The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Isolation and Purification of Bacteriophages
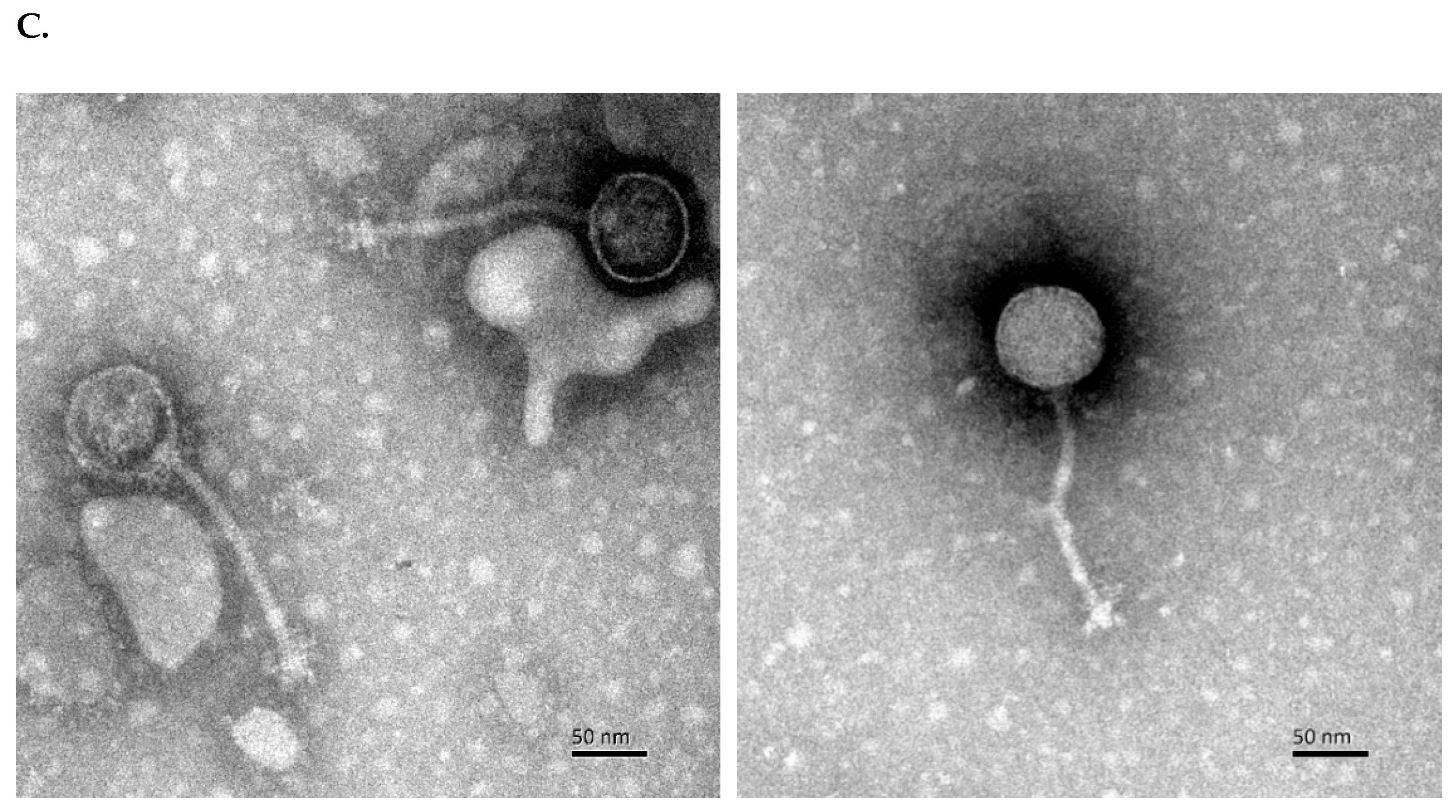
2.3. Preparation of Phages for Transmission Electron Microscopy
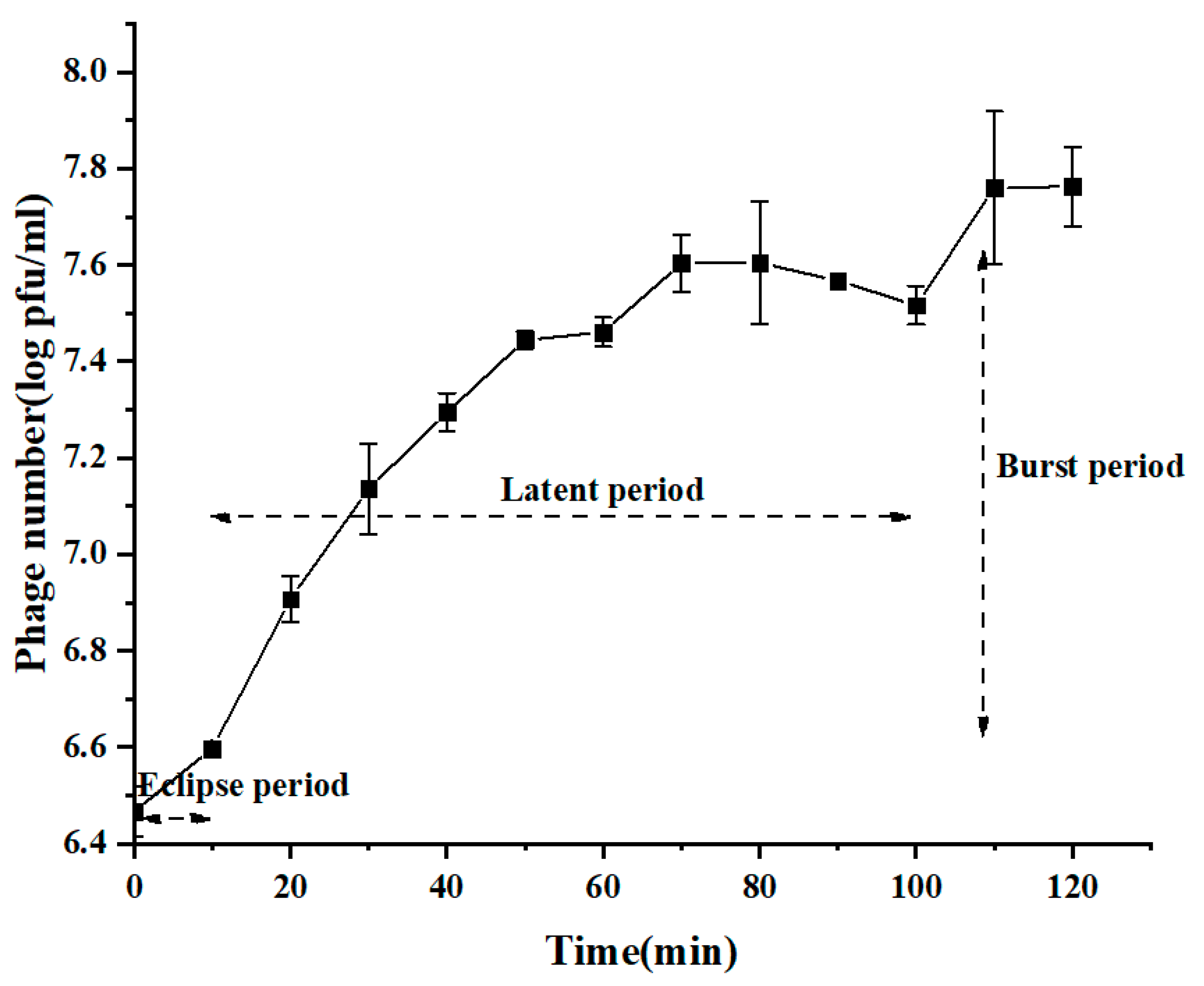
2.4. One-Step Growth Curve
2.5. Host Range Analysis
2.6. Bacterial Challenge Assays
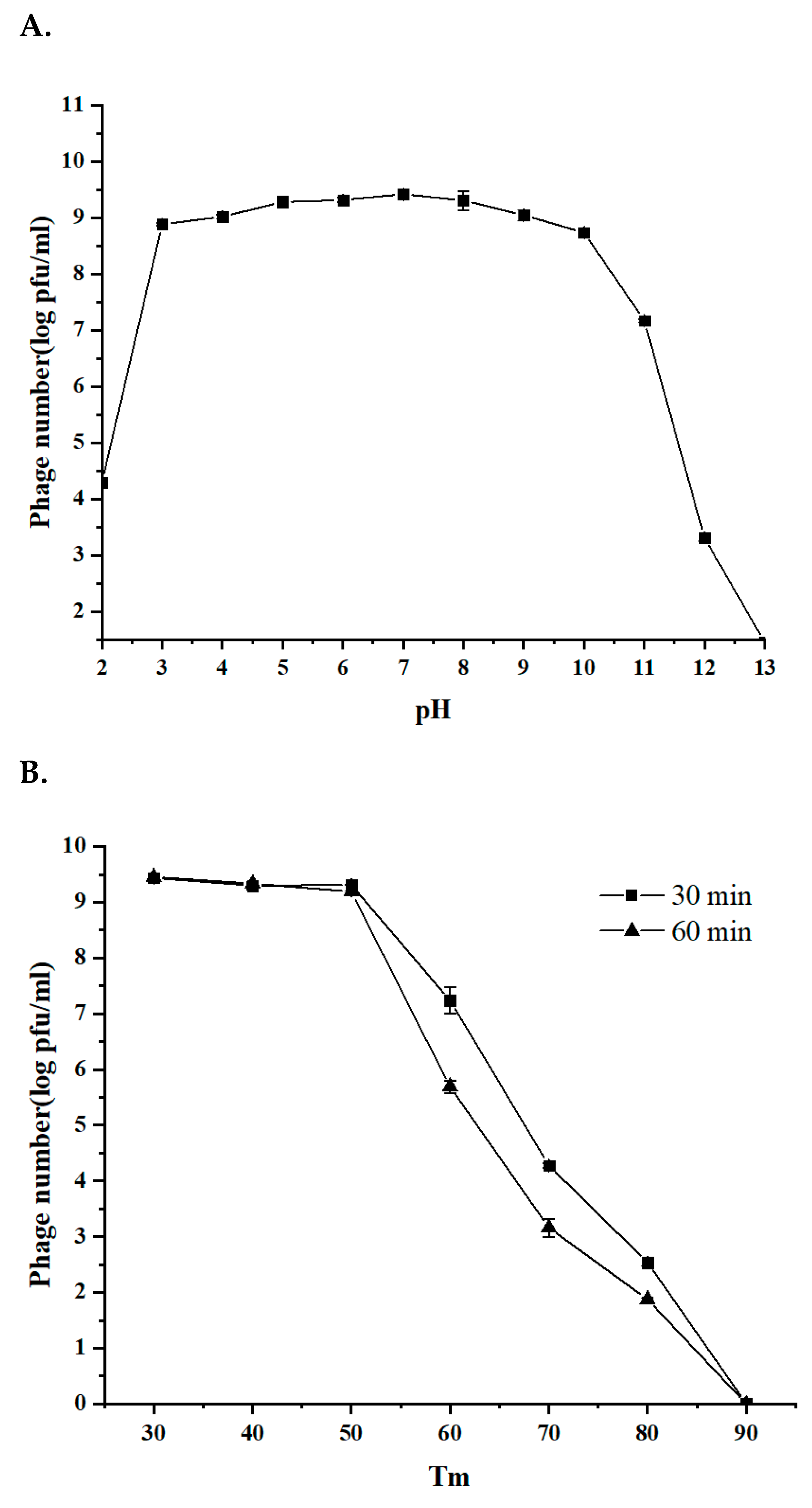
2.7. Thermal and pH Stability of the Phages
2.8. Phage Adsorption and Lytic Assays
2.9. Phage Genome Sequencing and Bioinformatics Analysis
2.10. Determination of Gene Transfer from Phage to Host Strain
2.11. Statistical Analysis
3. Results
3.1. Isolation and Characterization of C. perfringens Phage
3.2. Phage BG3P Has a Broad Host Range
3.3. The Stability of Phage BG3P in Response to Temperature and pH Is Suitable for Further Application
3.4. Quick Adsorption and Efficient Lytic Activity of BG3P
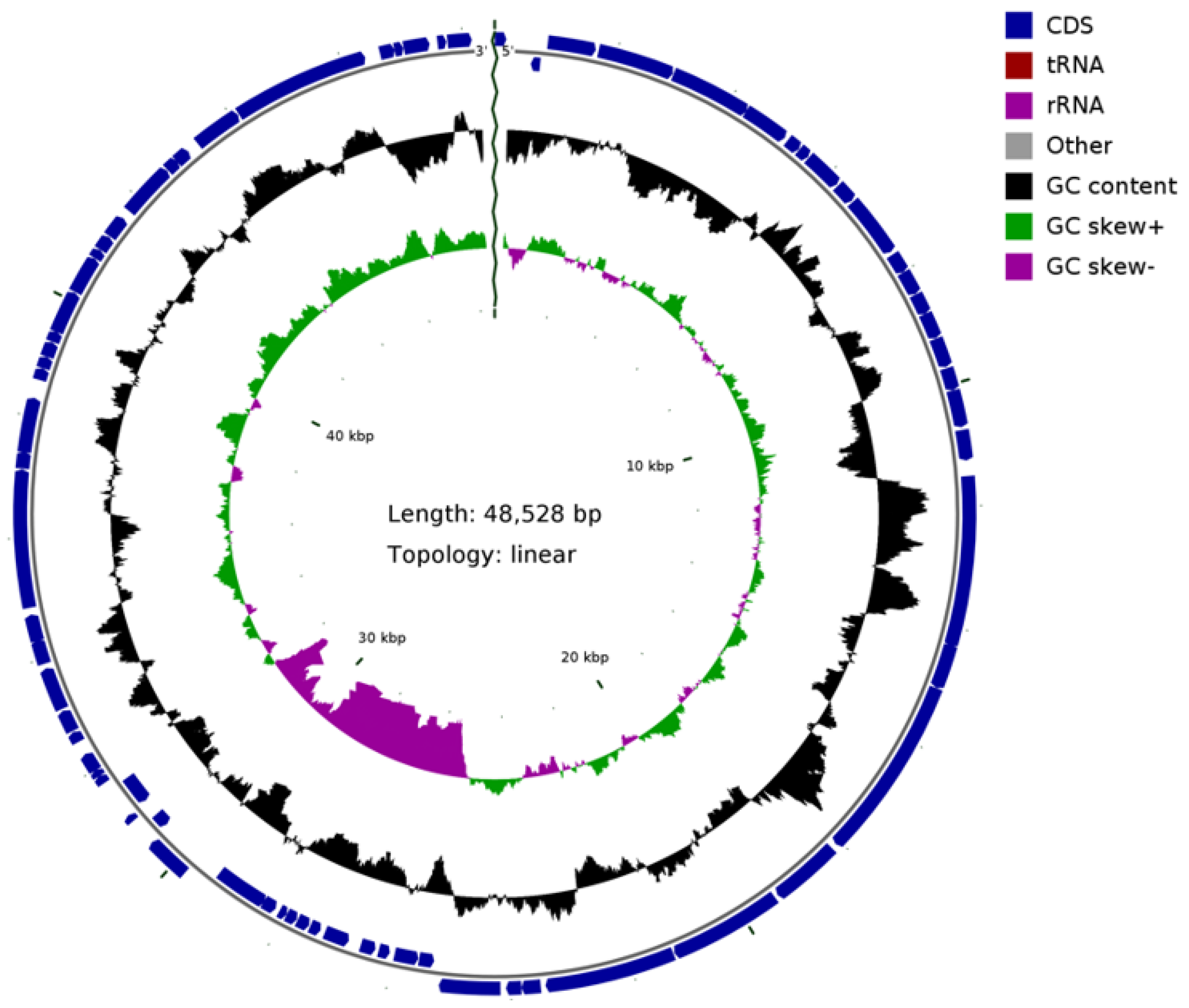
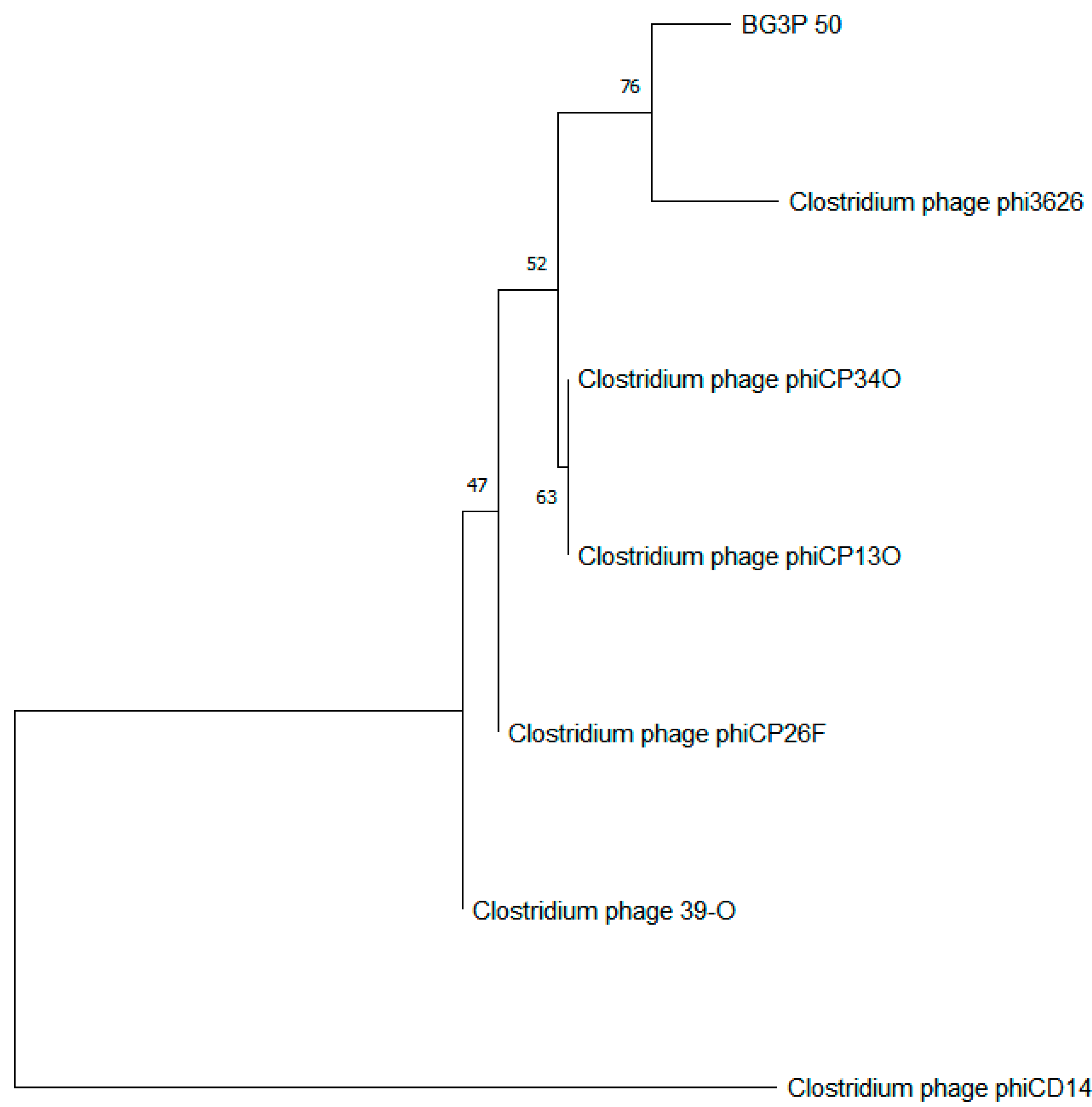
3.5. The Sequence Analysis of Phage BG3P Genome Revealed That It Is a New Phage
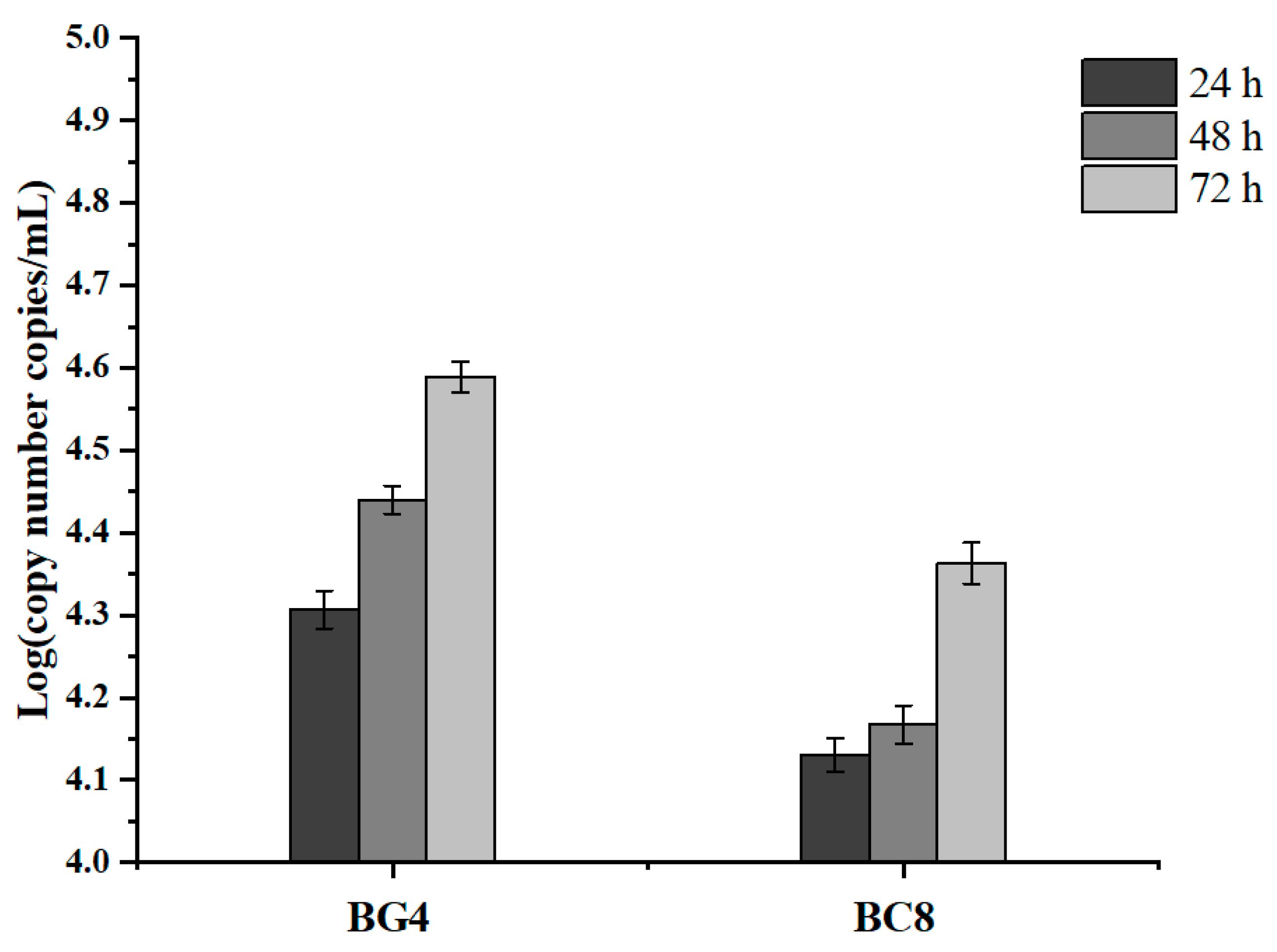
3.6. Chloramphenicol Resistance Gene Transfer
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van Immerseel, F.; De Buck, J.; Pasmans, F.; Huyghebaert, G.; Haesebrouck, F.; Ducatelle, R. Clostridium perfringens in poultry: An emerging threat for animal and public health. Avian Pathol. 2004, 33, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Timbermont, L.; Haesebrouck, F.; Ducatelle, R.; Immerseel, F.V. Necrotic enteritis in broilers: An updated review on the pathogenesis. Avian Pathol. 2011, 40, 341–347. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, C.; Hwang, C. Growth/no growth boundary of Clostridium perfringens from spores in cooked meat: A logistic analysis. Int. J. Food Microbiol. 2018, 266, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Huyghebaert, G.; Ducatelle, R.; Immerseel, F.V. An update on alternatives to antimicrobial growth promoters for broilers. Vet. J. 2011, 187, 182–188. [Google Scholar] [CrossRef] [Green Version]
- Millet, S.; Maertens, L. The European ban on antibiotic growth promoters in animal feed: From challenges to opportunities. Vet. J. 2011, 187, 143–144. [Google Scholar] [CrossRef]
- Gharib-Naseri, K.; Kheravii, S.K.; Keerqin, C.; Morgan, N.; Swick, R.A.; Choct, M.; Wu, S.B. Two different Clostridium perfringens strains produce different levels of necrotic enteritis in broiler chickens. Poult. Sci. 2019, 98, 6422–6432. [Google Scholar] [CrossRef]
- Richards, G.P. Bacteriophage remediation of bacterial pathogens in aquaculture: A review of the technology. Bacteriophage 2014, 4, e975540. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Dai, J.; Guo, M.; Li, J.; Zhou, X.; Li, F.; Gao, Y.; Qu, H.; Lu, H.; Jin, J.; et al. Pre-optimized phage therapy on secondary Acinetobacter baumannii infection in four critical COVID-19 patients. Emerg. Microbes Infect. 2021, 10, 612–618. [Google Scholar] [CrossRef]
