Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis
Abstract
:1. Introduction
2. Generation of D RNAs and Mechanism of Induction
3. Structure of D RNAs and Their Population
4. Cis- and Trans- Regulation of DI RNA
5. Effect of D RNAs
5.1. Interference with Virus Accumulation
5.2. Attenuation of Infection Symptoms
5.3. Other Effects of D RNAs
6. Diversity and Evolution of D RNAs
7. D RNA Detection Tools
8. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Sanjuán, R.; Domingo-Calap, P. Genetic Diversity and Evolution of Viral Populations. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2021; pp. 53–61. [Google Scholar]
- Michalakis, Y.; Blanc, S. The Curious Strategy of Multipartite Viruses. Annu. Rev. Virol. 2020, 7, 203–218. [Google Scholar] [CrossRef]
- Roossinck, M.J. Mechanisms of Plantvirus Evolution. Annu. Rev. Phytopathol. 1997, 35, 191–209. [Google Scholar] [CrossRef] [PubMed]
- von Magnus, P.; Gard, S. Studies on Interference in Experimental Influenza. Ark. Kemi. Mineral. Geol. 1947, 24, 4. [Google Scholar]
- von Magnus, P. Incomplete Forms of Influenza Virus. In Advances in Virus Research; Smith, K.M., Lauffer, M.A., Eds.; Academic Press: Cambridge, MA, USA, 1954; Volume 2, pp. 59–79. [Google Scholar]
- Cooper, P.D.; Bellett, A.J. A Transmissible Interfering Component of Vesicular Stomatitis Virus Preparations. J. Gen. Microbiol. 1959, 21, 485–497. [Google Scholar] [CrossRef] [Green Version]
- Bellett, A.J.D.; Cooper, P.D. Some Properties of the Transmissible Interfering Component of Vesicular Stomatitis Virus Preparations. J. Gen. Microbiol. 1959, 21, 498–509. [Google Scholar] [CrossRef] [Green Version]
- Sokol, F.; Neurath, A.R.; Vilcek, J. Formation of Incomplete Sendai Virus in Embryonated Eggs. Acta Virol. 1964, 8, 59–67. [Google Scholar] [PubMed]
- Huang, A.S.; Baltimore, D. Defective Viral Particles and Viral Disease Processes. Nature 1970, 226, 325–327. [Google Scholar] [CrossRef]
- Vignuzzi, M.; López, C.B. Defective Viral Genomes Are Key Drivers of the Virus-Host Interaction. Nat. Microbiol. 2019, 4, 1075–1087. [Google Scholar] [CrossRef]
- Noppornpanth, S.; Smits, S.L.; Lien, T.X.; Poovorawan, Y.; Osterhaus, A.D.M.E.; Haagmans, B.L. Characterization of Hepatitis C Virus Deletion Mutants Circulating in Chronically Infected Patients. J. Virol. 2007, 81, 12496–12503. [Google Scholar] [CrossRef] [Green Version]
- Pesko, K.N.; Fitzpatrick, K.A.; Ryan, E.M.; Shi, P.-Y.; Zhang, B.; Lennon, N.J.; Newman, R.M.; Henn, M.R.; Ebel, G.D. Internally Deleted WNV Genomes Isolated from Exotic Birds in New Mexico: Function in Cells, Mosquitoes, and Mice. Virology 2012, 427, 10–17. [Google Scholar] [CrossRef]
- Ziegler, C.M.; Botten, J.W. Defective Interfering Particles of Negative-Strand RNA Viruses. Trends Microbiol. 2020, 28, 554–565. [Google Scholar] [CrossRef] [PubMed]
- Adam, G.; Gaedigk, K.; Mundry, K.W. Alterations of a Plant Rhabdovirus during Successive Mechanical Transfers/Veränderungen Eines Pflanzen-Rhabdovirus Im Zuge Serieller Mechanischer Transfers. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 1983, 90, 28–35. [Google Scholar]
- Verkleij, F.N.; Peters, D. Characterization of a Defective Form of Tomato Spotted Wilt Virus. J. Gen. Virol. 1983, 64, 677–686. [Google Scholar] [CrossRef]
- Hillman, B.I.; Carrington, J.C.; Morris, T.J. A Defective Interfering RNA That Contains a Mosaic of a Plant Virus Genome. Cell 1987, 51, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Burgyán, J.; Grieco, F.; Russo, M. A Defective Interfering RNA Molecule in Cymbidium Ringspot Virus Infections. J. Gen. Virol. 1989, 70, 235–239. [Google Scholar] [CrossRef]
- Li, X.H.; Heaton, L.A.; Morris, T.J.; Simon, A.E. Turnip Crinkle Virus Defective Interfering RNAs Intensify Viral Symptoms and Are Generated de Novo. Proc. Natl. Acad. Sci. USA 1989, 86, 9173–9177. [Google Scholar] [CrossRef] [Green Version]
- Rochon, D.M. Rapid de Novo Generation of Defective Interfering RNA by Cucumber Necrosis Virus Mutants That Do Not Express the 20-KDa Nonstructural Protein. Proc. Natl. Acad. Sci. USA 1991, 88, 11153–11157. [Google Scholar] [CrossRef] [Green Version]
