Effect of Insertion and Deletion in the Meq Protein Encoded by Highly Oncogenic Marek’s Disease Virus on Transactivation Activity and Virulence
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells and Virus
2.3. Plasmids
2.4. Dual-Luciferase Reporter Assay
2.5. Generation of Recombinant Viruses
2.6. In Vitro Replication of the Recombinant Viruses
2.7. Confirmation of Meq Expression Levels by RT-PCR
2.8. In Vivo Characterization of Recombinant Viruses
2.8.1. Experimental Chickens
2.8.2. 1st Animal Experiment
- In vivo kinetics of recombinant viruses
- Pathogenicity of recombinant viruses in unvaccinated chickens
2.8.3. 2nd Animal Experiment: Pathogenicity of Recombinant Viruses in Vaccinated Chickens
2.9. DNA Extraction
2.10. qPCR
2.11. Statistical Analyses
3. Results
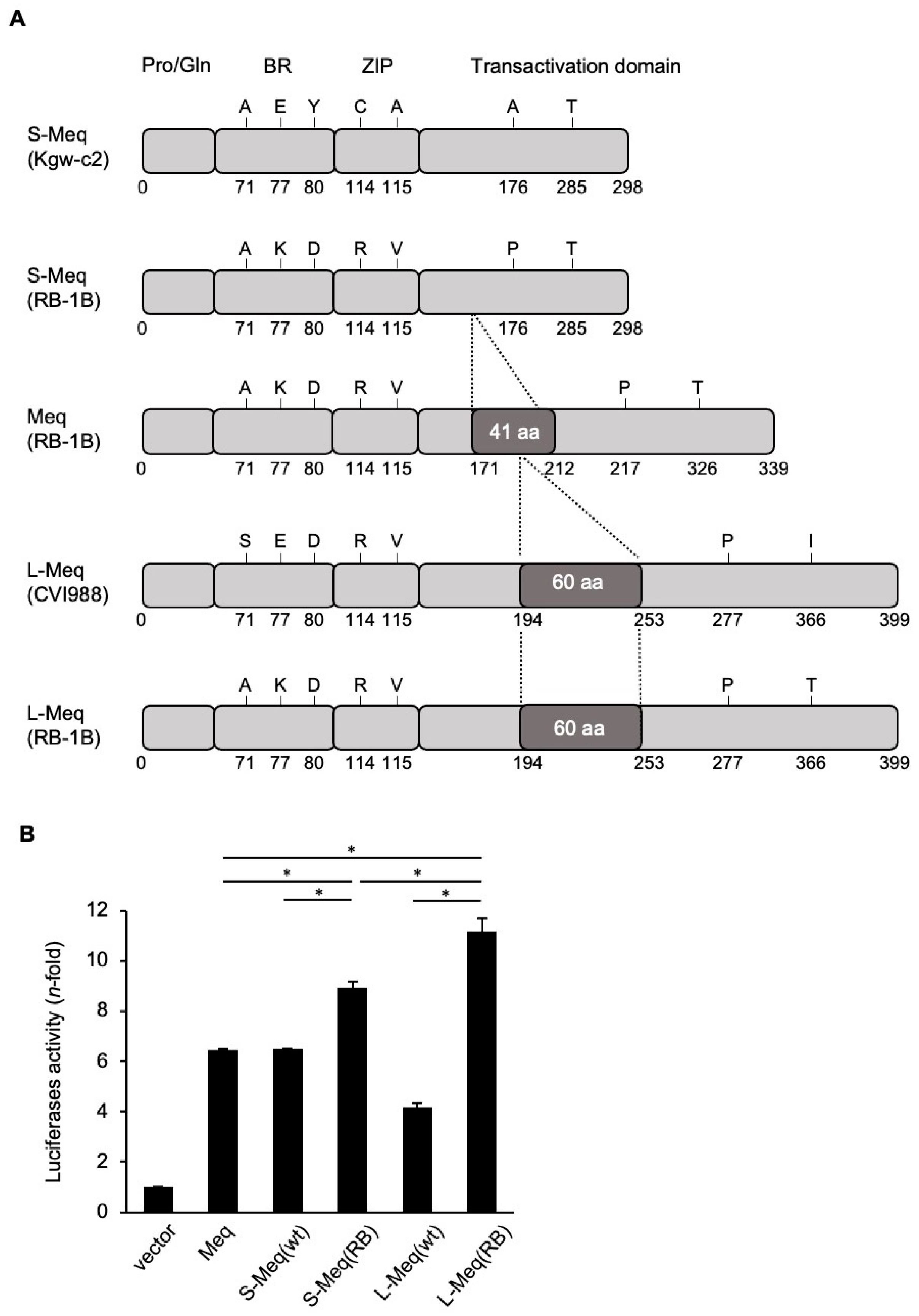
3.1. Transactivation Activity of Meq Isoforms
3.2. Generation of Recombinant Viruses
3.3. Characterization of rMDVs In Vitro
3.4. Replication of rMDVs In Vivo
3.5. Pathogenicity of rMDVs In Vivo
3.6. Pathogenicity of rMDVs in Vaccinated Chickens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Osterrieder, N.; Kamil, J.P.; Schumacher, D.; Tischer, B.K.; Trapp, S. Marek’s disease virus: From miasma to model. Nat. Rev. Microbiol. 2006, 4, 283–294. [Google Scholar] [CrossRef]
- Schat, K.A. History of the first-generation Marek’s disease vaccines: The science and little-known facts. Avian Dis. 2016, 60, 715–724. [Google Scholar] [CrossRef] [Green Version]
- Othman, I.; Aklilu, E. Marek’s disease herpesvirus serotype 1 in broiler breeder and layer chickens in Malaysia. Vet. World 2019, 12, 472–476. [Google Scholar] [CrossRef]
- Raja, A.; Raj, G.D.; Bhuvaneswari, P.; Balachandran, C.; Kumanan, K. Detection of virulent Marek’s disease virus in poultry in India. Acta Virol. 2009, 53, 255–260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, M.Y.; Li, M.; Wang, W.W.; Deng, Q.M.; Li, Q.H.; Gao, Y.L.; Wang, P.K.; Huang, T.; Wei, P. The emergence of a vv + MDV can break through the protections provided by the current vaccines. Viruses 2020, 12, 1048. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.R.; Zhang, Y.P.; Lv, H.C.; Zhou, L.Y.; Cui, H.Y.; Gao, Y.L.; Qi, X.L.; Wang, Y.Q.; Li, K.; Gao, L.; et al. A Chinese variant Marek’s disease virus strain with divergence between virulence and vaccine resistance. Viruses 2017, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Sung, H.W. Recent increase of Marek’s disease in Korea related to the virulence increase of the virus. Avian Dis. 2002, 46, 517–524. [Google Scholar] [CrossRef]