- Bae, D.; Lee, J.W.; Chae, J.P.; Kim, J.W.; Eun, J.S.; Lee, K.W.; Seo, K.H. Characterization of a novel bacteriophage phiCJ22 and its prophylactic and inhibitory effects on necrotic enteritis and Clostridium perfringens in broilers. Poult. Sci. 2021, 100, 302–313. [Google Scholar] [CrossRef]
- Heo, S.; Kim, M.G.; Kwon, M.; Lee, H.S.; Kim, G.B. Inhibition of Clostridium perfringens using bacteriophages and bacteriocin producing strains. Korean J. Food Sci. Anim. Resour. 2018, 38, 88–98. [Google Scholar]
- Seal, B.S. Characterization of bacteriophages virulent for Clostridium perfringens and identification of phage lytic enzymes as alternatives to antibiotics for potential control of the bacterium. Poult. Sci. 2013, 92, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.W.; Skinner, E.J.; Sulakvelidze, A.; Mathis, G.F.; Hofacre, C.L. Bacteriophage therapy for control of necrotic enteritis of broiler chickens experimentally infected with Clostridium perfringens. Avian Dis. 2010, 54, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, M.; Scherer, S.; Loessner, M.J. Genomic analysis of Clostridium perfringens bacteriophage phi3626, which integrates into guaA and possibly affects sporulation. J. Bacteriol. 2002, 184, 4359–4368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiya, H.; Okada, M.; Tamai, E.; Shimamoto, T.; Shimamoto, T.; Nariya, H. A Putative Amidase Endolysin Encoded by Clostridium perfringens St13 Exhibits Specific Lytic Activity and Synergizes with the Muramidase Endolysin Psm. Antibiotics 2021, 10, 245. [Google Scholar] [CrossRef] [PubMed]
- Cho, J.H.; Kwon, J.G.; O’Sullivan, D.J.; Ryu, S.; Lee, J.H. Development of an endolysin enzyme and its cell wall-binding domain protein and their applications for biocontrol and rapid detection of Clostridium perfringens in food. Food Chem. 2021, 345, 128562. [Google Scholar] [CrossRef] [PubMed]
- Swift, S.M.; Waters, J.J.; Rowley, D.T.; Oakley, B.B.; Donovan, D.M. Characterization of two glycosyl hydrolases, putative prophage endolysins, that target Clostridium perfringens. FEMS Microbiol. Lett. 2018, 365, 179. [Google Scholar] [CrossRef]
- Gervasi, T.; Curto, R.L.; Minniti, E.; Narbad, A.; Mayer, M.J. Application of Lactobacillus johnsonii expressing phage endolysin for control of Clostridium perfringens. Lett. Appl. Microbiol. 2014, 59, 355–361. [Google Scholar] [CrossRef]
- Van Asten, A.J.; van der Wiel, C.W.; Nikolaou, G.; Houwers, D.J.; Gröne, A. A multiplex PCR for toxin typing of Clostridium perfringens isolates. Vet. Microbiol. 2009, 136, 411–412. [Google Scholar] [CrossRef]