- Graves, M.V.; Roossinck, M.J. Characterization of Defective RNAs Derived from RNA 3 of the Fny Strain of Cucumber Mosaic Cucumovirus. J. Virol. 1995, 69, 4746–4751. [Google Scholar] [CrossRef] [Green Version]
- Knapp, E.; Danyluk, G.M.; Achor, D.; Lewandowski, D.J. A Bipartite Tobacco Mosaic Virus-Defective RNA (D RNA) System to Study the Role of the N-Terminal Methyl Transferase Domain in Cell-to-Cell Movement of D RNAs. Virology 2005, 341, 47–58. [Google Scholar] [CrossRef]
- Czosnek, H.; Ber, R.; Navot, N.; Antignus, Y.; Cohen, S.; Zamir, D. Tomato Yellow Leaf Curl Virus DNA Forms in the Viral Capside, in Infected Plants And in The Insect Vector. J. Phytopathol. 1989, 125, 47–54. [Google Scholar] [CrossRef]
- Casado, C.G.; Javier Ortiz, G.; Padron, E.; Bean, S.J.; McKenna, R.; Agbandje-McKenna, M.; Boulton, M.I. Isolation and Characterization of Subgenomic DNAs Encapsidated in “Single” T = 1 Isometric Particles of Maize Streak Virus. Virology 2004, 323, 164–171. [Google Scholar] [CrossRef] [PubMed]
- Patil, B.L.; Dasgupta, I. Defective Interfering DNAs of Plant Viruses. Crit. Rev. Plant Sci. 2006, 25, 47–64. [Google Scholar] [CrossRef]
- Knorr, D.A.; Mullin, R.H.; Hearne, P.Q.; Morris, T.J. De Novo Generation of Defective Interfering RNAs of Tomato Bushy Stunt Virus by High Multiplicity Passage. Virology 1991, 181, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Law, M.D.; Morris, T.J. De Novo Generation and Accumulation of Tomato Bushy Stunt Virus Defective Interfering RNAs without Serial Host Passage. Virology 1994, 198, 377–380. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.C.; Borja, M.; Scholthof, H.B.; Jackson, A.O.; Morris, T.J. Host Effects and Sequences Essential for Accumulation of Defective Interfering RNAs of Cucumber Necrosis and Tomato Bushy Stunt Tombusviruses. Virology 1995, 210, 41–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burgyán, J.; Rubino, L.; Russo, M. De Novo Generation of Cymbidium Ringspot Virus Defective Interfering RNA. J. Gen. Virol. 1991, 72, 505–509. [Google Scholar] [CrossRef]
- Kollàr, A.; Dalmay, T.; Burgyán, J. Defective Interfering RNA-Mediated Resistance against Cymbidium Ringspot Tombusvirus in Transgenic Plants. Virology 1993, 193, 313–318. [Google Scholar] [CrossRef]
- Havelda, Z.; Dalmay, T.; Burgyán, J. Localization of Cis-Acting Sequences Essential for Cymbidium Ringspot Tombusvirus Defective Interfering RNA Replication. J. Gen. Virol. 1995, 76, 2311–2316. [Google Scholar] [CrossRef]
- Resende, R.d.O.; de Haan, P.; van de Vossen, E.; de Avila, A.C.; Goldbach, R.; Peters, D. Defective Interfering L RNA Segments of Tomato Spotted Wilt Virus Retain Both Virus Genome Termini and Have Extensive Internal Deletions. J. Gen. Virol. 1992, 73, 2509–2516. [Google Scholar] [CrossRef]
- Resende, R.d.O.; de Haan, P.; de Avila, A.C.; Kitajima, E.W.; Kormelink, R.; Goldbach, R.; Peters, D. Generation of Envelope and Defective Interfering RNA Mutants of Tomato Spotted Wilt Virus by Mechanical Passage. J. Gen. Virol. 1991, 72, 2375–2383. [Google Scholar] [CrossRef]
- Che, X.; Mawassi, M.; Bar-Joseph, M. A Novel Class of Large and Infectious Defective RNAs of Citrus Tristeza Virus. Virology 2002, 298, 133–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mawassi, M.; Gafny, R.; Gagliardi, D.; Bar-Joseph, M. 1995 Populations of Citrus Tristeza Virus Contain Smaller-than-Full-Length Particles Which Encapsidate Sub-Genomic RNA Molecules. J. Gen. Virol. 2022, 76, 651–659. [Google Scholar] [CrossRef]
- Mawassi, M.; Karasev, A.V.; Mietkiewska, E.; Gafny, R.; Lee, R.F.; Dawson, W.O.; Bar-Joseph, M. Defective RNA Molecules Associated with Citrus Tristeza Virus. Virology 1995, 208, 383–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mawassi, M.; Mietkiewska, E.; Hilf, M.E.; Ashoulin, L.; Karasev, A.V.; Gafny, R.; Lee, R.F.; Garnsey, S.M.; Dawson, W.O.; Bar-joseph, M. Multiple Species of Defective RNAs in Plants Infected with Citrus Tristeza Virus. Virology 1995, 214, 264–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernandez, C.; Carette, J.E.; Brown, D.J.; Bol, J.F. Serial Passage of Tobacco Rattle Virus under Different Selection Conditions Results in Deletion of Structural and Nonstructural Genes in RNA 2. J. Virol. 1996, 70, 4933–4940. [Google Scholar] [CrossRef] [Green Version]
