Development and Large-Scale Testing of a Novel One-Step Triplex RT-qPCR Assay for Simultaneous Detection of “Neurotropic” Porcine Sapeloviruses, Teschoviruses (Picornaviridae) and Type 3 Porcine Astroviruses (Astroviridae) in Various Samples including Nasal Swabs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Background and RNA Extraction
2.2. RT-PCR and Dye-Terminator Sequencing
2.3. Design of Oligonucleotide Primers and Hydrolysis Probes for the RT-qPCR-Based Detection of PTV, PSV and PoAstV-3
2.4. Production of PSV, PTV and PoAstV-3 RNA Standards
2.5. RT-qPCR and Data Analyses
2.6. Analytical Performance Assays and Statistical Calculations
3. Results
3.1. Design of Oligonucleotide Primers and Hydrolysis Probes for PTV, PSV and PoAstV-3
3.2. Performance and Analytical Sensitivity of Singleplex and Triplex RT-qPCR Assays
3.3. Comparison of Triplex RT-qPCR and RT-PCR Assay Diagnostic Sensitivity in Selected Samples
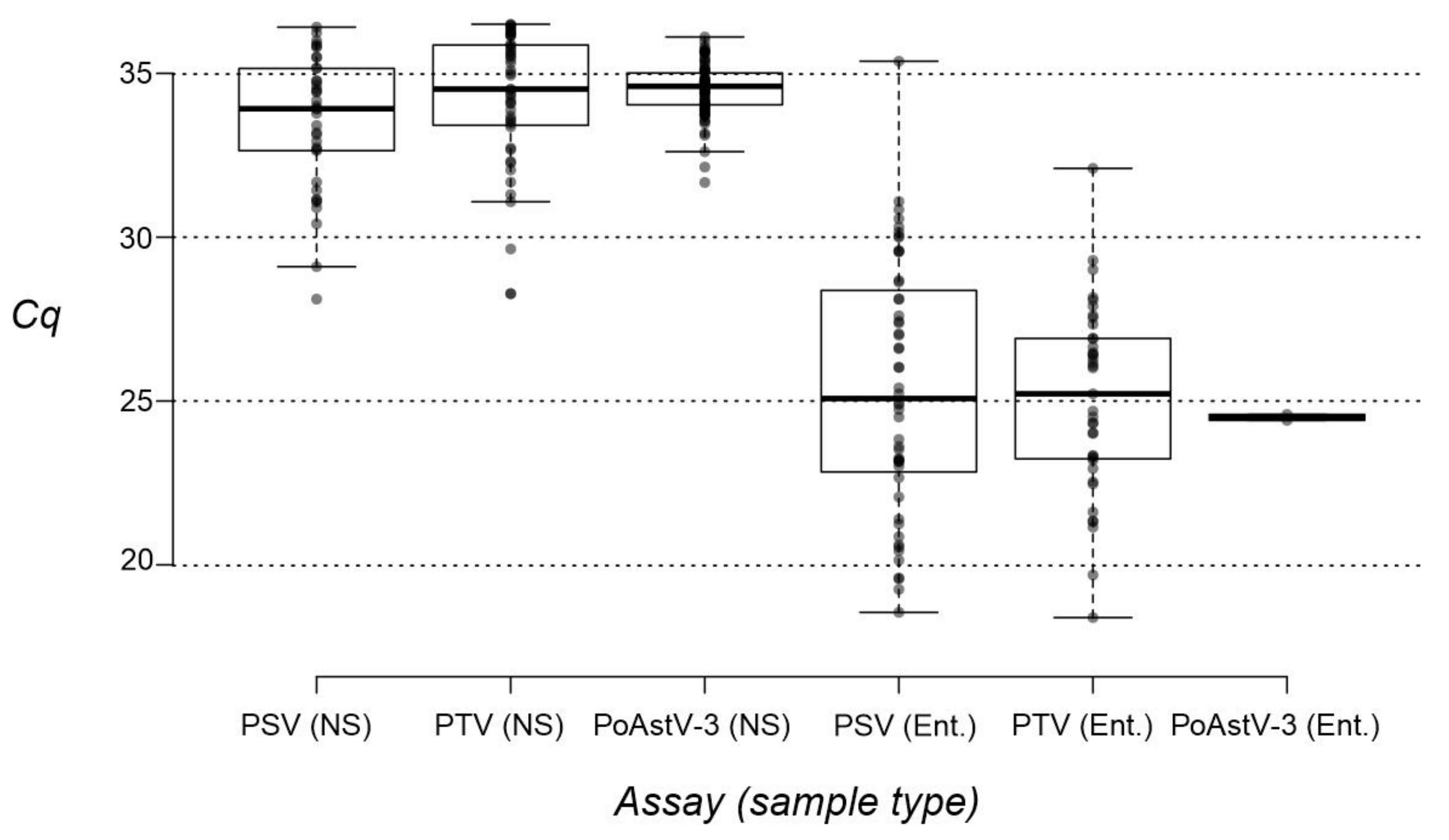
3.4. Investigation of Nasal Swab and Central Nervous System Samples by RT-qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schock, A.; Gurrala, R.; Fuller, H.; Foyle, L.; Dauber, M.; Martelli, F.; Scholes, S.; Roberts, L.; Steinbach, F.; Dastjerdi, A. Investigation into an outbreak of encephalomyelitis caused by a neuroinvasive porcine sapelovirus in the United Kingdom. Vet. Microbiol. 2014, 172, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Arruda, P.A.; Hensch, M.; Chen, Q.; Zheng, Y.; Yang, C.; Gatto, I.R.H.; Li, G. Porcine astrovirus type 3 in central nervous system of swine with polioencephalomyelitis. Emerg. Infect. Dis. 2017, 23, 2097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arruda, P.H.E.; Arruda, B.L.; Schwartz, K.J.; Vannucci, F.; Resende, T.; Rovira, A.; Sundberg, P.; Nietfeld, J.; Hause, B.M. Detection of a novel sapelovirus in central nervous tissue of pigs with polioencephalomyelitis in the USA. Transbound. Emerg. Dis. 2017, 64, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Matias Ferreyra, F.; Arruda, B.; Stevenson, G.; Schwartz, K.; Madson, D.; Yoon, K.J.; Arruda, P. Development of polioencephalomyelitis in cesarean-derived colostrum-deprived pigs following experimental inoculation with either teschovirus a serotype 2 or serotype 11. Viruses 2017, 9, 179. [Google Scholar] [CrossRef] [Green Version]