- Clokie, M.R.J.; Kropinski, A.M.; Lavigne, R. Bacteriophages: Methods and protocols. Methods Mol. Biol. 2009, 1681. [Google Scholar]
- Lewis, G.D.; Metcalf, T.G. Polyethylene glycol precipitation for recovery of pathogenic viruses, including hepatitis A virus and human rotavirus, from oyster, water, and sediment samples. Appl. Environ. Microbiol. 1988, 54, 1983–1988. [Google Scholar] [CrossRef] [Green Version]
- Hyman, P.; Abedon, S.T. Practical methods for determining phage growth parameters. Methods Mol. Biol. 2009, 501, 175–202. [Google Scholar]
- Lu, Z.; Breidt, F.; Fleming, H.P.; Altermann, E.; Klaenhammer, T.R. Isolation and characterization of a Lactobacillus plantarum bacteriophage, phiJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003, 84, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, L.; Zhao, Z.; Peng, D.; Zhou, X. Polar flagella rotation in Vibrio parahaemolyticus confers resistance to bacteriophage infection. Sci. Rep. 2016, 6, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Blake, J.D.; Cohen, F.E. Pairwise sequence alignment below the twilight zone. J. Mol. Biol. 2001, 307, 721–735. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Kalvari, I.; Argasinska, J.; Quinones-Olvera, N.; Nawrocki, E.P.; Rivas, E.; Eddy, S.R.; Bateman, A.; Finn, R.D.; Petrov, A.I. Rfam 13.0: Shifting to a genome-centric resource for non-coding RNA families. Nucleic Acids Res. 2018, 46, D335–D342. [Google Scholar] [CrossRef]
- Mora, Z.V.; Macías-Rodríguez, M.E.; Arratia-Quijada, J.; Gonzalez-Torres, Y.S.; Nuño, K.; Villarruel-López, A. Clostridium perfringens as Foodborne Pathogen in Broiler Production: Pathophysiology and Potential Strategies for Controlling Necrotic Enteritis. Animals 2020, 10, 1718. [Google Scholar] [CrossRef]
- Volozhantsev, N.V.; Verevkin, V.V.; Bannov, V.A.; Krasilnikova, V.M.; Myakinina, V.P.; Zhilenkov, E.L.; Svetoch, E.A.; Stern, N.J.; Oakley, B.B.; Seal, B.S. The genome sequence and proteome of bacteriophage ΦCPV1 virulent for Clostridium perfringens. Virus Res. 2011, 155, 433–439. [Google Scholar] [CrossRef]
- Morales, C.A.; Oakley, B.B.; Garrish, J.K.; Siragusa, G.R.; Ard, M.B.; Seal, B.S. Complete genome sequence of the podoviral bacteriophage ΦCP24R, which is virulent for Clostridium perfringens. Arch. Virol. 2012, 157, 769–772. [Google Scholar] [CrossRef]
- Seal, B.S.; Fouts, D.E.; Simmons, M.; Garrish, J.K.; Kuntz, R.L.; Woolsey, R.; Schegg, K.M.; Kropinski, A.M.; Ackermann, H.W.; Siragusa, G.R. Clostridium perfringens bacteriophages ΦCP39O and ΦCP26F: Genomic organization and proteomic analysis of the virions. Arch. Virol. 2011, 156, 25–35. [Google Scholar] [CrossRef] [Green Version]