- Visser, P.B.; Brown, D.J.F.; Brederode, F.T.; Bol, J.F. Nematode Transmission of Tobacco Rattle Virus Serves as a Bottleneck to Clear the Virus Population from Defective Interfering RNAs. Virology 1999, 263, 155–165. [Google Scholar] [CrossRef] [Green Version]
- White, K.A.; Bancroft, J.B.; Mackie, G.A. Defective RNAs of Clover Yellow Mosaic Virus Encode Nonstructural/Coat Protein Fusion Products. Virology 1991, 183, 479–486. [Google Scholar] [CrossRef]
- White, K.A.; Bancroft, J.B.; Mackie, G.A. Coding Capacity Determines in Vivo Accumulation of a Defective RNA of Clover Yellow Mosaic Virus. J. Virol. 1992, 66, 3069–3076. [Google Scholar] [CrossRef] [Green Version]
- Calvert, L.A.; Cuervo, M.I.; Ospina, M.D.; Fauquet, C.M.; Ramirez, B.C. Characterization of Cassava Common Mosaic Virus and a Defective RNA Species. J. Gen. Virol. 1996, 77, 525–530. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Nagano, H.; Mise, K.; Furusawa, I.; Okuno, T. Brome Mosaic Virus Defective RNAs Generated during Infection of Barley Plants. J. Gen. Virol. 1999, 80, 2511–2518. [Google Scholar] [CrossRef]
- Damayanti, T.A.; Nagano, H.; Mise, K.; Furusawa, I.; Okuno, T. Positional Effect of Deletions on Viability, Especially on Encapsidation, of Brome Mosaic Virus D-RNA in Barley Protoplasts. Virology 2002, 293, 314–319. [Google Scholar] [CrossRef] [Green Version]
- Romero, J.; Huang, Q.; Pogany, J.; Bujarski, J.J. Characterization of Defective Interfering RNA Components That Increase Symptom Severity of Broad Bean Mottle Virus Infections. Virology 1993, 194, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Llamas, S.; Sandoval, C.; Babin, M.; Pogany, J.; Bujarski, J.J.; Romero, J. Effect of the Host and Temperature on the Formation of Defective RNAs Associated with Broad Bean Mottle Virus Infection. Phytopathology 2004, 94, 69–75. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, I.B.; Lee, K.-C.; Canto, T.; Wong, S.-M.; Palukaitis, P. Host-Specific Encapsidation of a Defective RNA 3 of Cucumber Mosaic Virus. J. Gen. Virol. 2004, 85, 3757–3763. [Google Scholar] [CrossRef] [PubMed]
- Rubio, L.; Yeh, H.H.; Tian, T.; Falk, B.W. A Heterogeneous Population of Defective RNAs Is Associated with Lettuce Infectious Yellows Virus. Virology 2000, 271, 205–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubio, L.; Tian, T.; Yeh, H.-H.; Livieratos, Y.; Falk, B.W. De Novo Generation of Lettuce Infectious Yellows Virus Defective RNAs in Protoplasts. Mol. Plant. Pathol. 2002, 3, 321–327. [Google Scholar] [CrossRef] [Green Version]
- Rymelska, N.; Borodynko, N.; Pospieszny, H.; Hasiów-Jaroszewska, B. Analysis of the Biological and Molecular Variability of the Polish Isolates of Tomato Black Ring Virus (TBRV). Virus Genes 2013, 47, 338–346. [Google Scholar] [CrossRef]
- Budzyńska, D.; Minicka, J.; Hasiów-Jaroszewska, B.; Elena, S.F. Molecular Evolution of Tomato Black Ring Virus and de Novo Generation of a New Type of Defective RNAs during Long-term Passaging in Different Hosts. Plant Pathol. 2020, 69, 1767–1776. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Borodynko, N.; Figlerowicz, M.; Pospieszny, H. Two Types of Defective RNAs Arising from the Tomato Black Ring Virus Genome. Arch. Virol. 2012, 157, 569–572. [Google Scholar] [CrossRef]
- Lukhovitskaya, N.I.; Thaduri, S.; Garushyants, S.K.; Torrance, L.; Savenkov, E.I. Deciphering the Mechanism of Defective Interfering RNA (DI RNA) Biogenesis Reveals That a Viral Protein and the DI RNA Act Antagonistically in Virus Infection. J. Virol. 2013, 87, 6091–6103. [Google Scholar] [CrossRef] [Green Version]