- Boros, Á.; Albert, M.; Pankovics, P.; Bíró, H.; Pesavento, P.A.; Phan, T.G.; Delwart, E.; Reuter, G. Outbreaks of neuroinvasive astrovirus associated with encephalomyelitis, weakness, and paralysis among weaned pigs, Hungary. Emerg. Infect. Dis. 2017, 23, 1982–1993. [Google Scholar] [CrossRef]
- Carnero, J.; Prieto, C.; Polledo, L.; Martínez-Lobo, F.J. Detection of teschovirus type 13 from two swine herds exhibiting nervous clinical signs in growing pigs. Transbound. Emerg. Dis. 2018, 65, e489–e493. [Google Scholar] [CrossRef]
- Vreman, S.; Caliskan, N.; Harders, F.; Boonstra, J.; Peperkamp, K.; Ho, C.K.; Kuller, W.; Kortekaas, J. Two novel porcine teschovirus strains as the causative agents of encephalomyelitis in the Netherlands. BMC Vet. Res. 2020, 16, 51. [Google Scholar] [CrossRef] [Green Version]
- Hammerschmitt, M.E.; de Almeida, P.R.; de Cecco, B.S.; Lorenzett, M.P.; Schwertz, C.I.; da Cruz, R.A.S.; Caprioli, R.A.; Schuh, D.T.; Demoliner, M.; Eisen, A.K.A.; et al. Swine polioencephalomyelitis in Brazil: Identification of Teschovirus A, Sapelovirus A, and Enterovirus G in a farm from Southern Brazil. Braz. J. Microbiol. 2021, 52, 1617–1622. [Google Scholar] [CrossRef]
- Zell, R.; Delwart, E.; Gorbalenya, A.; Hovi, T.; King, A.M.Q.; Knowles, N.J.; Lindberg, A.M.; Pallansch, M.A.; Palmenberg, A.C.; Reuter, G.; et al. ICTV Virus Taxonomy Profile: Picornaviridae. J. Gen. Virol. 2017, 98, 2421–2422. [Google Scholar] [CrossRef]
- Boros, Á.; László, Z.; Pankovics, P.; Marosi, A.; Albert, M.; Cságola, A.; Bíró, H.; Fahsbender, E.; Delwart, E.; Reuter, G. High prevalence, genetic diversity and a potentially novel genotype of Sapelovirus A (Picornaviridae) in enteric and respiratory samples in Hungarian swine farms. J. Gen. Virol. 2020, 101, 609–621. [Google Scholar] [CrossRef]
- The Picornavirus Pages. Available online: https://www.picornaviridae.com/ (accessed on 25 January 2021).