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens Virulent Bacteriophage CPS2 and Its Thermostable Endolysin LysCPS2. Viruses 2018, 10, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ha, E.; Chun, J.; Kim, M.; Ryu, S. Capsular Polysaccharide Is a Receptor of a Clostridium perfringens Bacteriophage CPS1. Viruses 2019, 11, 1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Myers, G.S.; Rasko, D.A.; Cheung, J.K.; Ravel, J.; Seshadri, R.; DeBoy, R.T.; Ren, Q.; Varga, J.; Awad, M.M.; Brinkac, L.M.; et al. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 2006, 16, 1031–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oakley, B.B.; Talundzic, E.; Morales, C.A.; Hiett, K.L.; Siragusa, G.R.; Volozhantsev, N.V.; Seal, B.S. Comparative genomics of four closely related Clostridium perfringens bacteriophages reveals variable evolution among core genes with therapeutic potential. BMC Genom. 2011, 12, 282. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Wu, L.; Lu, R.; Bao, H.; Zhou, Y.; Pang, M.; Brown, J.; Wang, J.; Wang, R.; Zhang, H. Virulent phage vB_CpeP_HN02 inhibits Clostridium perfringens on the surface of the chicken meat. Int. J. Food Microbiol. 2022, 363, 109514. [Google Scholar] [CrossRef] [PubMed]
- Abraham, L.J.; Rood, J.I. The Clostridium perfringens chloramphenicol resistance transposon Tn4451 excises precisely in Escherichia coli. Plasmid 1988, 19, 164–168. [Google Scholar] [CrossRef]
- Magot, M. Physical characterization of the Clostridium perfringens tetracycline-chloramphenicol resistance plasmid pIP401. Ann. Microbiol. 1984, 135, 269–282. [Google Scholar] [CrossRef] [PubMed]
- Petty, N.K.; Foulds, I.J.; Pradel, E.; Ewbank, J.J.; Salmond, G.P.C. A generalized transducing phage (phiIF3) for the genomically sequenced Serratia marcescens strain Db11: A tool for functional genomics of an opportunistic human pathogen. Microbiology 2006, 152, 1701–1708. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roberts, M.D.; Drexler, H. Isolation and genetic characterization of T1-transducing mutants with increased transduction frequency. Virology 1981, 112, 662–669. [Google Scholar] [CrossRef]
- Young, K.K.; Edlin, G.J.; Wilson, G.G. Genetic analysis of bacteriophage T4 transducing bacteriophages. J. Virol. 1982, 41, 345–347. [Google Scholar] [CrossRef] [Green Version]
- Cerquetti, M.C.; Hooke, A.M. Vi I typing phage for generalized transduction of Salmonella typhi. J. Bacteriol. 1993, 175, 5294–5296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dzhusupova, A.B.; Plotnikova, T.G.; Krylov, V.N. Detection of transduction by virulent bacteriophage phi KZ of Pseudomonas aeruginosa chromosomal markers in the presence of plasmid RMS148. Genetika 1982, 18, 1799–1802. [Google Scholar] [PubMed]