- Torrance, L.; Cowan, G.H.; Sokmen, M.A.; Reavy, B. A Naturally Occurring Deleted Form of RNA 2 of Potato Mop-Top Virus. J. Gen. Virol. 1999, 80, 2211–2215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, K.A.; Morris, T.J. Defective and Defective Interfering RNAs of Monopartite Plus-Strand RNA Plant Viruses. Curr. Top. Microbiol. Immunol. 1999, 239, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Inoue-Nagata, A.K.; Kormelink, R.; Nagata, T.; Kitajima, E.W.; Goldbach, R.; Peters, D. Effects of Temperature and Host on the Generation of Tomato Spotted Wilt Virus Defective Interfering RNAs. Phytopathology 1997, 87, 1168–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Omarov, R.T.; Rezende, J.A.M.; Scholthof, H.B. Host-Specific Generation and Maintenance of Tomato Bushy Stunt Virus Defective Interfering RNAs. Mol. Plant. Microbe. Interact. 2004, 17, 195–201. [Google Scholar] [CrossRef] [Green Version]
- Qiu, W.; Park, J.W.; Jackson, A.O.; Scholthof, H.B. Retention of a Small Replicase Gene Segment in Tomato Bushy Stunt Virus Defective RNAs Inhibits Their Helper-Mediated Trans-Accumulation. Virology 2001, 281, 51–60. [Google Scholar] [CrossRef]
- Szittya, G.; Salamon, P.; Burgyán, J. The Complete Nucleotide Sequence and Synthesis of Infectious RNA of Genomic and Defective Interfering RNAs of TBSV-P. Virus. Res. 2000, 69, 131–136. [Google Scholar] [CrossRef]
- Kirkwood, T.B.; Bangham, C.R. Cycles, Chaos, and Evolution in Virus Cultures: A Model of Defective Interfering Particles. Proc. Natl. Acad. Sci. USA 1994, 91, 8685–8689. [Google Scholar] [CrossRef] [Green Version]
- Willemsen, A.; Zwart, M.P. On the Stability of Sequences Inserted into Viral Genomes. Virus Evol. 2019, 5, vez045. [Google Scholar] [CrossRef] [Green Version]
- White, K.A. Formation and Evolution Of Tombusvirus Defective Interfering RNAs. Semin. Virol. 1996, 7, 409–416. [Google Scholar] [CrossRef]
- Lazzarini, R.A.; Keene, J.D.; Schubert, M. The Origins of Defective Interfering Particles of the Negative-Strand RNA Viruses. Cell 1981, 26, 145–154. [Google Scholar] [CrossRef]
- Pathak, K.B.; Nagy, P.D. Defective Interfering RNAs: Foes of Viruses and Friends of Virologists. Viruses 2009, 1, 895–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasiów-Jaroszewska, B.; Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Elena, S.F. Defective RNA Particles Derived from Tomato Black Ring Virus Genome Interfere with the Replication of Parental Virus. Virus Res. 2018, 250, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Rubino, L.; Burgyán, J.; Grieco, F.; Russo, M. Sequence Analysis of Cymbidium Ringspot Virus Satellite and Defective Interfering RNAs. J. Gen. Virol. 1990, 71, 1655–1660. [Google Scholar] [CrossRef] [PubMed]
- Dalmay, T.; Szittya, G.; Burgyán, J. Generation of Defective Interfering RNA Dimers of Cymbidium Ringspot Tombusvirus. Virology 1995, 207, 510–517. [Google Scholar] [CrossRef] [Green Version]
- Szittya, G.; Silhavy, D.; Dalmay, T.; Burgyán, J. Size-Dependent Cell-to-Cell Movement of Defective Interfering RNAs of Cymbidium Ringspot Virus. J. Gen. Virol. 2002, 83, 1505–1510. [Google Scholar] [CrossRef] [Green Version]
- Quito-Avila, D.F.; Ibarra, M.A.; Alvarez, R.; Peralta, E.L.; Martin, R.R. A Raspberry Bushy Dwarf Virus Isolate from Ecuadorean Rubus Glaucus Contains an Additional RNA That Is a Rearrangement of RNA-2. Arch. Virol. 2014, 159, 2519–2521. [Google Scholar] [CrossRef]
- Figlerowicz, M.; Nagy, P.D.; Bujarski, J.J. A Mutation in the Putative RNA Polymerase Gene Inhibits Nonhomologous, but Not Homologous, Genetic Recombination in an RNA Virus. Proc. Natl. Acad. Sci. USA 1997, 94, 2073–2078. [Google Scholar] [CrossRef] [Green Version]
- Graves, M.V.; Pogany, J.; Romero, J. Defective Interfering RNAs and Defective Viruses Associated with Multipartite RNA Viruses of Plants. Semin. Virol. 1996, 7, 399–408. [Google Scholar] [CrossRef]
- Pogany, J.; Romero, J.; Huang, Q.; Sgro, J.-Y.; Shang, H.; Bujarski, J.J. De Novo Generation of Defective Interfering-like RNAs in Broad Bean Mottle Bromovirus. Virology 1995, 212, 574–586. [Google Scholar] [CrossRef] [Green Version]
- Russo, M.; Burgyán, J.; Martelli, G.P. Molecular Biology of Tombusviridae. In Advances in Virus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: Cambridge, MA, USA; Volume 44, pp. 381–428.
