Exploring New Functional Aspects of HTLV-1 RNA-Binding Protein Rex: How Does Rex Control Viral Replication?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Transcriptome and Alternative Splicing Analysis in Rex-Expressing CEM Cells
2.1.1. Construction of Retroviral Plasmid for Rex Expression
2.1.2. Establishment of Rex-Expressing CEM Using Retrovirus Vector Expression System
2.1.3. Gene Expression and Exon Microarray Analysis
2.1.4. Biological Annotation Analysis of the Microarray Data and the Interactome Data
2.2. Rex Interactome Analysis
2.2.1. Preparation of His-Halo-Rex Expression Plasmid
2.2.2. Identification of Interacting Proteins by Tandem Affinity Purification of His-Halo-Rex
2.3. Abnormal PD-L1 mRNA Splicing by Rex
2.3.1. Identification of vPD-L1 mRNA Sequence in CEM-Rex
2.3.2. Real-Time q-PCR with Primers Specific for vPD-L1 mRNA
2.3.3. NanoLuc Reporter Secretion Assay for sPD-L1
2.3.4. Amount of Secreted PD-L1 in HTLV-1-Genome-Expressing Cells
2.4. Biological Significance of the Interaction between Rex and NONO
2.4.1. Construction of the GST-Rex Expression Plasmid
2.4.2. GST-Rex Pulldown Assay
2.4.3. NONO Knockdown
2.4.4. NMD Luciferase Reporter Assay
2.4.5. RxRE Reporter Assay
2.4.6. Viral Reproduction Assay
2.5. Cell Culture
2.6. Statistical Analysis
3. Results
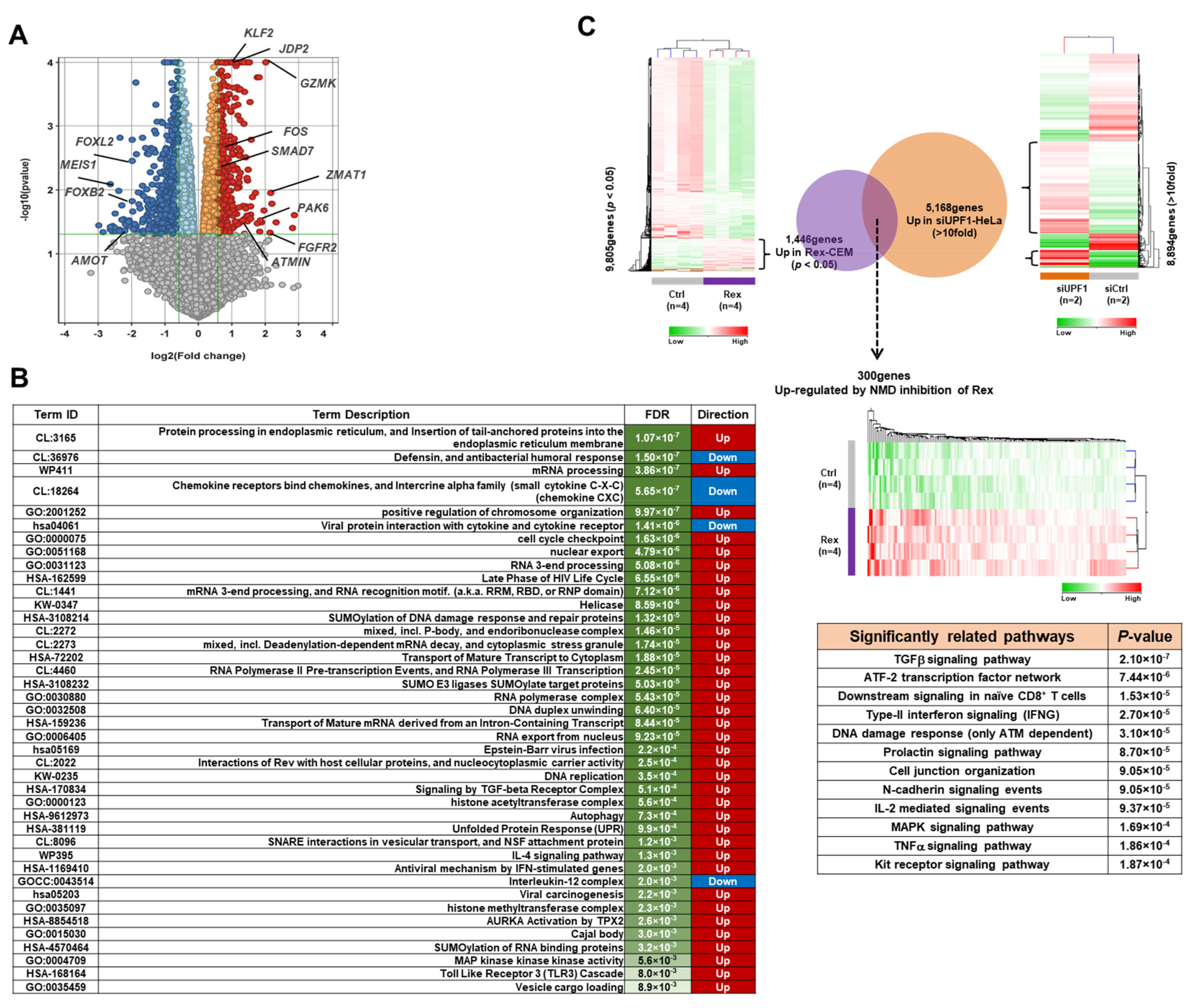
3.1. The Effect of Rex on the Gene Expression Profile of T Cells
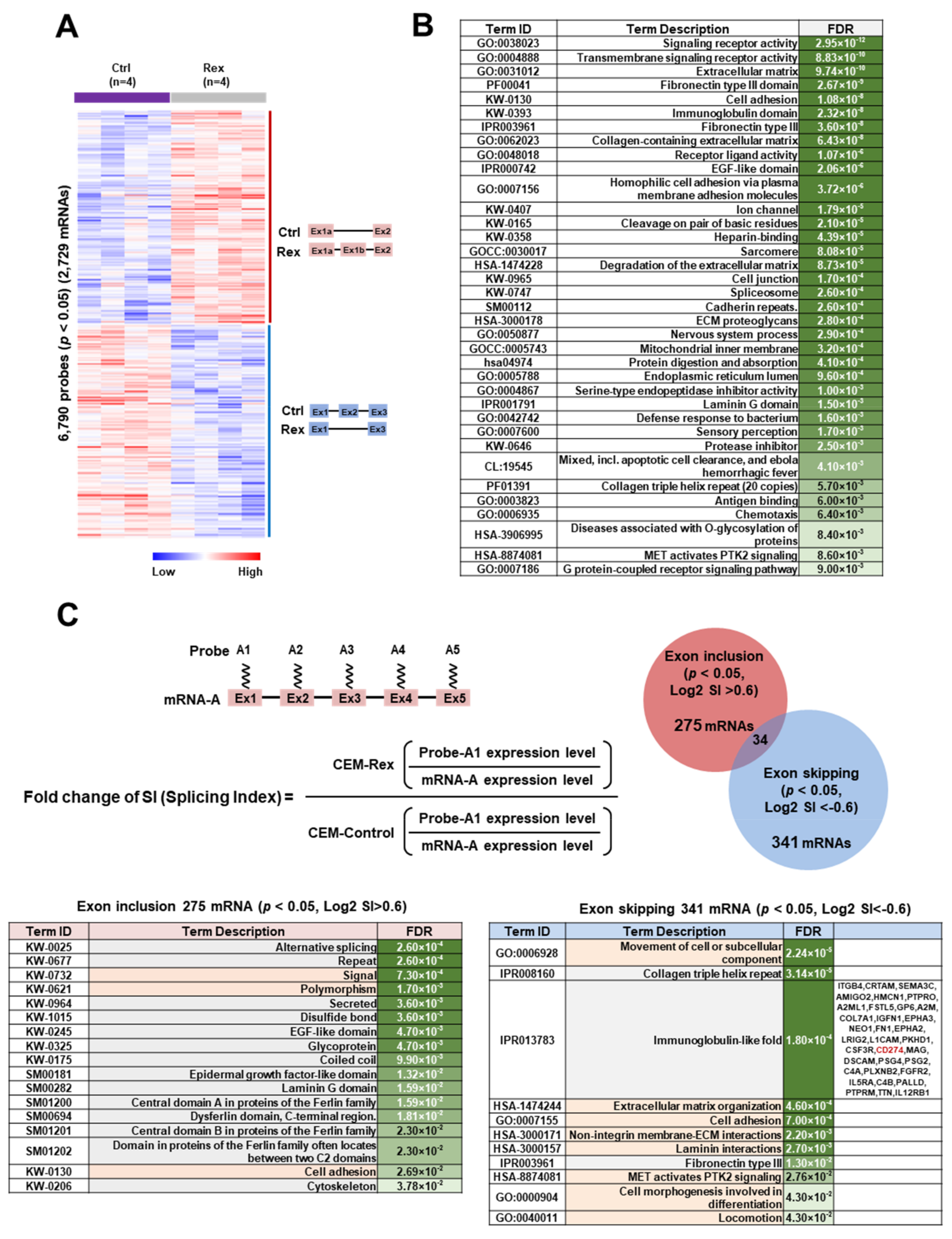
3.2. Effect of Rex on the mRNA Splicing Patterns in T cells
3.3. Abnormal CD274 (PD-L1) mRNA Splicing by Rex
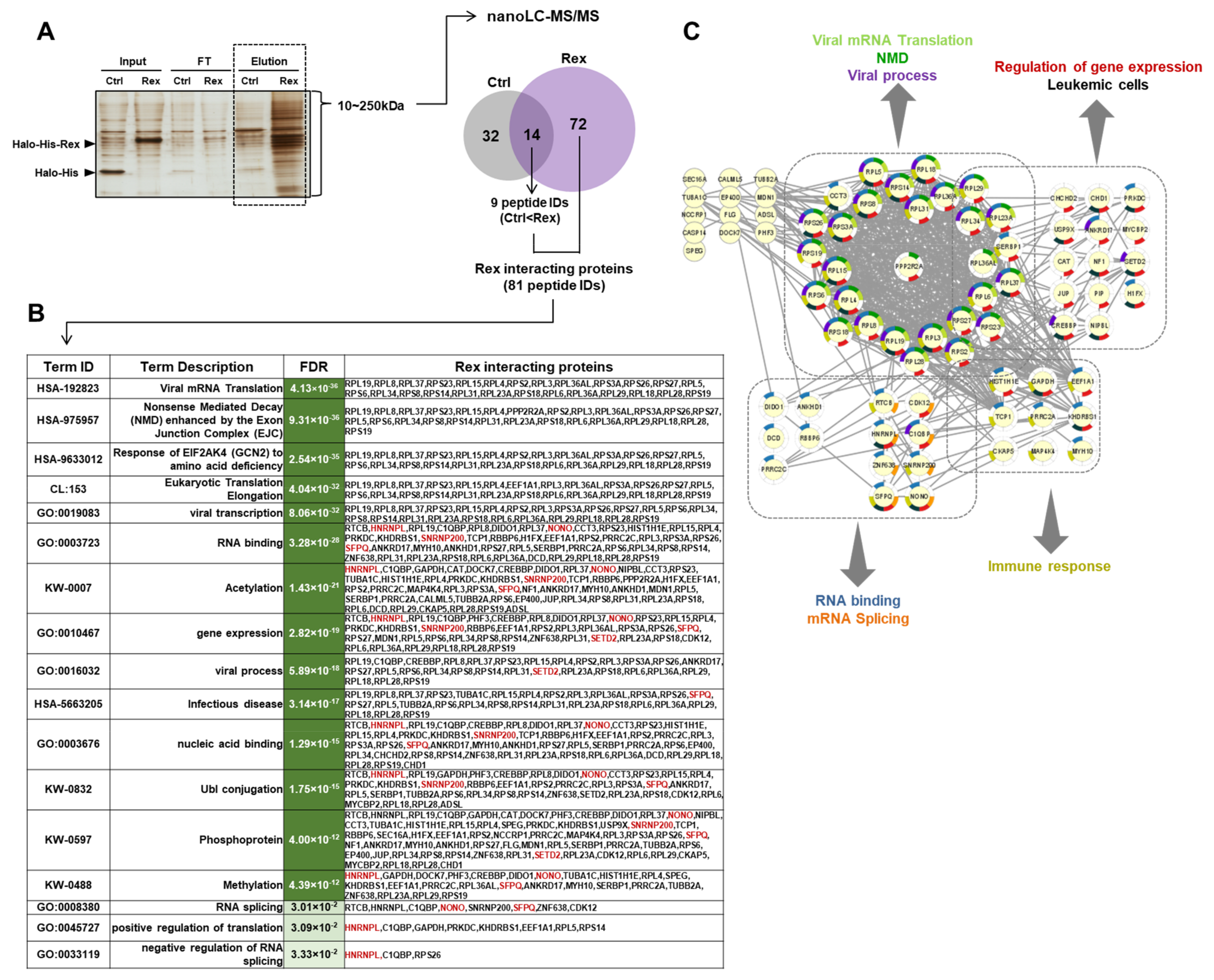
3.4. Human Proteins Interacting with Rex in HEK293FT Cells
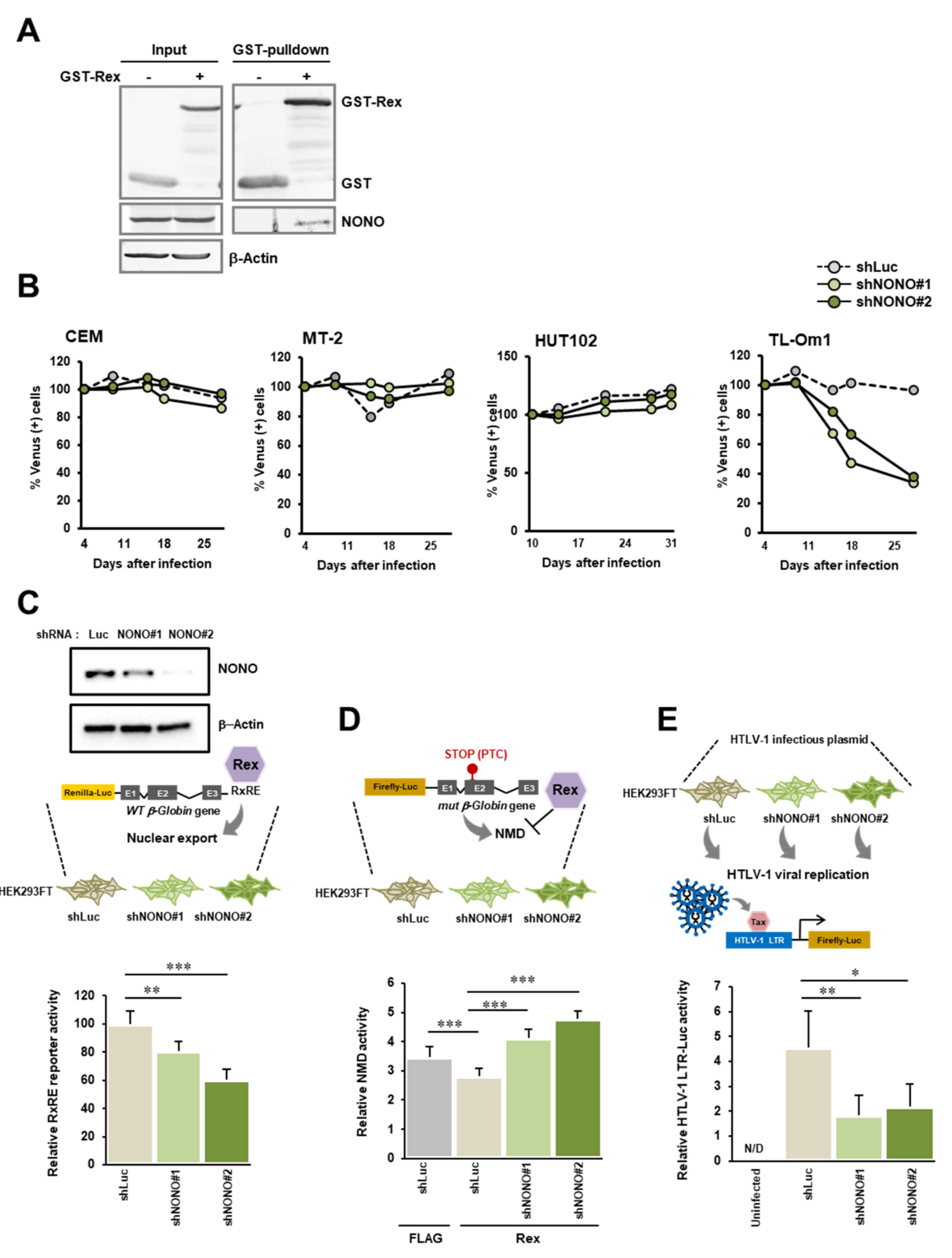
3.5. Interaction between Rex and NONO
4. Discussion