- Lencastre, H.D.; Archer, L.J. Characterization of bacteriophage SPP1 transducing particles. J. Gen. Microbiol. 1980, 117, 347–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goodridge, L.D. Designing phage therapeutics. Curr. Pharm. Biotechnol. 2010, 11, 15–27. [Google Scholar] [CrossRef]
- Balogh, B.; Jones, J.B.; Iriarte, F.B.; Momol, M.T. Phage therapy for plant disease control. Curr. Pharm. Biotechnol. 2010, 11, 48–57. [Google Scholar] [CrossRef]

 ), 1 × 109 (
), 1 × 109 (  ), 1 × 108 (
), 1 × 108 (  ), 1 × 107 (
), 1 × 107 (  ), 1 × 106 (
), 1 × 106 (  ), 1 × 105 (
), 1 × 105 (  ), 1 × 104 (
), 1 × 104 (  ) log PFU/mL. The initial concentration of C. perfringens BG3 is 107 CFU/mL (
) log PFU/mL. The initial concentration of C. perfringens BG3 is 107 CFU/mL (  ). Values are the mean of three determinations. (C) Inhibitory effect of phage BG3P against C. perfringens ATCC 13,124 at different MOIs. 1 × 1010 (
). Values are the mean of three determinations. (C) Inhibitory effect of phage BG3P against C. perfringens ATCC 13,124 at different MOIs. 1 × 1010 (  ), 1 × 109 (
), 1 × 109 (  ), 1 × 108 (
), 1 × 108 (  ), 1 × 107 (
), 1 × 107 (  ), 1 × 106 (
), 1 × 106 (  ), 1 × 105 (
), 1 × 105 (  ), 1 × 104 (
), 1 × 104 (  ) log PFU/mL. The initial concentration of C. perfringens ATCC 13,124 is 107 CFU/mL (
) log PFU/mL. The initial concentration of C. perfringens ATCC 13,124 is 107 CFU/mL (  ). Values are the mean of three determinations.
). Values are the mean of three determinations.
 ), 1 × 109 (
), 1 × 109 (  ), 1 × 108 (
), 1 × 108 (  ), 1 × 107 (
), 1 × 107 (  ), 1 × 106 (
), 1 × 106 (  ), 1 × 105 (
), 1 × 105 (  ), 1 × 104 (
), 1 × 104 (  ) log PFU/mL. The initial concentration of C. perfringens BG3 is 107 CFU/mL (
) log PFU/mL. The initial concentration of C. perfringens BG3 is 107 CFU/mL (  ). Values are the mean of three determinations. (C) Inhibitory effect of phage BG3P against C. perfringens ATCC 13,124 at different MOIs. 1 × 1010 (
). Values are the mean of three determinations. (C) Inhibitory effect of phage BG3P against C. perfringens ATCC 13,124 at different MOIs. 1 × 1010 (  ), 1 × 109 (
), 1 × 109 (  ), 1 × 108 (
), 1 × 108 (  ), 1 × 107 (
), 1 × 107 (  ), 1 × 106 (
), 1 × 106 (  ), 1 × 105 (
), 1 × 105 (  ), 1 × 104 (
), 1 × 104 (  ) log PFU/mL. The initial concentration of C. perfringens ATCC 13,124 is 107 CFU/mL (
) log PFU/mL. The initial concentration of C. perfringens ATCC 13,124 is 107 CFU/mL (  ). Values are the mean of three determinations.
). Values are the mean of three determinations.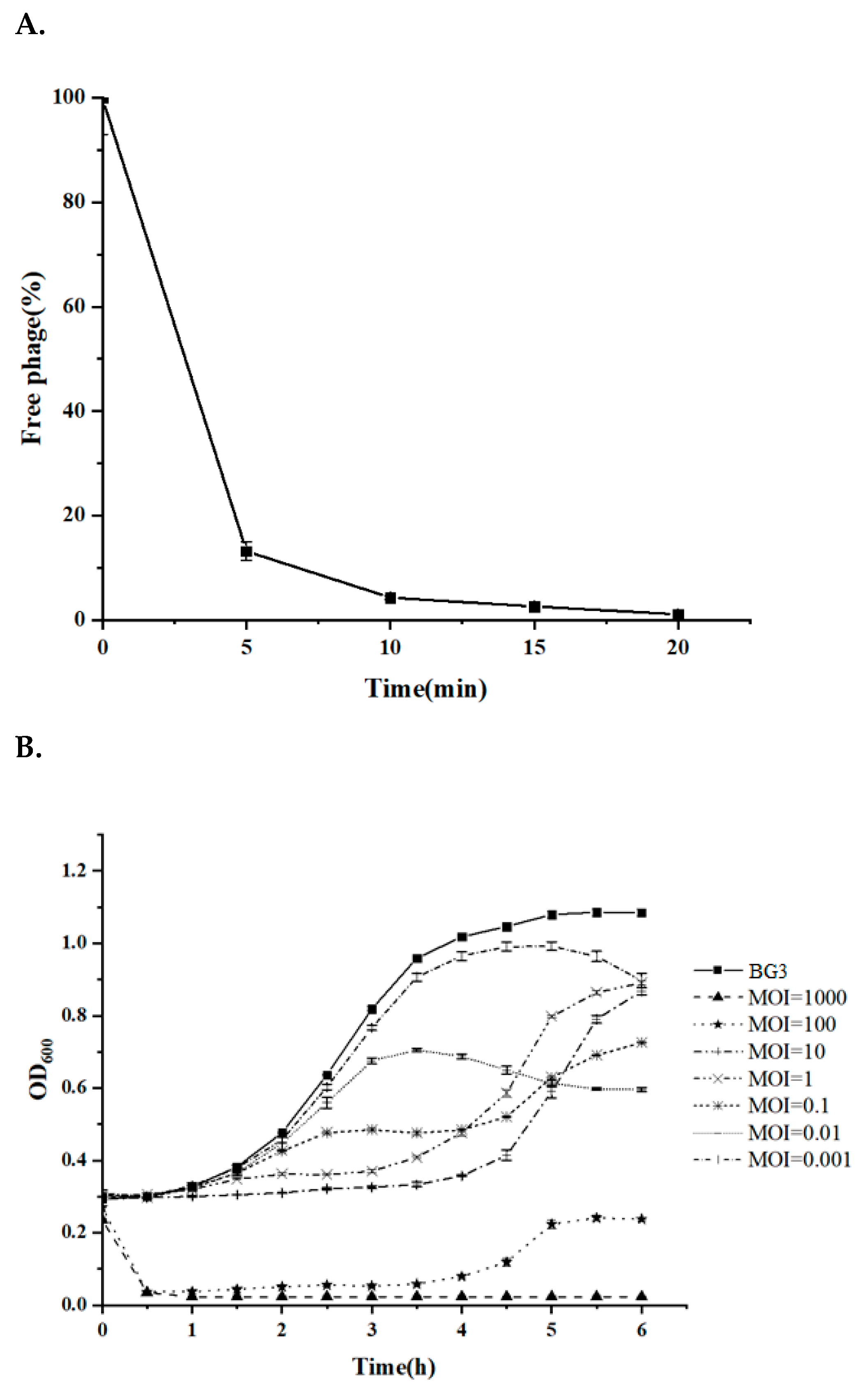

| Strain | Type | BG3P | Source | orf 15 |
|---|---|---|---|---|
| BX3 | A | ++ | Chicken heart feces | + |
| BG9-1 | A (NetB toxin positive) | + | Chicken liver feces | + |
| ZA2 | A | ++ | Chicken intestinal feces | + |
| BG5 | A | ++ | Chicken liver feces | - |