- Reuter, G.; Pankovics, P.; Boros, Á. Nonsuppurative (aseptic) meningoencephalomyelitis associated with neurovirulent astrovirus infections in humans and animals. Clin. Microbiol. Rev. 2018, 31, e00040-18. [Google Scholar] [CrossRef] [Green Version]
- Chiu, S.C.; Hu, S.C.; Chang, C.C.; Chang, C.Y.; Huang, C.C.; Pang, V.F.; Wang, F.I. The role of porcine teschovirus in causing diseases in endemically infected pigs. Vet. Microbiol. 2012, 161, 88–95. [Google Scholar] [CrossRef]
- Tsai, A.T.H.; Kuo, C.C.; Kuo, Y.C.; Yang, J.L.; Chang, C.Y.; Wang, F.I. The urinary shedding of porcine teschovirus in endemic field situations. Vet. Microbiol. 2016, 182, 150–155. [Google Scholar] [CrossRef]
- Kim, D.S.; Kang, M.I.; Son, K.Y.; Bak, G.Y.; Park, J.G.; Hosmillo, M.; Seo, J.Y.; Kim, J.Y.; Alfajaro, M.M.; Soliman, M.; et al. Pathogenesis of Korean Sapelovirus A in piglets and chicks. J. Gen. Virol. 2016, 97, 2566. [Google Scholar] [CrossRef]
- Rawal, G.; Ferreyra, F.M.; Macedo, N.R.; Bradner, L.K.; Harmon, K.M.; Allison, G.; Arruda, B.L. Ecology of Porcine Astrovirus Type 3 in a Herd with Associated Neurologic Disease. Viruses 2020, 12, 992. [Google Scholar] [CrossRef]
- Zell, R.; Dauber, M.; Krumbholz, A.; Henke, A.; Birch-Hirschfeld, E.; Stelzner, A.; Prager, D.; Wurm, R. Porcine teschoviruses comprise at least eleven distinct serotypes: Molecular and evolutionary aspects. J. Virol. 2001, 75, 1620–1631. [Google Scholar] [CrossRef] [Green Version]
- Krumbholz, A.; Dauber, M.; Henke, A.; Birch-Hirschfeld, E.; Knowles, N.J.; Stelzner, A.; Zell, R. Sequencing of porcine enterovirus groups II and III reveals unique features of both virus groups. J. Virol. 2002, 76, 5813–5821. [Google Scholar] [CrossRef] [Green Version]
- Xiao, C.T.; Halbur, P.G.; Opriessnig, T. Complete genome sequence of a newly identified porcine astrovirus genotype 3 strain US-MO123. J. Virol. 2012, 86, 13126. [Google Scholar] [CrossRef] [Green Version]
- Cortez, V.; Meliopoulos, V.A.; Karlsson, E.A.; Hargest, V.; Johnson, C.; Schultz-Cherry, S. Astrovirus biology and pathogenesis. Annu. Rev. Virol. 2017, 29, 327–348. [Google Scholar] [CrossRef]