- Zhuang, X.; Zou, H.; Shi, H.; Shao, H.; Ye, J.; Miao, J.; Wu, G.; Qin, A. Outbreak of Marek’s disease in a vaccinated broiler breeding flock during its peak egg-laying period in China. BMC Vet. Res. 2015, 11, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrow, C.; Fehler, F. Marek’s disease: A worldwide problem. In Marek’s Disease: An Evolving Problem; Davison, F., Nair, V., Eds.; Elsevier: Amsterdam, The Netherlands, 2004; pp. 49–61. [Google Scholar]
- Trimpert, J.; Groenke, N.; Jenckel, M.; He, S.; Kunec, D.; Szpara, M.L.; Spatz, S.J.; Osterrieder, N.; McMahon, D.P. A phylogenomic analysis of Marek’s disease virus reveals independent paths to virulence in Eurasia and North America. Evol. Appl. 2017, 10, 1091–1101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Padhi, A.; Parcells, M.S. Positive selection drives rapid evolution of the meq oncogene of Marek’s disease virus. PLoS ONE 2016, 11, e0162180. [Google Scholar]
- Murata, S.; Machida, Y.; Isezaki, M.; Maekawa, N.; Okagawa, T.; Konnai, S.; Ohashi, K. Genetic characterization of a Marek’s disease virus strain isolated in Japan. Virol. J. 2020, 17, 186. [Google Scholar] [CrossRef]
- Witter, R. Increased virulence of Marek’s disease virus field isolates. Avian Dis. 1997, 41, 149–163. [Google Scholar] [CrossRef]
- Liu, J.L.; Kung, H.J. Marek’s disease herpesvirus transforming protein MEQ: A c-Jun analogue with an alternative life style. Virus Genes 2000, 21, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Qian, Z.; Brunovskis, P.; Lee, L.; Vogt, P.K.; Kung, H. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J. Virol. 1996, 70, 7161–7170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, A.M.; Gilad, O.; Xia, L.; Izumiya, Y.; Choi, J.; Tsalenko, A.; Yakhini, Z.; Witter, R.; Lee, L.; Cardona, C.J.; et al. Marek’s disease virus Meq transforms chicken cells via the v-Jun transcriptional cascade: A converging transforming pathway for avian oncoviruses. Proc. Natl. Acad. Sci. USA 2005, 102, 14831–14836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.-L.; Ye, Y.; Lee, L.F.; Kung, H.-J. Transforming potential of the herpesvirus oncoprotein MEQ: Morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 1998, 71, 388–395. [Google Scholar] [CrossRef] [Green Version]
- Lupiani, B.; Lee, L.F.; Cui, X.; Gimeno, I.; Anderson, A.; Morgan, R.W.; Silva, R.F.; Witter, R.L.; Kung, H.-J.; Reddy, S.M. Marek’s disease virus-encoded Meq gene is involved in transformation of lymphocytes but is dispensable for replication. Proc. Natl. Acad. Sci. USA 2004, 101, 11815–11820. [Google Scholar] [CrossRef] [Green Version]