| RZ-5 | A (NetB toxin positive) | ++ | Chicken intestinal feces | - |
| BP1 | A | ++ | Chicken spleen feces | + |
| BX4 | A | ++ | Chicken heart feces | + |
| BC5 | A | + | Chicken intestinal feces | + |
| BX1 | A | ++ | Chicken heart feces | + |
| BC7 | A (NetB toxin positive) | + | Chicken intestinal feces | + |
| ZA4 | A | + | Chicken intestinal feces | + |
| BG1 | A | ++ | Chicken liver feces | - |
| BG3 | A | ++ | Chicken liver feces | - |
| G1-12 | A | - | Chicken intestinal feces | + |
| BC8 | A | ++ | Chicken intestinal feces | - |
| G2-4 | A | + | Chicken intestinal feces | + |
| BX5 | A | + | Chicken liver feces | - |
| ZA3 | A (NetB toxin positive) | ++ | Chicken intestinal feces | + |
| ZA1 | A (NetB toxin positive) | + | Chicken intestinal feces | + |
| BC4 | A | + | Chicken intestinal feces | + |
| BP3 | A | ++ | Chicken spleen feces | + |
| BG7 | A | + | Chicken liver feces | + |
| RZ-1 | A | + | Chicken intestinal feces | + |
| BG4 | A | ++ | Chicken liver feces | - |
| G3-2 | A | + | Chicken intestinal feces | + |
| BX6 | A | + | Chicken liver feces | + |
| BX9-1 | A (NetB toxin positive) | + | Chicken liver feces | - |
| BP5 | A | + | Chicken spleen feces | + |
| BC3 | A | - | Chicken intestinal feces | + |
| G2-2 | A | + | Chicken intestinal feces | + |
| RZ-4 | A | + | Chicken intestinal feces | + |
| BP9-1 | A (NetB toxin positive) | - | Chicken spleen feces | + |
| C. perfringens ATCC 13124 | A | + | Our collection or Reference | - |
| C. perfringens ATCC 64722 | A | + | ||
| C. perfringens ATCC 64723 | A | + | ||
| C. perfringens ATCC 64724 | A | + | ||
| Staphylococcus aureus ATCC 25923 | - | - | - | |
| Salmonella enteritidis ATCC 13076 | - | - | - | |
| Escherichia coli ATCC 25922 | - | - | - | |
| Listeria monocytogenes ATCC 19114 | - | - | - |
| Primer Sequence 5′-3′ | Size (bp) | |
|---|---|---|
| α (cpa)-F | GCTAATGTTACTGCCGTTGA | 324 |
| α (cpa)-R | CCTCTGATACATCGTGTAAG | |
| β (cpb)-F | GCGAATATGCTGAATCATCTA | 196 |
| β (cpb)-R | GCAGGAACATTAGTATATCTTC | |
| ε (etx)-F | GCGGTGATATCCATCTATTC | 655 |
| ε (etx)-R | CCACTTACTTGTCCTACTAAC | |
| ι (iap)-F | ACTACTCTCAGACAAGACAG | 446 |
| ι (iap)-R | CTTTCCTTCTATTACTATACG |
| Gene | Sequence 5′-3′ | Amplicon Size (bp) |
|---|---|---|
| orf 5 | CGACCGAAGGATACGCATT | 736 |
| GCAAGAATTTGGGCAGTCATTA | ||
| orf 11 | TTACTGGAGCGAACGGACTT | 776 |
| ACCAACCATCTTAGCTTCTTCC | ||
| orf 15 | GGGAATTCCATATGAAAAAGTGTCCGCATTTAGAA | 435 |
| CCGCTCGAGCTGCGCTTTATCAATATCTGT | ||
| orf 25 | TACTAATGCTGCTGAAGGTGAA | 1924 |
| TGCAAGCGGATTGCTTAACA | ||
| orf 28 | AATGCTAAGAGCTGGATATGGT | 862 |
| GCCAATAAAGTCTGAGTCTACC |
| Gene ID | Sbjct ID | Organism | Hit_Description | Identity(%) | E Value |
|---|---|---|---|---|---|
| BG3P_1 | None | ||||
| BG3P_2 | None | ||||
