b | ||||||
| ALSV, Miass515 b | ||||||
| ALSV, Miass519 | ||||||
| ALSV, Miass523 b | ||||||
| ALSV, Miass527 | ||||||
| 52.468017, 60.226217 | D. reticulatus | 102 | 26 | 4 | 4.18 (1.31–9.45) | YGTV, Bredy15-T22173 |
| YGTV, Bredy15-T22181 | ||||||
| YGTV, Bredy15-T22188 | ||||||
| YGTV, Bredy15-T22189 | ||||||
| D. marginatus | 24 | 7 | 0 | 0 | - | |
| 52.494667, 60.011817 | D. reticulatus | 14 | 4 | 0 | 0 | - |
| D. marginatus | 27 | 7 | 1 | 3.93 (0.23–16.22) | YGTV, Bredy15-T22208 | |
| 52.970983, 60.604833 | D. reticulatus | 15 | 4 | 1 | 7.17 (0.42–28.07) c | YGTV, Kartaly15-T22141 |
| D. marginatus | 16 | 4 | 0 | 0 | - | |
| 54.39845, 60.783167 | D. reticulatus | 130 | 34 | 2 | 1.58 (0.26–4.78) | YGTV, Plast15-T22436 |
| YGTV, Plast15-T22438 | ||||||
| D. marginatus | 15 | 4 | 1 | 7.17 (0.42–28.07) c | YGTV, Plast15-T22415 | |
| 54.569117, 60.288267 | D. reticulatus | 24 | 6 | 0 | 0 | - |
| D. marginatus | 118 | 30 | 1 | 0.86 (0.05–3.72) | YGTV, Mir15-T22470 | |
| 54.527867, 60.334233 | D. reticulatus | 5 | 2 | 0 | 0 | - |
| D. marginatus | 61 | 16 | 1 | 1.68 (0.10–7.19) | YGTV, Zaozernyy15-T22264 | |
| I. persulcatus | 4 | 2 | 0 | - | ||
| 55.209033, 59.571467 | I. persulcatus | 133 | 26 | 1 | 0.76 (0.04–3.32) | TBEV, Zlatoust15-T22637 |
| 2 | 1.56 (0.26–4.73) | Gomselga, GOM-Che-T22634 b | ||||
| Gomselga, GOM-Che-T22645 b | ||||||
| 55.22005, 59.56005 | I. persulcatus | 154 | 29 | 1 | 0.66 (0.04–2.87) | YGTV, Gubenka15-T22237 |
| 1 | 0.66 (0.04–2.87) | TBEV, Zlatoust15-T22241 | ||||
| 1 | 0.66 (0.04–2.87) | Gomselga, GOM-Che-T22248 b | ||||
| 55.021583, 60.169783 | I. persulcatus | 76 | 14 | 2 | 2.78 (0.47–8.35) | ALSV, Miass15-T22516 |
| ALSV, Miass15-T22517 | ||||||
| 55.357683, 59.51585 | I. persulcatus | 46 | 9 | 1 | 2.28 (0.13–9.64) | TBEV, Kusa15-T22532 |
| 56.168967, 60.471233 | I. persulcatus | 90 | 18 | 1 | 1.13 (0.06–4.88) | ALSV, Salma15-T22545 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kholodilov, I.S.; Belova, O.A.; Ivannikova, A.Y.; Gadzhikurbanov, M.N.; Makenov, M.T.; Yakovlev, A.S.; Polienko, A.E.; Dereventsova, A.V.; Litov, A.G.; Gmyl, L.V.; et al. Distribution and Characterisation of Tick-Borne Flavi-, Flavi-like, and Phenuiviruses in the Chelyabinsk Region of Russia. Viruses 2022, 14, 2699. https://doi.org/10.3390/v14122699
Kholodilov IS, Belova OA, Ivannikova AY, Gadzhikurbanov MN, Makenov MT, Yakovlev AS, Polienko AE, Dereventsova AV, Litov AG, Gmyl LV, et al. Distribution and Characterisation of Tick-Borne Flavi-, Flavi-like, and Phenuiviruses in the Chelyabinsk Region of Russia. Viruses. 2022; 14(12):2699. https://doi.org/10.3390/v14122699
Chicago/Turabian StyleKholodilov, Ivan S., Oxana A. Belova, Anna Y. Ivannikova, Magomed N. Gadzhikurbanov, Marat T. Makenov, Alexander S. Yakovlev, Alexandra E. Polienko, Alena V. Dereventsova, Alexander G. Litov, Larissa V. Gmyl, and et al. 2022. "Distribution and Characterisation of Tick-Borne Flavi-, Flavi-like, and Phenuiviruses in the Chelyabinsk Region of Russia" Viruses 14, no. 12: 2699. https://doi.org/10.3390/v14122699