- Ray, D.; White, K.A. Enhancer-like Properties of an RNA Element That Modulates Tombusvirus RNA Accumulation. Virology 1999, 256, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Jupin, I.; Richards, K.; Jonard, G.; Guilley, H.; Pleij, C.W.A. Mapping Sequences Required for Productive Replication of Beet Necrotic Yellow Vein Virus RNA 3. Virology 1990, 178, 273–280. [Google Scholar] [CrossRef]
- Flobinus, A.; Chevigny, N.; Charley, P.A.; Seissler, T.; Klein, E.; Bleykasten-Grosshans, C.; Ratti, C.; Bouzoubaa, S.; Wilusz, J.; Gilmer, D. Beet Necrotic Yellow Vein Virus Noncoding Rna Production Depends on a 5′→3′ Xrn Exoribonuclease Activity. Viruses 2018, 10, 137. [Google Scholar] [CrossRef] [PubMed]
- Pathak, K.B.; Pogany, J.; Xu, K.; White, K.A.; Nagy, P.D. Defining the Roles of Cis-Acting RNA Elements in Tombusvirus Replicase Assembly In Vitro. J. Virol. 2012, 86, 156–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, K.A.; Morris, T.J. RNA Determinants of Junction Site Selection in RNA Virus Recombinants and Defective Interfering RNAs. RNA 1995, 1, 1029–1040. [Google Scholar] [PubMed]
- Wu, B.; White, K.A. Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J. Virol. 1998, 72, 9897–9905. [Google Scholar] [CrossRef] [Green Version]
- Chandrika, R.; Rabindran, S.; Lewandowski, D.J.; Manjunath, K.L.; Dawson, W.O. Full-Length Tobacco Mosaic Virus RNAs and Defective RNAs Have Different 3′ Replication Signals. Virology 2000, 273, 198–209. [Google Scholar] [CrossRef] [Green Version]
- Kollár, Á.; Burgyán, J. Evidence That ORF 1 and 2 Are the Only Virus-Encoded Replicase Genes of Cymbidium Ringspot Tombusvirus. Virology 1994, 201, 169–172. [Google Scholar] [CrossRef]
- Takeshita, M.; Matsuo, Y.; Yoshikawa, T.; Suzuki, M.; Furuya, N.; Tsuchiya, K.; Takanami, Y. Characterization of a Defective RNA Derived from RNA 3 of the Y Strain of Cucumber Mosaic Virus. Arch. Virol. 2008, 153, 579–583. [Google Scholar] [CrossRef]
- Takeshita, M.; Matsuo, Y.; Suzuki, M.; Furuya, N.; Tsuchiya, K.; Takanami, Y. Impact of a Defective RNA 3 from Cucumber Mosaic Virus on Helper Virus Infection Dynamics. Virology 2009, 389, 59–65. [Google Scholar] [CrossRef]
- Scholthof, K.B.; Scholthof, H.B.; Jackson, A.O. The Effect of Defective Interfering RNAs on the Accumulation of Tomato Bushy Stunt Virus Proteins and Implications for Disease Attenuation. Virology 1995, 211, 324–328. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.W.; Jackson, A.O.; Morris, T.J. Defective-Interfering RNAs and Elevated Temperatures Inhibit Replication of Tomato Bushy Stunt Virus in Inoculated Protoplasts. Virology 1990, 176, 539–545. [Google Scholar] [CrossRef]