- Zell, R.; Krumbholz, A.; Henke, A.; Birch-Hirschfeld, E.; Stelzner, A.; Doherty, M.; Dauber, M.; Prager, D.; Wurm, R. Detection of porcine enteroviruses by nRT–PCR: Differentiation of CPE groups I–III with specific primer sets. J. Virol. Methods 2000, 88, 205–218. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, W.; Wang, D.; Zhu, X.; Chen, Y.; Ouyang, K.; Huang, W. Establishment of a multiplex RT-PCR method for the detection of five known genotypes of porcine astroviruses. Front. Vet. Sci. 2021, 8, 684279. [Google Scholar] [CrossRef]
- Cano-Gómez, C.; Buitrago, D.; Fernández-Pinero, J.; Fernández-Pacheco, P.; Mansilla, C.; Agüero, M.; Jiménez-Clavero, M.A. Evaluation of a fluorogenic real-time reverse transcription-polymerase chain reaction method for the specific detection of all known serotypes of porcine teschoviruses. J. Virol. Methods 2011, 176, 131–134. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, Z.; Hu, F.; Liu, Y.; Qiu, Z.; Zhou, S.; Cui, S.; Wang, M. The survey of porcine teschoviruses in field samples in China with a universal rapid probe real-time RT-PCR assay. Trop. Anim. Health. Prod. 2013, 45, 1057–1061. [Google Scholar] [CrossRef]
- Yang, T.; Zhang, L.; Lu, Y.; Guo, M.; Zhang, Z.; Lin, A. Characterization of porcine sapelovirus prevalent in western Jiangxi, China. BMC Vet. Res. 2021, 17, 273. [Google Scholar] [CrossRef]
- Rawal, G.; Matias Ferreyra, F.; Macedo, N.R.; Bradner, L.K.; Harmon, K.M.; Mueller, A.; Allison, G.; Linhares, D.C.L.; Arruda, B.L. Detection and cellular tropism of porcine astrovirus type 3 on breeding farms. Viruses 2019, 11, 1051. [Google Scholar] [CrossRef] [Green Version]
- Stäubli, T.; Rickli, C.I.; Torgerson, P.R.; Fraefel, C.; Lechmann, J. Porcine teschovirus, sapelovirus, and enterovirus in Swiss pigs: Multiplex RT-PCR investigation of viral frequencies and disease association. J. Vet. Diagn. Investig. 2021, 33, 864–874. [Google Scholar] [CrossRef]
- Boros, Á.; Nemes, C.; Pankovics, P.; Kapusinszky, B.; Delwart, E.; Reuter, G. Porcine teschovirus in wild boars in Hungary. Arch. Virol. 2012, 157, 1573–1578. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Boros, Á.; Pankovics, P.; Simmonds, P.; Reuter, G. Novel positive-sense, single-stranded RNA (+ssRNA) virus with di-cistronic genome from intestinal content of freshwater carp (Cyprinus carpio). PLoS ONE 2011, 6, e29145. [Google Scholar]