4.1. Impact of Rex on Gene Expression Profiles Icluding AP-1 Family Proteins
4.2. Effect of Rex on mRNA Splicing Patterns in T Cells
4.3. Physiological Effect of Abnormal Splicing of PD-L1 mRNA by Rex
4.4. Rex May Affect the Gene Expression Profile and mRNA Splicing Patterns via Interation with Various Cellular Proteins
4.5. The Biological Importance of the Interaction between Rex and NONO
4.6. Summary: New Possibilities in the Rex Functional Pathways in HTLV-1-Infected T Cells
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hirons, A.; Khoury, G.; Purcell, D.F.J. Human T-cell lymphotropic virus type-1: A lifelong persistent infection, yet never truly silent. Lancet Infect. Dis. 2021, 21, e2–e10. [Google Scholar] [CrossRef]
- Tagaya, Y.; Matsuoka, M.; Gallo, R. 40 years of the human T-cell leukemia virus: Past, present, and future. F1000Research 2019, 8, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Futsch, N.; Mahieux, R.; Dutartre, H. HTLV-1, the other pathogenic yet neglected human retrovirus: From transmission to therapeutic treatment. Viruses 2018, 10, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, K.; Watanabe, T. HTLV-1 Rex tunes the cellular environment favorable for viral replication. Viruses 2016, 8, 58. [Google Scholar] [CrossRef] [Green Version]
- Nakano, K.; Watanabe, T. HTLV-1 Rex: The courier of viral messages making use of the host vehicle. Front. Microbiol. 2012, 3, 330. [Google Scholar] [CrossRef] [Green Version]
- Rende, F.; Cavallari, I.; Corradin, A.; Silic-Benussi, M.; Toulza, F.; Toffolo, G.M.; Tanaka, Y.; Jacobson, S.; Taylor, G.P.; D’Agostino, D.M.; et al. Kinetics and intracellular compartmentalization of HTLV-1 gene expression: Nuclear retention of HBZ mRNAs. Blood 2011, 117, 4855–4859. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Ando, T.; Yamagishi, M.; Yokoyama, K.; Ishida, T.; Ohsugi, T.; Tanaka, Y.; Brighty, D.W.; Watanabe, T. Viral interference with host mRNA surveillance, the nonsense-mediated mRNA decay (NMD) pathway, through a new function of HTLV-1 Rex: Implications for retroviral replication. Microbe Infect. 2013, 15, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Weil, R.; Levraud, J.P.; Dodon, M.D.; Bessia, C.; Hazan, U.; Kourilsky, P.; Israël, A. Altered expression of tyrosine kinases of the Src and Syk families in human T-cell leukemia virus type 1-infected T-cell lines. J. Virol. 1999, 73, 3709–3717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gröne, M.; Koch, C.; Grassmann, R. The HTLV-1 Rex protein induces nuclear accumulation of unspliced viral RNA by avoiding intron excision and degradation. Virology 1996, 218, 316–325. [Google Scholar] [CrossRef] [Green Version]
- Ishida, T.; Mizushima, S.I.; Azuma, S.; Kobayashi, N.; Tojo, T.; Suzuki, K.; Aizawa, S.; Watanabe, T.; Mosialos, G.; Kieff, E.; et al. Identification of TRAF6, a novel tumor necrosis factor receptor- associated factor protein that mediates signaling from an amino-terminal domain of the CD40 cytoplasmic region. J. Biol. Chem. 1996, 271, 28745–28748. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, T.; Tojo, T.; Aoki, T.; Kobayashi, N.; Ohishi, T.; Watanabe, T.; Yamamoto, T.; Inoue, J.I. TRAF5, a novel tumor necrosis factor receptor-associated factor family protein, mediates CD40 signaling. Proc. Natl. Acad. Sci. USA 1996, 93, 9437–9442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, K.; Uchimaru, K.; Utsunomiya, A.; Yamaguchi, K.; Watanabe, T. Dysregulation of c-Myb pathway by aberrant expression of proto-oncogene MYB provides the basis for malignancy in adult T-cell leukemia/lymphoma cells. Clin. Cancer Res. 2016, 22, 5915–5928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef] [Green Version]