- Vaucheret, H.; Béclin, C.; Fagard, M. Post-Transcriptional Gene Silencing in Plants. J. Cell Sci. 2001, 114, 3083–3091. [Google Scholar] [CrossRef]
- Silhavy, D.; Molnár, A.; Lucioli, A.; Szittya, G.; Hornyik, C.; Tavazza, M.; Burgyán, J. A Viral Protein Suppresses RNA Silencing and Binds Silencing-Generated, 21- to 25-Nucleotide Double-Stranded RNAs. EMBO J. 2002, 21, 3070–3080. [Google Scholar] [CrossRef]
- Szittya, G.; Molnár, A.; Silhavy, D.; Hornyik, C.; Burgyán, J. Short Defective Interfering RNAs of Tombusviruses Are Not Targeted but Trigger Post-Transcriptional Gene Silencing against Their Helper Virus. The Plant Cell 2002, 14, 359–372. [Google Scholar] [CrossRef]
- Simon, A.E.; Roossinck, M.J.; Havelda, Z. Plant Virus Satellite and Defective Interfering RNAs: New Paradigms for a New Century. Annu. Rev. Phytopathol. 2004, 42, 415–437. [Google Scholar] [CrossRef] [Green Version]
- Havelda, Z.; Szittya, G.; Burgyán, J. Characterization of the Molecular Mechanism of Defective Interfering RNA-Mediated Symptom Attenuation in Tombusvirus-Infected Plants. J. Virol. 1998, 72, 6251–6256. [Google Scholar] [CrossRef] [Green Version]
- Burgyán, J.; Hornyik, C.; Szittya, G.; Silhavy, D.; Bisztray, G. The ORF1 Products of Tombusviruses Play a Crucial Role in Lethal Necrosis of Virus-Infected Plants. J. Virol. 2000, 74, 10873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospieszny, H.; Hasiów-Jaroszewska, B.; Borodynko-Filas, N.; Elena, S.F. Effect of Defective Interfering RNAs on the Vertical Transmission of Tomato Black Ring Virus. Plant Prot. Sci. 2020, 56, 261–267. [Google Scholar] [CrossRef]
- Rubino, L.; Carrington, J.C.; Russo, M. Biologically Active Cymbidium Ringspot Virus Satellite RNA in Transgenic Plants Suppresses Accumulation of DI RNA. Virology 1992, 188, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Havelda, Z.; Dalmay, T.; Burgyán, J. Secondary Structure-Dependent Evolution of Cymbidium Ringspot Virus Defective Interfering RNA. J. Gen. Virol. 1997, 78, 1227–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, A.E.; Bujarski, J.J. Rna-Rna Recombination and Evolution in Virus-Infected Plants. Annu. Rev. Phytopathol. 1994, 32, 337–362. [Google Scholar] [CrossRef]
- Hasiów-Jaroszewska, B.; Boezen, D.; Zwart, M.P. Metagenomic Studies of Viruses in Weeds and Wild Plants: A Powerful Approach to Characterise Variable Virus Communities. Viruses 2021, 13, 1939. [Google Scholar] [CrossRef]