- Spitzer, M.; Wildenhain, J.; Rappsilber, J.; Tyers, M. BoxPlotR: A web tool for generation of box plots. Nat. Methods 2014, 11, 121–122. [Google Scholar] [CrossRef]
- Debode, F.; Marien, A.; Janssen, É.; Bragard, C.; Berben, G. The influence of amplicon length on real-time PCR results. Biotechnol. Agron. Soc. Environ. 2017, 21, 3–11. [Google Scholar] [CrossRef]
- Matias Ferreyra, F.; Harmon, K.; Bradner, L.; Burrough, E.; Derscheid, R.; Magstadt, D.R.; Arruda, B. Comparative Analysis of Novel Strains of Porcine Astrovirus Type 3 in the USA. Viruses 2021, 13, 1859. [Google Scholar] [CrossRef] [PubMed]
- Kralik, P.; Ricchi, M. A basic guide to real time PCR in microbial diagnostics: Definitions, parameters, and everything. Front. Microbiol. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: M inimum I nformation for Publication of Q uantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [Green Version]
- Blomström, A.L.; Ye, X.; Fossum, C.; Wallgren, P.; Berg, M. Characterisation of the virome of tonsils from conventional pigs and from specific pathogen-free pigs. Viruses 2018, 10, 382. [Google Scholar] [CrossRef] [Green Version]



| Target Virus (Genome Region) | Oligonucleotide ID | Sequence (5′–3′) | Length (nt) | Tm (°C) | PCR Product Size (bp) |
|---|---|---|---|---|---|
| Sapelovirus A (3D-RdRp) | pan-PSV-3D-qPCR-R | CTC CTC ACC ATT GTG CCA TGC | 21 | 65 | 132 |
| pan-PSV-3D-qPCR-F | TAC CCC TTC CTG TGT CAT CCT | 21 | 64 | ||
| pan-PSV-3D-qPCR-PR | /6-FAM/CAA TTA GAT GGA CCA AGA ATG CCA G/IABkFQ/ | 25 | 65 | ||
| Teschovirus A&B (5′UTR) | pan-PTV-5UTR-qPCR-R | TGA CTA TAC AAA GTA CAG ACG GCC | 24 | 64 | 172 |
| pan-PTV-5UTR-qPCR-F | GTA CAG AAG AGC AAG TAC TCC TGA | 24 | 64 | ||
| pan-PTV-5UTR-qPCR-PR | /SUN/GGA GTT CCT GTA TGG GAA TGC A/IABkFQ/ | 22 | 65 | ||
| Porcine astrovirus type 3 | PoAstV3-CAP-qPCR-R | TCC TTG GCA ACC TCC TTA GC | 20 | 64 | 178 |
| (ORF1b-ORF2 junction) | PoAstV3-RdRp-qPCR-F | GGT GGC CTT GGA GGT TGT T | 19 | 64 | |
| PoAstV3-qPCR-PR | /Cy5/CTG ATG AGA TGC TTG ACC GGC/IAbRQSp/ | 21 | 65 |