- Shamblin, C.E.; Greene, N.; Arumugaswami, V.; Dienglewicz, R.L.; Parcells, M.S. Comparative analysis of Marek’s disease virus (MDV) glycoprotein-, lytic antigen pp38- and transformation antigen Meq-encoding genes: Association of meq mutations with MDVs of high virulence. Vet. Microbiol. 2004, 102, 147–167. [Google Scholar] [CrossRef]
- Murata, S.; Yamamoto, E.; Sakashita, N.; Maekawa, N.; Okagawa, T.; Konnai, S.; Ohashi, K. Research Note: Characterization of S-Meq containing the deletion in Meq protein’s transactivation domain in a Marek’s disease virus strain in Japan. Poult. Sci. 2021, 100, 101461. [Google Scholar] [CrossRef]
- Murata, S.; Okada, T.; Kano, R.; Hayashi, Y.; Hashiguchi, T.; Onuma, M.; Konnai, S.; Ohashi, K. Analysis of transcriptional activities of the Meq proteins present in highly virulent Marek’s disease virus strains, RB1B and Md5. Virus Genes 2011, 43, 66–71. [Google Scholar] [CrossRef]
- Murata, S.; Hashiguchi, T.; Hayashi, Y.; Yamamoto, Y.; Matsuyama-Kato, A.; Takasaki, S.; Isezaki, M.; Onuma, M.; Konnai, S.; Ohashi, K. Characterization of Meq proteins from field isolates of Marek’s disease virus in Japan. Infect. Genet. Evol. 2013, 16, 137–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conradie, A.M.; Bertzbach, L.D.; Trimpert, J.; Patria, J.N.; Murata, S.; Parcells, M.S.; Kaufer, B.B. Distinct polymorphisms in a single herpesvirus gene are capable of enhancing virulence and mediating vaccinal resistance. PLoS Pathog. 2020, 16, e1009104. [Google Scholar] [CrossRef] [PubMed]
- Renz, K.G.; Cooke, J.; Clarke, N.; Cheetham, B.F.; Hussain, Z.; Islam, A.F.M.F.; Tannock, G.A.; Walkden-Brown, S.W. Pathotyping of Australian isolates of Marek’s disease virus and association of pathogenicity with meq gene polymorphism. Avian Pathol. 2012, 41, 161–176. [Google Scholar] [CrossRef] [PubMed]
- Mescolini, G.; Lupini, C.; Felice, V.; Guerrini, A.; Silveira, F.; Cecchinato, M.; Catelli, E. Molecular characterization of the meq gene of Marek’s disease viruses detected in unvaccinated backyard chickens reveals the circulation of low- and high-virulence strains. Poult. Sci. 2019, 98, 3130–3137. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Shi, M.; Li, Q.; Wang, P.; Li, M.; Wang, W.; Gao, Y.; Li, H.; Lin, L.; Huang, T.; et al. Analysis of the evolution and transmission dynamics of the field MDV in China during the years 1995–2020, indicating the emergence of a unique cluster with the molecular characteristics of vv+ MDV that has become endemic in southern China. Transbound. Emerg. Dis. 2021, 68, 3574–3587. [Google Scholar] [CrossRef]