- Minicka, J.; Zarzyńska-Nowak, A.; Budzyńska, D.; Borodynko-Filas, N.; Hasiów-Jaroszewska, B. High-Throughput Sequencing Facilitates Discovery of New Plant Viruses in Poland. Plants 2020, 9, 820. [Google Scholar] [CrossRef]
- Ma, Y.; Fort, T.; Marais, A.; Lefebvre, M.; Theil, S.; Vacher, C.; Candresse, T. Leaf-Associated Fungal and Viral Communities of Wild Plant Populations Differ between Cultivated and Natural Ecosystems. Plant-Environ. Interact. 2021, 2, 87–99. [Google Scholar] [CrossRef]
- Thapa, V.; McGlinn, D.J.; Melcher, U.; Palmer, M.W.; Roossinck, M.J. Determinants of Taxonomic Composition of Plant Viruses at the Nature Conservancy’s Tallgrass Prairie Preserve, Oklahoma. Virus Evol. 2015, 1, vev007. [Google Scholar] [CrossRef]
- Maree, H.J.; Fox, A.; Al Rwahnih, M.; Boonham, N.; Candresse, T. Application of HTS for Routine Plant Virus Diagnostics: State of the Art and Challenges. Front. Plant Sci. 2018, 9, 1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Routh, A.; Johnson, J.E. Discovery of Functional Genomic Motifs in Viruses with ViReMa-a Virus Recombination Mapper-for Analysis of next-Generation Sequencing Data. Nucleic. Acids Res. 2014, 42, e11. [Google Scholar] [CrossRef] [Green Version]
- Beauclair, G.; Mura, M.; Combredet, C.; Tangy, F.; Jouvenet, N.; Komarova, A.V. DI-Tector: Defective Interfering Viral Genomes’ Detector for next-Generation Sequencing Data. RNA 2018, 24, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bosma, T.J.; Karagiannis, K.; Santana-Quintero, L.; Ilyushina, N.; Zagorodnyaya, T.; Petrovskaya, S.; Laassri, M.; Donnelly, R.P.; Rubin, S.; Simonyan, V.; et al. Identification and Quantification of Defective Virus Genomes in High Throughput Sequencing Data Using DVG-Profiler, a Novel Post-Sequence Alignment Processing Algorithm. PLoS ONE 2019, 14, e0216944. [Google Scholar] [CrossRef] [Green Version]
- Olmo-Uceda, M.J.; Muñoz-Sánchez, J.C.; Lasso-Giraldo, W.; Arnau, V.; Díaz-Villanueva, W.; Elena, S.F. DVGfinder: A Metasearch Tool for Identifying Defective Viral Genomes in RNA-Seq Data. Viruses 2022, 14, 1114. [Google Scholar] [CrossRef] [PubMed]
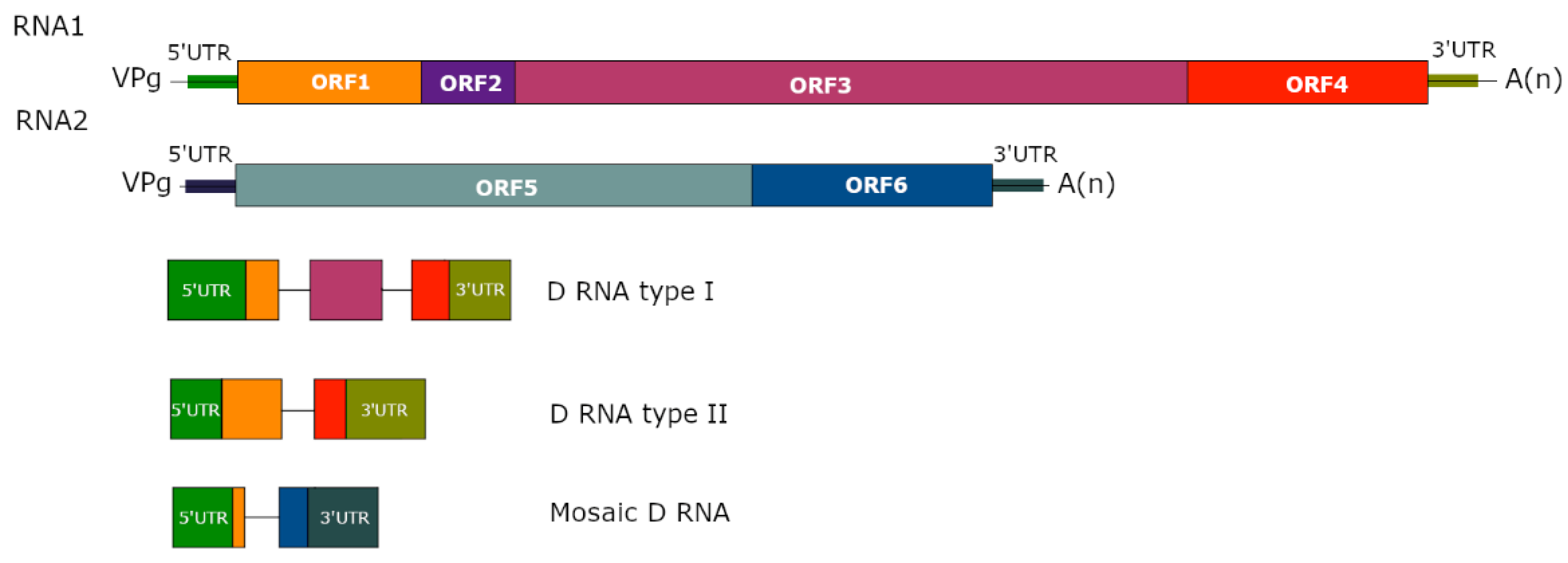
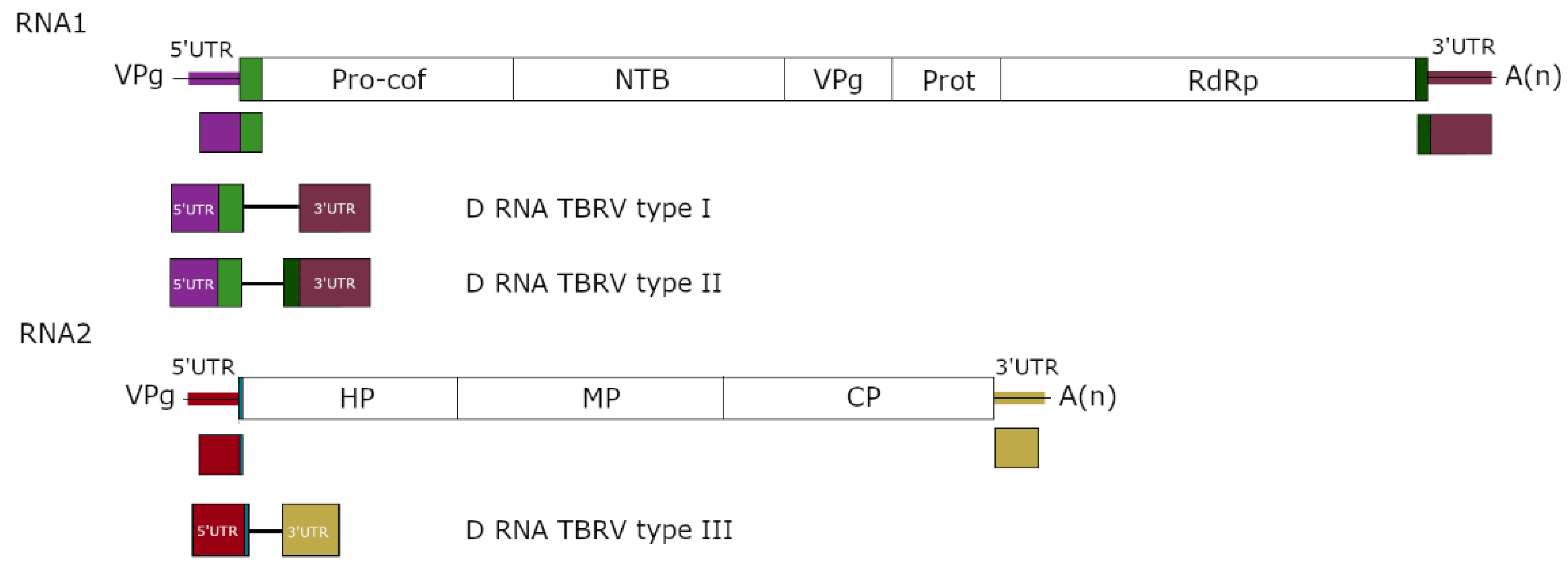
| Genus | Virus | Origin/Mutation | Characteristics | References |
|---|---|---|---|---|
| Tombusvirus | tomato bushy stunt virus (TBSV) | serial passaging/single round of infection/multiple deletions | attenuation of infection symptoms/reduced virus accumulation | [25,26,27] |