| Copies/Reaction | Mean Cq (SD) | Detected/Tested (%) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Singleplex | Triplex | Singleplex | Triplex | |||||||||
| PSV | PTV | PoAstV-3 | PSV | PTV | PoAstV-3 | PSV | PTV | PoAstV-3 | PSV | PTV | PoAstV-3 | |
| 10 | 35.92 (0.63) | 34.45 (1.31) | 36.21 (1.30) | 34.82 (1.20) | 35.75 (0.95) | 34.56 (0.63) | 9/11 (82) | 4/11 (36) | 5/11 (46) | 7/11 (64) | 5/11 (46) | 6/11 (55) |
| 100 | 32.61 (0.50) | 31.50 (1.04) | 31.87 (0.91) | 31.06 (0.59) | 31.23 (0.53) | 30.87 (0.31) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) |
| 1000 | 29.02 (0.56) | 28.33 (0.25) | 28.31 (0.83) | 28.25 (0.36) | 28.29 (0.35) | 28.22 (0.24) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) | 11/11 (100) |
| 10,000 | 25.77 (0.43) | 25.48 (0.31) | 25.11 (0.36) | 25.88 (0.22) | 25.86 (0.23) | 25.63 (0.41) | 6/6 (100) | 6/6 (100) | 6/6 (100) | 6/6 (100) | 6/6 (100) | 6/6 (100) |
| Intra-Assay Variation (CV%) | Inter-Assay Variation (CV%) | |||||||
|---|---|---|---|---|---|---|---|---|
| Singleplex | Triplex | Singleplex | Triplex | |||||
| Virus | 1 × 102 | 1 × 103 | 1 × 102 | 1 × 103 | 1 × 102 | 1 × 103 | 1 × 102 | 1 × 103 |
| PSV | 0.05–1.55 | 0.12–1.87 | 0.03–1.75 | 0.13–0.32 | 1.53 | 1.92 | 1.91 | 1.26 |
| PTV | 0.03–4.62 | 0.10–0.88 | 0.13–1.62 | 0.11–0.91 | 3.29 | 0.87 | 1.70 | 1.22 |
| PoAstV-3 | 0.63–3.58 | 0.29–3.38 | 0.10–0.88 | 0.20–1.53 | 2.87 | 2.95 | 1.02 | 0.86 |
| Results of tRT-qPCR vs. RT-PCR (No. of Samples (Seq+)) | Performance of tRT-qPCR Compared to RT-PCR | ||||||
|---|---|---|---|---|---|---|---|
| Virus | +/+ | +/− | −/+ | −/− | Sensitivity (%) | Specificity (%) | Concordance (%) |
| PSV | 57 | 7 (5) | 6 (3) | 72 | 95.59 | 97.30 | 90.85 |
| PTV | 42 | 5 (4) | 7 (4) | 88 | 92.59 | 98.88 | 91.55 |
| PoAstV-3 | 18 | 2 (2) | 7 (2) | 115 | 91.67 | 100.00 | 93.66 |
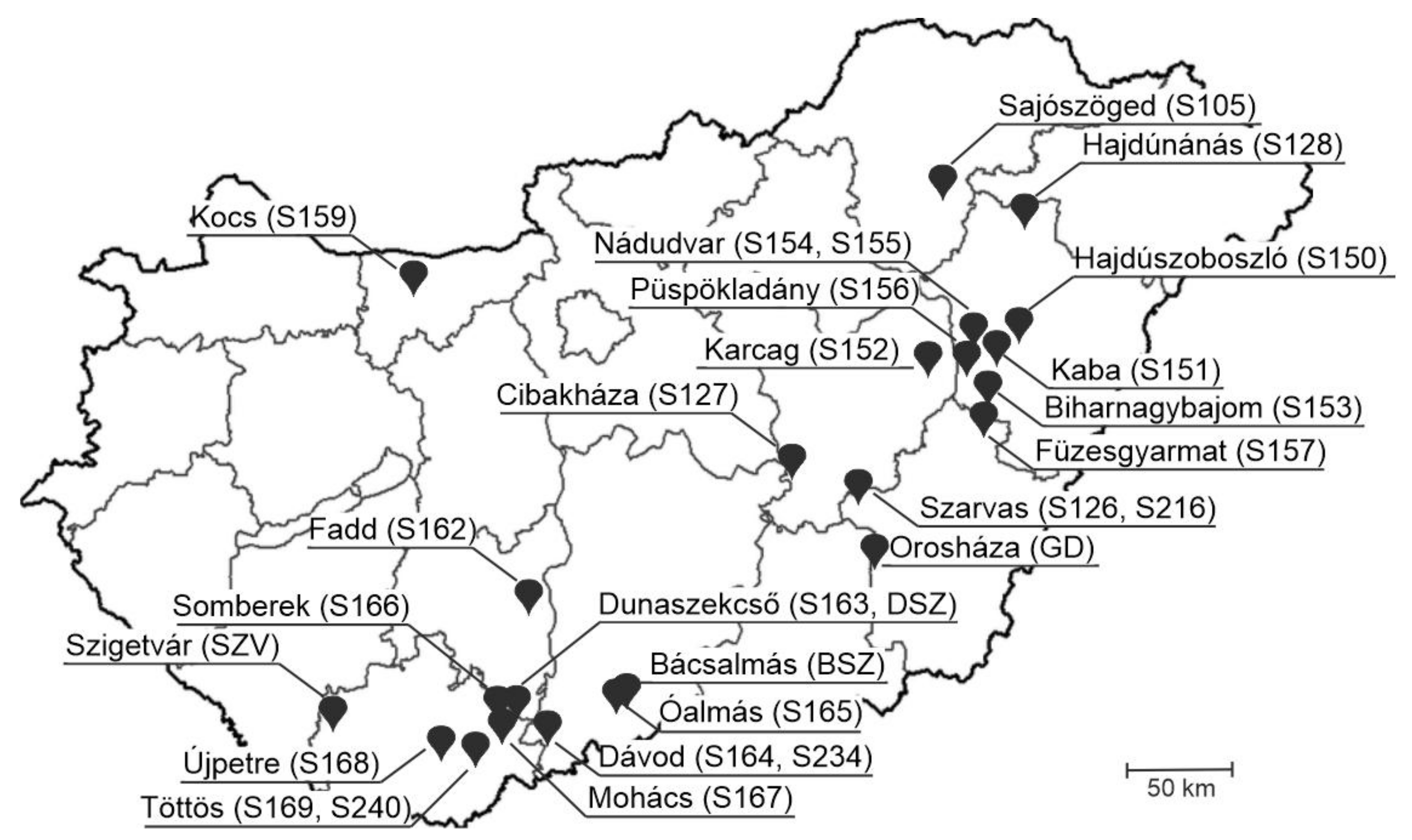
| Farm ID | Location | PSV | PTV | PoAstV-3 | PSV &PTV | PSV& | PTV& | Triple Infection |
|---|---|---|---|---|---|---|---|---|
| PoAstV-3 | PoAstV-3 | |||||||
| S105 | Sajószöged | 0/13 | 1/13 (7.7%) | 4/13 | 0/13 | 0/13 | 1/13 | 0/13 |
| (30.8%) | (7.7%) | |||||||
| S126 | Szarvas | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 | 0/13 |
| S127 | Cibakháza | 0/10 | 1/10 | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| (10.0%) | (30.0%) | |||||||