- Spatz, S.J.; Petherbridge, L.; Zhao, Y.; Nair, V. Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek’s disease virus. J. Gen. Virol. 2007, 88, 1080–1096. [Google Scholar] [CrossRef]
- Wajid, S.J.; Katz, M.E.; Renz, K.G.; Walkden-Brown, S.W. Prevalence of Marek’s disease virus in different chicken populations in Iraq and indicative virulence based on sequence variation in the ecoRI-q (meq) gene. Avian Dis. 2013, 57, 562–568. [Google Scholar] [CrossRef]
- Lee, S.I.; Takagi, M.; Ohashi, K.; Sugimoto, C.; Onuma, M. Difference in the meq gene between oncogenic and attenuated strains of Marek’s disease virus serotype 1. J. Vet. Med. Sci. 2000, 62, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Conradie, A.M.; Bertzbach, L.D.; Bhandari, N.; Parcells, M.; Kaufer, B.B. A common live-attenuated avian herpesvirus vaccine expresses a very potent oncogene. mSphere 2019, 4, e00658-19. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, M.H.A.; El-Sabagh, I.M.; Al-Habeeb, M.A.; Al-Hammady, Y.M. Diversity of Meq gene from clinical Marek’s disease virus infection in Saudi Arabia. Vet. World 2016, 9, 572–578. [Google Scholar] [CrossRef]
- Osterrieder, N. Sequence and initial characterization of the U(L)10 (glycoprotein M) and U(L)11 homologous genes of serotype 1 Marek’s Disease Virus. Arch. Virol. 1999, 144, 1853–1863. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, D.; Tischer, B.K.; Fuchs, W.; Osterrieder, N. Reconstitution of Marek’s disease virus serotype 1 (MDV-1) from DNA cloned as a bacterial artificial chromosome and characterization of a glycoprotein B-negative MDV-1 mutant. J. Virol. 2000, 74, 11088–11098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jarosinski, K.W.; Schat, K.A. Multiple alternative splicing to exons II and III of viral interleukin-8 (vIL-8) in the Marek’s disease virus genome: The importance of vIL-8 exon I. Virus Genes 2007, 34, 9–22. [Google Scholar] [CrossRef] [PubMed]
- Engel, A.T.; Selvaraj, R.K.; Kamil, J.P.; Osterrieder, N.; Kaufer, B.B. Marek’s disease viral interleukin-8 promotes lymphoma formation through targeted recruitment of B cells and CD4+ CD25+ T cells. J. Virol. 2012, 86, 8536–8545. [Google Scholar] [CrossRef] [Green Version]
- Tischer, B.K.; Kaufer, B.B. Viral bacterial artificial chromosomes: Generation, mutagenesis, and removal of mini-F sequences. J. Biomed. Biotechnol. 2012, 2012, 472537. [Google Scholar] [CrossRef] [Green Version]
- Tischer, B.K.; von Einem, J.; Kaufer, B.; Osterrieder, N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 2006, 40, 191–197. [Google Scholar]