- Knott, G.J.; Bond, C.S.; Fox, A.H. The DBHS proteins SFPQ, NONO and PSPC1: A multipurpose molecular scaffold. Nucleic Acids Res. 2016, 44, 3989–4004. [Google Scholar] [CrossRef]
- Shaulian, E.; Karin, M. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 2002, 4, E131–E136. [Google Scholar] [CrossRef]
- Katagiri, T.; Kameda, H.; Nakano, H.; Yamazaki, S. Regulation of T cell differentiation by the AP-1 transcription factor JunB. Immunol. Med. 2021, 44, 197–203. [Google Scholar] [CrossRef]
- Eferl, R.; Wagner, E.F. AP-1: A double-edged sword in tumorigenesis. Nat. Rev. Cancer 2003, 3, 859–868. [Google Scholar] [CrossRef]
- Atsaves, V.; Leventaki, V.; Rassidakis, G.Z.; Claret, F.X. AP-1 transcription factors as regulators of immune responses in cancer. Cancers 2019, 11, 1037. [Google Scholar] [CrossRef] [Green Version]
- Yukawa, M.; Jagannathan, S.; Vallabh, S.; Kartashov, A.V.; Chen, X.; Weirauch, M.T.; Weirauch, M.T.; Weirauch, M.T.; Weirauch, M.T.; Barski, A.; et al. AP-1 activity induced by co-stimulation is required for chromatin opening during T cell activation. J. Exp. Med. 2020, 217, e20182009. [Google Scholar] [CrossRef]
- Vierbuchen, T.; Ling, E.; Cowley, C.J.; Couch, C.H.; Wang, X.; Harmin, D.A.; Roberts, C.W.M.; Greenberg, M.E. AP-1 Transcription Factors and the BAF Complex Mediate Signal-Dependent Enhancer Selection. Mol. Cell 2017, 68, 1067–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirzaei, H.; Khodadad, N.; Karami, C.; Pirmoradi, R.; Khanizadeh, S. The AP-1 pathway; A key regulator of cellular transformation modulated by oncogenic viruses. Rev. Med. Virol. 2020, 30, e2088. [Google Scholar] [CrossRef]