| S128 | Hajdúnánás | 0/12 | 0/12 | 4/12 | 0/12 | 0/12 | 0/12 | 0/12 |
| (33.3%) | ||||||||
| S150 | Hajdószoboszló | 0/15 | 7/15 | 3/15 | 0/15 | 0/15 | 1/15 | 0/15 |
| (46.6%) | (20.0%) | (6.6%) | ||||||
| S151 | Kaba | 2/11 | 7/11 | 3/11 | 0/11 | 0/11 | 2/11 | 1/11 |
| (18.1%) | (63.6%) | (27.3%) | (18.2%) | (9.1%) | ||||
| S152 | Karcag | 0/15 | 0/15 | 2/15 | 0/15 | 0/15 | 0/15 | 0/15 |
| (13.3%) | ||||||||
| S153 | Biharnagybajom | 0/15 | 0/15 | 6/15 | 0/15 | 0/15 | 0/15 | 0/15 |
| (40.0%) | ||||||||
| S154 | Nádudvar | 0/15 | 3/15 | 3/15 | 0/15 | 0/15 | 1/15 | 0/15 |
| (20.0%) | (20.0%) | (6.7%) | ||||||
| S155 | Nádudvar | 0/14 | 1/14 | 4/14 | 0/14 | 0/14 | 0/14 | 0/14 |
| (7.1%) | (28.6%) | |||||||
| S156 | Püspökladány | 0/15 | 0/15 | 2/15 | 0/15 | 0/15 | 0/15 | 0/15 |
| (13.3%) | ||||||||
| S157 | Füzesgyarmat | 8/15 | 6/15 | 3/15 | 3/15 | 1/15 | 2/15 | 0/15 |
| (53.3%) | (40.0%) | (20.0%) | (20.0%) | (6.7%) | (13.3%) | |||
| S159 | Kocs | 0/14 | 0/14 | 3/14 | 0/14 | 0/14 | 0/14 | 0/14 |
| (21.4%) | ||||||||
| S162 | Fadd | 4/9 | 2/9 | 3/9 | 1/9 | 1/9 | 0/9 | 1/9 |
| (44.4%) | (22.2%) | (33.3%) | (11.1%) | (11.1%) | (11.1%) | |||
| S163 | Dunaszekcső | 3/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 |
| (30.0%) | ||||||||
| S164 | Dávod | 2/15 | 2/15 | 5/15 | 1/15 | 1/15 | 1/15 | 0/15 |
| (13.3%) | (13.3%) | (33.3%) | (6.7%) | (6.7%) | (6.7%) | |||
| S165 | Óalmás | 0/15 | 7/15 | 5/15 | 0/15 | 0/15 | 2/15 | 0/15 |
| (46.7%) | (33.3%) | (13.3%) | ||||||
| S166 | Somberek | 0/15 | 0/15 | 0/15 | 0/15 | 0/15 | 0/15 | 0/15 |
| S167 | Mohács | 0/14 | 2/14 | 3/14 | 0/14 | 0/14 | 0/14 | 0/14 |
| (14.3%) | (21.4%) | |||||||
| S168 | Újpetre | 0/13 | 0/13 | 2/13 | 0/13 | 0/13 | 0/13 | 0/13 |
| (15.4%) | ||||||||
| S169 | Töttös | 0/15 | 2/15 | 2/15 | 0/15 | 0/15 | 1/15 | 0/15 |
| (13.3%) | (13.3%) | (6.7%) | ||||||
| S216 | Szarvas | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 | 0/9 |
| S234 | Dávod | 2/30 | 1/30 | 3/30 | 0/30 | 0/30 | 1/30 | 0/30 |
| (6.7%) | (3.3%) | (10.0%) | (3.3%) | |||||
| S240 | Töttös | 1/30 | 0/30 | 3/30 | 0/30 | 0/30 | 0/30 | 0/30 |
| (3.3%) | (10.0%) | |||||||