| BG3P_3 | YP_001426191.1 | Chlorella virus FR483 | hypothetical protein FR483_N559R | 32.23 | 2.50E-10 |
| BG3P_4 | YP_007237242.1 | Clostridium phage phi8074-B1 | putative terminase large subunit | 52.00 | 3.00E-53 |
| BG3P_5 | ARB07045.1 | Clostridioides phage phiSemix9P1 | portal protein | 47.79 | 4.10E-98 |
| BG3P_6 | ARB07046.1 | Clostridioides phage phiSemix9P1 | head morphogenesis protein | 31.47 | 1.30E-19 |
| BG3P_7 | None | ||||
| BG3P_8 | None | ||||
| BG3P_9 | ARB07047.1 | Clostridioides phage phiSemix9P1 | hypothetical protein Semix9P1_phi04 | 33.00 | 9.60E-21 |
| BG3P_10 | None | ||||
| BG3P_11 | YP_008858642.1 | Brevibacillus phage Davies | major capsid protein | 45.35 | 4.30E-71 |
| BG3P_12 | None | ||||
| BG3P_13 | YP_009214137.1 | Clostridium phage phiCD146 | conserved protein of unknown function | 36.30 | 5.80E-19 |
| BG3P_14 | None | ||||
| BG3P_15 | WP_111946556.1 | Clostridium perfringens strain CPI 103K-3 plasmid pCPCPI103K-3_3 | chloramphenicol resistance protein | 97.92 | 1E-99 |
| BG3P_16 | None | ||||
| BG3P_17 | None | ||||
| BG3P_18 | None | ||||
| BG3P_19 | YP_009215039.1 | Brevibacillus phage Osiris | hypothetical protein OSIRIS_25 | 36.36 | 1.30E-07 |
| BG3P_20 | YP_006383517.1 | Clostridium phage phiS63 | Clostridium phage phiS63 gp13 | 35.66 | 4.00E-95 |
| BG3P_21 | None | ||||
| BG3P_22 | YP_006383519.1 | Clostridium phage phiS63 | Clostridium phage phiS63 gp15 | 53.85 | 7.00E-16 |
| BG3P_23 | ARQ95837.1 | Staphylococcus phage qdsa001 | hypothetical protein qdsa001_81 | 30.99 | 6.50E-23 |
| BG3P_24 | YP_006383520.1 | Clostridium phage phiS63 | Clostridium phage phiS63 gp16 | 83.25 | 0.00E+00 |
| BG3P_25 | YP_009188471.1 | Streptococcus phage Str-PAP-1 | neck passage structure protein | 31.93 | 4.40E-41 |
| BG3P_26 | None | ||||
| BG3P_27 | YP_009219438.1 | Clostridium phage phiCT9441A | holin | 53.09 | 1.30E-16 |
| BG3P_28 | YP_699978.1 | Clostridium phage phiSM101 | autolytic lysozyme | 48.26 | 5.70E-76 |
| BG3P_29 | YP_699977.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0049 | 95.35 | 1.80E-37 |
| BG3P_30 | YP_699976.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0048 | 97.08 | 6.20E-69 |
| BG3P_31 | YP_699975.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0047 | 100.00 | 2.70E-27 |
| BG3P_32 | YP_699974.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0046 | 92.94 | 9.30E-39 |
| BG3P_33 | YP_699973.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0045 | 94.12 | 2.20E-61 |
| BG3P_34 | YP_699972.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0044 | 100.00 | 7.50E-30 |
| BG3P_35 | None | ||||
| BG3P_36 | None | ||||
| BG3P_37 | None | ||||
| BG3P_38 | None | ||||
| BG3P_39 | None | ||||
| BG3P_40 | YP_699968.1 | Clostridium phage phiSM101 | hypothetical protein CPR_C0040 | 44.79 | 2.50E-41 |