| cymbidium ringspot virus (CymRSV) | serial passaging/multiple deletions | attenuation of infection symptoms/reduced virus accumulation | [17,28,29,30] | |
| cucumber necrosis virus (CNV) | serial passaging/ | attenuation of infection symptoms/reduced virus accumulation | [19,27] | |
| turnip crinkle virus (TCV) | serial passaging/ | exacerbation of infection symptoms | [18] | |
| Orthotospovirus | tomato spotted wilt virus (TSWV) | serial passaging/single deletion | modulate virulence | [31,32] |
| Closterovirus | citrus tristeza virus (CTV) | serial passaging/single deletion | no interference observed | [33,34,35,36] |
| Tobravirus | tobacco rattle virus (TRV) | serial passaging/deletions and/or rearrangements—mosaic of RNA1 and RNA2 of the HV | reduced virus accumulation | [37,38] |
| Potexvirus | clover yellow mosaic virus (ClMV) | serial passaging/single deletion | no interference observed | [39,40] |
| casava common mosaic virus (CsCMV) | serial passaging/single deletion | no interference observed | [41] | |
| Bromovirus | brome mosaic virus (BMV) | prolonged single round of infection | - | [42,43] |
| broad bean mottle virus (BBMV) | prolonged single round of infection/single deletion | exacerbation of infection symptoms | [44,45] | |
| Cucumovirus | cucumber mosaic virus (CMV) | serial passaging/single deletion | - | [20,46] |
| Crinivirus | lettuce infectious yellows virus (LlYV) | single round of infection | - | [47,48] |
| Nepovirus | tomato black ring virus (TBRV) | serial passaging/single deletion | attenuation or enhancement of infection symptoms/suppression of virus accumulation | [49,50,51] |
| Pomovirus | potato mop-top virus (PMTV) | serial passaging/single deletion | attenuation of infection symptoms | [52,53] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Budzyńska, D.; Zwart, M.P.; Hasiów-Jaroszewska, B. Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis. Viruses 2022, 14, 2814. https://doi.org/10.3390/v14122814
Budzyńska D, Zwart MP, Hasiów-Jaroszewska B. Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis. Viruses. 2022; 14(12):2814. https://doi.org/10.3390/v14122814
Chicago/Turabian StyleBudzyńska, Daria, Mark P. Zwart, and Beata Hasiów-Jaroszewska. 2022. "Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis" Viruses 14, no. 12: 2814. https://doi.org/10.3390/v14122814
APA StyleBudzyńska, D., Zwart, M. P., & Hasiów-Jaroszewska, B. (2022). Defective RNA Particles of Plant Viruses—Origin, Structure and Role in Pathogenesis. Viruses, 14(12), 2814. https://doi.org/10.3390/v14122814