- Matsumoto, J.; Ohshima, T.; Isono, O.; Shimotohno, K. HTLV-1 HBZ suppresses AP-1 activity by impairing both the DNA-binding ability and the stability of c-Jun protein. Oncogene 2005, 24, 1001–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuoka, M.; Mesnard, J.M. HTLV-1 bZIP factor: The key viral gene for pathogenesis. Retrovirology 2020, 17, 4–11. [Google Scholar] [CrossRef] [PubMed]
- Gazon, H.; Barbeau, B.; Mesnard, J.M.; Peloponese, J.M. Hijacking of the AP-1 signaling pathway during development of ATL. Front. Microbiol. 2018, 8, 2686. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 2017, 18, 655–670. [Google Scholar] [CrossRef]
- Liu, Y.; Gonzàlez-Porta, M.; Santos, S.; Brazma, A.; Marioni, J.C.; Aebersold, R.; Venkitaraman, A.R.; Wickramasinghe, V.O. Impact of Alternative Splicing on the Human Proteome. Cell Rep. 2017, 20, 1229–1241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Scotti, M.M.; Swanson, M.S. RNA mis-splicing in disease. Nat. Rev. Genet. 2016, 17, 19–32. [Google Scholar] [CrossRef]
- El Marabti, E.; Younis, I. The cancer spliceome: Reprograming of alternative splicing in cancer. Front. Mol. Biosci. 2018, 5, 80. [Google Scholar] [CrossRef]
- Frankiw, L.; Baltimore, D.; Li, G. Alternative mRNA splicing in cancer immunotherapy. Nat. Rev. Immunol. 2019, 19, 675–687. [Google Scholar] [CrossRef]
- Bonnal, S.C.; López-Oreja, I.; Valcárcel, J. Roles and mechanisms of alternative splicing in cancer — implications for care. Nat. Rev. Clin. Oncol. 2020, 17, 457–474. [Google Scholar] [CrossRef]