- Borenshtain, R.; Davidson, I. Marek’s disease virus genome separation from feather tip extracts by pulsed field gel electrophoresis. J. Virol. Methods 2002, 101, 169–174. [Google Scholar] [CrossRef]
- Bello, N.; Francino, O.; Sánchez, A. Isolation of genomic DNA from feathers. J. Vet. Diagn. Investig. 2001, 13, 162–164. [Google Scholar] [CrossRef] [Green Version]
- Baigent, S.J.; Kgosana, L.B.; Gamawa, A.A.; Smith, L.P.; Read, A.F.; Nair, V.K. Relationship between levels of very virulent MDV in poultry dust and in feather tips from vaccinated chickens. Avian Dis. 2013, 57, 440–447. [Google Scholar] [CrossRef] [Green Version]
- Li, K.; Liu, Y.; Xu, Z.; Zhang, Y.; Luo, D.; Gao, Y.; Qian, Y.; Bao, C.; Liu, C.; Zhang, Y.; et al. Avian oncogenic herpesvirus antagonizes the cGAS-STING DNA-sensing pathway to mediate immune evasion. PLoS Pathog. 2019, 15, e1007999. [Google Scholar] [CrossRef]
- Deng, X.; Li, X.; Shen, Y.; Qiu, Y.; Shi, Z.; Shao, D.; Jin, Y.; Chen, H.; Ding, C.; Li, L.; et al. The Meq oncoprotein of Marek’s disease virus interacts with p53 and inhibits its transcriptional and apoptotic activities. Virol. J. 2010, 7, 348. [Google Scholar] [CrossRef] [Green Version]
- Subramaniam, S.; Johnston, J.; Preeyanon, L.; Brown, C.T.; Kung, H.-J.; Cheng, H.H. Integrated analyses of genome-wide DNA occupancy and expression profiling identify key genes and pathways involved in cellular transformation by a Marek’s disease virus oncoprotein, Meq. J. Virol. 2013, 87, 9016–9029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qian, Z.; Brunovskis, P.; Rauscher, F., III; Lee, L.; Kung, H. Transactivation activity of Meq, a Marek’s disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 1995, 69, 4037–4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molouki, A.; Ghalyanchilangeroudi, A.; Abdoshah, M.; Shoushtari, A.; Abtin, A.; Eshtartabadi, F.; Akhijahani, M.M.; Ziafatikafi, Z.; Babaeimarzango, S.S.; Allahyari, E.; et al. Report of a new meq gene size: The first study on genetic characterisation of Marek’s disease viruses circulating in Iranian commercial layer and backyard chicken. Br. Poult. Sci. 2021, 20, 1–8. [Google Scholar] [CrossRef] [PubMed]





| Position in Meq | Substitution | Primers | |
|---|---|---|---|
| 71 | serine–to–alanine | F | 5′-GAA TCG TGA CGC CGC TCG GAG AAG ACG-3′ |
| R | 5′-CGT CTT CTC CGA GCG GCG TCA CGA TTC-3′ | ||
| 77, 80 | glutamic-acid–to–lysine, tyrosine–to–aspartic-acid | F | 5′-CGG AGA AGA CGC AGG AAG CAG ACG GAC TAT GTA GAC AAA C-3′ |
| R | 5′-GTT TGT CTA CAT AGT CCG TCT GCT TCC TGC GTC TTC TCC G-3′ | ||