| SZV | Szigetvár | 7/29 | 0/29 | 2/29 | 0/29 | 0/29 | 0/29 | 0/29 |
| (24.1%) | (6.9%) | |||||||
| BSZ | Bácsalmás | 0/42 | 2/42 | 7/42 | 0/42 | 0/42 | 1/42 | 0/42 |
| (4.8%) | (16.7%) | (2.4%) | ||||||
| DSZ | Dunaszekcső | 0/31 | 1/31 | 6/31 | 0/31 | 0/31 | 0/31 | 0/31 |
| (3.2%) | (19.3%) | |||||||
| GD | Gádoros | 9/19 | 2/19 | 3/19 | 2/19 | 3/19 | 0/19 | 0/19 |
| (47.4%) | (10.5%) | (15.8%) | (10.5%) | (15.8%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
László, Z.; Pankovics, P.; Reuter, G.; Cságola, A.; Bodó, K.; Gáspár, G.; Albert, M.; Bíró, H.; Boros, Á. Development and Large-Scale Testing of a Novel One-Step Triplex RT-qPCR Assay for Simultaneous Detection of “Neurotropic” Porcine Sapeloviruses, Teschoviruses (Picornaviridae) and Type 3 Porcine Astroviruses (Astroviridae) in Various Samples including Nasal Swabs. Viruses 2022, 14, 513. https://doi.org/10.3390/v14030513
László Z, Pankovics P, Reuter G, Cságola A, Bodó K, Gáspár G, Albert M, Bíró H, Boros Á. Development and Large-Scale Testing of a Novel One-Step Triplex RT-qPCR Assay for Simultaneous Detection of “Neurotropic” Porcine Sapeloviruses, Teschoviruses (Picornaviridae) and Type 3 Porcine Astroviruses (Astroviridae) in Various Samples including Nasal Swabs. Viruses. 2022; 14(3):513. https://doi.org/10.3390/v14030513
Chicago/Turabian StyleLászló, Zoltán, Péter Pankovics, Gábor Reuter, Attila Cságola, Kornélia Bodó, Gábor Gáspár, Mihály Albert, Hunor Bíró, and Ákos Boros. 2022. "Development and Large-Scale Testing of a Novel One-Step Triplex RT-qPCR Assay for Simultaneous Detection of “Neurotropic” Porcine Sapeloviruses, Teschoviruses (Picornaviridae) and Type 3 Porcine Astroviruses (Astroviridae) in Various Samples including Nasal Swabs" Viruses 14, no. 3: 513. https://doi.org/10.3390/v14030513
APA StyleLászló, Z., Pankovics, P., Reuter, G., Cságola, A., Bodó, K., Gáspár, G., Albert, M., Bíró, H., & Boros, Á. (2022). Development and Large-Scale Testing of a Novel One-Step Triplex RT-qPCR Assay for Simultaneous Detection of “Neurotropic” Porcine Sapeloviruses, Teschoviruses (Picornaviridae) and Type 3 Porcine Astroviruses (Astroviridae) in Various Samples including Nasal Swabs. Viruses, 14(3), 513. https://doi.org/10.3390/v14030513