- Agirre, E.; Oldfield, A.J.; Bellora, N.; Segelle, A.; Luco, R.F. Splicing-associated chromatin signatures: A combinatorial and position-dependent role for histone marks in splicing definition. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Jubel, J.M.; Barbati, Z.R.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. The Role of PD-1 in Acute and Chronic Infection. Front. Immunol. 2020, 11, 487. [Google Scholar] [CrossRef] [Green Version]
- Schönrich, G.; Raftery, M.J. The PD-1/PD-L1 axis and virus infections: A delicate balance. Front. Cell. Infect. Microbiol. 2019, 9, 207. [Google Scholar] [CrossRef] [Green Version]
- Leung, C.S.; Douglass, S.M.; Morselli, M.; Obusan, M.B.; Pavlyukov, M.S.; Pellegrini, M.; Johnson, T.L. H3K36 Methylation and the Chromodomain Protein Eaf3 Are Required for Proper Cotranscriptional Spliceosome Assembly. Cell Rep. 2019, 27, 3760–3769. [Google Scholar] [CrossRef] [Green Version]
- Hu, Q.; Greene, C.S.; Heller, E.A. Specific histone modifications associate with alternative exon selection during mammalian development. Nucleic Acids Res. 2020, 48, 4709–4724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, S.; Levy, M.J.; Zhang, N.; Li, H.; Florens, L.; Washburn, M.P.; Workman, J.L. The methyltransferase SETD2 couples transcription and splicing by engaging mRNA processing factors through its SHI domain. Nat. Commun. 2021, 12, 1–16. [Google Scholar] [CrossRef]
- Yuan, H.; Han, Y.; Wang, X.; Li, N.; Liu, Q.; Yin, Y.; Wang, H.; Pan, L.; Li, L.; Song, K.; et al. SETD2 Restricts Prostate Cancer Metastasis by Integrating EZH2 and AMPK Signaling Pathways. Cancer Cell 2020, 38, 350–365. [Google Scholar] [CrossRef] [PubMed]
- Gautam, D.; Johnson, B.A.; Mac, M.; Moody, C.A. SETD2-dependent H3K36me3 plays a critical role in epigenetic regulation of the HPV31 life cycle. PLoS Pathog. 2018, 14, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Feng, P.; Li, L.; Deng, T.; Liu, Y.; Ling, N.; Qiu, S.; Zhang, L.; Peng, B.; Xiong, W.; Cao, L.; et al. NONO and tumorigenesis: More than splicing. J. Cell. Mol. Med. 2020, 24, 4368–4376. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Goyama, S.; Asada, S.; Fujino, T.; Yonezawa, T.; Sato, N.; Takeda, R.; Tsuchiya, A.; Fukuyama, T.; Tanaka, Y.; et al. A histone modifier, ASXL1, interacts with NONO and is involved in paraspeckle formation in hematopoietic cells. Cell Rep. 2021, 36, 109576. [Google Scholar] [CrossRef]
- Petti, E.; Buemi, V.; Zappone, A.; Schillaci, O.; Broccia, P.V.; Dinami, R.; Matteoni, S.; Benetti, R.; Schoeftner, S. SFPQ and NONO suppress RNA:DNA-hybrid-related telomere instability. Nat. Commun. 2019, 10, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Ho, T.-T.; Huang, J.; Zhou, N.; Zhang, Z.; Koirala, P.; Zhou, X.; Wu, F.; Ding, X.; Mo, Y.-Y. Regulation of PCGEM1 by p54/nrb in prostate cancer. Sci. Rep. 2016, 6, 34529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Z.; Zhao, X.; Zhao, L.; Yang, H.; Liu, L.; Li, J.; Wu, J.; Yang, F.; Huang, G.; Liu, J. p54nrb/NONO regulates lipid metabolism and breast cancer growth through SREBP-1A. Oncogene 2016, 35, 1399–1410. [Google Scholar] [CrossRef] [PubMed]
- Landeras-Bueno, S.; Jorba, N.; Pérez-Cidoncha, M.; Ortín, J. The splicing factor proline-glutamine rich (SFPQ/PSF) is involved in influenza virus transcription. PLoS Pathog. 2011, 7, e1002397. [Google Scholar] [CrossRef]
- Lee, N.; Yario, T.A.; Gao, J.S.; Steitz, J.A. EBV noncoding RNA EBER2 interacts with host RNA-binding proteins to regulate viral gene expression. Proc. Natl. Acad. Sci. USA 2016, 113, 201601773. [Google Scholar] [CrossRef] [Green Version]