| 114, 115 | cysteine–to–arginine, alanine–to–valine | F | 5′-GAG TGC ACG TCC CTG CGT GTA CAG TTG GCT TGT CA-3′ |
| R | 5′-TGA CAA GCC AAC TGT ACA CGC AGG GAC GTG CAC TC-3′ | ||
| 217 | alanine–to–proline | F | 5′-ATC TGT ACC CCC CCT CCT CCC GAT G-3′ |
| R | 5′-CAT CGG GAG GAG GGG GGG TAC AGA T-3′ | ||
| 326 | isoleucine–to–threonine | F | 5′-GTT TCC CTC GGA TAC TCA GTC TAC GGT CT-3′ |
| R | 5′-AGA CCG TAG ACT GAG TAT CCG AGG GAA AC-3′ | ||
| Gene | Sequence | Application |
|---|---|---|
| meq | F 5′-AGT TGG CTT GTC ATG AGC CAG-3′ | RT-PCR |
| R 5′-TGT TCG GGA TCC TCG GTA AGA-3′ | ||
| ICP4 | F 5′-GCA TCG ACA AGC ACT TAC GG-3′ | qPCR |
| R 5′-CGA GAG CGT CGT ATT GTT TGG-3′ | ||
| iNOS | F 5′-GAG TGG TTT AAG GAG TTG GAT CTG A-3′ | qPCR |
| R 5′-TTC CAG ACC TCC CAC CTC AA-3′ |
| rMDV | Survival Rate | Tumor Incidence |
|---|---|---|
| vRB-1B_S-Meq | 90.9% (10/11) | 18.2% (2/11) |
| vRB-1B_Meq | 41.7% (5/12) | 58.3% (7/12) |
| vRB-1B_L-Meq | 0% (11/11) | 100% (11/11) |
| rMDV | Survival Rate | Tumor Incidence |
|---|---|---|
| vRB-1B_S-Meq | 100% (8/8) | 12.5% (1/8) |
| vRB-1B_Meq | 100% (9/9) | 0% (0/9) |
| vRB-1B_L-Meq | 83.3% (10/12) | 33.3% (4/12) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, J.; Murata, S.; Yang, Z.; Kaufer, B.B.; Fujisawa, S.; Seo, H.; Maekawa, N.; Okagawa, T.; Konnai, S.; Osterrieder, N.; et al. Effect of Insertion and Deletion in the Meq Protein Encoded by Highly Oncogenic Marek’s Disease Virus on Transactivation Activity and Virulence. Viruses 2022, 14, 382. https://doi.org/10.3390/v14020382
Sato J, Murata S, Yang Z, Kaufer BB, Fujisawa S, Seo H, Maekawa N, Okagawa T, Konnai S, Osterrieder N, et al. Effect of Insertion and Deletion in the Meq Protein Encoded by Highly Oncogenic Marek’s Disease Virus on Transactivation Activity and Virulence. Viruses. 2022; 14(2):382. https://doi.org/10.3390/v14020382
Chicago/Turabian StyleSato, Jumpei, Shiro Murata, Zhiyuan Yang, Benedikt B. Kaufer, Sotaro Fujisawa, Hikari Seo, Naoya Maekawa, Tomohiro Okagawa, Satoru Konnai, Nikolaus Osterrieder, and et al. 2022. "Effect of Insertion and Deletion in the Meq Protein Encoded by Highly Oncogenic Marek’s Disease Virus on Transactivation Activity and Virulence" Viruses 14, no. 2: 382. https://doi.org/10.3390/v14020382
APA StyleSato, J., Murata, S., Yang, Z., Kaufer, B. B., Fujisawa, S., Seo, H., Maekawa, N., Okagawa, T., Konnai, S., Osterrieder, N., Parcells, M. S., & Ohashi, K. (2022). Effect of Insertion and Deletion in the Meq Protein Encoded by Highly Oncogenic Marek’s Disease Virus on Transactivation Activity and Virulence. Viruses, 14(2), 382. https://doi.org/10.3390/v14020382