- Cao, S.; Moss, W.; O’Grady, T.; Concha, M.; Strong, M.J.; Wang, X.; Yu, Y.; Baddoo, M.; Zhang, K.; Fewell, C.; et al. New Noncoding Lytic Transcripts Derived from the Epstein-Barr Virus Latency Origin of Replication, oriP, Are Hyperedited, Bind the Paraspeckle Protein, NONO/p54nrb, and Support Viral Lytic Transcription. J. Virol. 2015, 89, 7120–7132. [Google Scholar] [CrossRef] [Green Version]
- St. Gelais, C.; Roger, J.; Wu, L. Non-POU Domain-Containing Octamer-Binding Protein Negatively Regulates HIV-1 Infection in CD4 + T Cells. AIDS Res. Hum. Retrovir. 2015, 31, 806–816. [Google Scholar] [CrossRef] [Green Version]
- Kula, A.; Gharu, L.; Marcello, A. HIV-1 pre-mRNA commitment to Rev mediated export through PSF and Matrin 3. Virology 2013, 435, 329–340. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nakano, K.; Yokoyama, K.; Shin, S.; Uchida, K.; Tsuji, K.; Tanaka, M.; Uchimaru, K.; Watanabe, T. Exploring New Functional Aspects of HTLV-1 RNA-Binding Protein Rex: How Does Rex Control Viral Replication? Viruses 2022, 14, 407. https://doi.org/10.3390/v14020407
Nakano K, Yokoyama K, Shin S, Uchida K, Tsuji K, Tanaka M, Uchimaru K, Watanabe T. Exploring New Functional Aspects of HTLV-1 RNA-Binding Protein Rex: How Does Rex Control Viral Replication? Viruses. 2022; 14(2):407. https://doi.org/10.3390/v14020407
Chicago/Turabian StyleNakano, Kazumi, Koichi Yokoyama, Shuichi Shin, Koki Uchida, Kazuki Tsuji, Marie Tanaka, Kaoru Uchimaru, and Toshiki Watanabe. 2022. "Exploring New Functional Aspects of HTLV-1 RNA-Binding Protein Rex: How Does Rex Control Viral Replication?" Viruses 14, no. 2: 407. https://doi.org/10.3390/v14020407
APA StyleNakano, K., Yokoyama, K., Shin, S., Uchida, K., Tsuji, K., Tanaka, M., Uchimaru, K., & Watanabe, T. (2022). Exploring New Functional Aspects of HTLV-1 RNA-Binding Protein Rex: How Does Rex Control Viral Replication? Viruses, 14(2), 407. https://doi.org/10.3390/v14020407