| BG3P_41 | None | ||||
| BG3P_42 | None | ||||
| BG3P_43 | ARB07097.1 | Clostridioides phage phiSemix9P1 | y4mF family transcriptional regulator | 42.37 | 7.90E-18 |
| BG3P_44 | None | ||||
| BG3P_45 | None | ||||
| BG3P_46 | None | ||||
| BG3P_47 | None | ||||
| BG3P_48 | YP_006990357.1 | Streptococcus phage phiNJ2 | hypothetical protein phiNJ2_0038 | 41.41 | 1.60E-12 |
| BG3P_49 | APC46471.1 | Aeribacillus phage AP45 | hypothetical protein | 49.33 | 4.50E-51 |
| BG3P_50 | YP_002265460.1 | Clostridium phage phiCP9O | single-stranded DNA-binding protein | 59.56 | 4.10E-33 |
| BG3P_51 | None | ||||
| BG3P_52 | APC46462.1 | Aeribacillus phage AP45 | nucleoside triphosphate hydrolase | 38.79 | |
| BG3P_53 | None | ||||
| BG3P_54 | APC46463.1 | Aeribacillus phage AP45 | recombinase | 44.44 | 1.80E-60 |
| BG3P_55 | None | ||||
| BG3P_56 | None | ||||
| BG3P_57 | None | ||||
| BG3P_58 | YP_009223784.1 | Geobacillus virus E3 | hypothetical protein E3_077 | 50.94 | 3.50E-13 |
| BG3P_59 | YP_009151501.1 | Bacillus phage Pascal | Metal-dependent hydrolase | 44.26 | 9.60E-46 |
| BG3P_60 | YP_002265457.1 | Clostridium phage phiCP39-O Clostridium phage phiCP13-O | hypothetical protein 39-O_gp49 hypothetical protein 13-O_gp46 | 41.87 58.1 | 1.60E-39 |
| BG3P_61 | None | ||||
| BG3P_62 | None | ||||
| BG3P_63 | None | ||||
| BG3P_64 | AFB75778.1 | unidentified phage | hypothetical protein 2019_scaffold132_00014 | 65.40 | 1.10E-07 |
| BG3P_65 | YP_006907549.1 | Bacillus phage Bastille | hypothetical protein | 43.66 | 1.80E-08 |
| BG3P_66 | None | ||||
| BG3P_67 | Q9PD82 | 37.93 | 3.00E-12 | ||
| BG3P_68 | CUQ99459.1 | Lactobacillus phage EV3 | phage primase | 36.57 | 4.00E-77 |
| BG3P_69 | None | ||||
| BG3P_70 | None | ||||
| BG3P_71 | None | ||||
| BG3P_72 | None | ||||
| BG3P_73 | YP_007005016.1 | Clostridium phage phiCP13O | hypothetical protein phi13O_gp1 | 44.26 | 8.70E-20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, S.; Tian, Y.; Wang, Y.; García, P.; Liu, B.; Lu, R.; Wu, L.; Bao, H.; Pang, M.; Zhou, Y.; et al. The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth. Viruses 2022, 14, 676. https://doi.org/10.3390/v14040676
Huang S, Tian Y, Wang Y, García P, Liu B, Lu R, Wu L, Bao H, Pang M, Zhou Y, et al. The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth. Viruses. 2022; 14(4):676. https://doi.org/10.3390/v14040676
Chicago/Turabian StyleHuang, Sisi, Yuan Tian, Yongjuan Wang, Pilar García, Banhong Liu, Rui Lu, Liting Wu, Hongduo Bao, Maoda Pang, Yan Zhou, and et al. 2022. "The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth" Viruses 14, no. 4: 676. https://doi.org/10.3390/v14040676
APA StyleHuang, S., Tian, Y., Wang, Y., García, P., Liu, B., Lu, R., Wu, L., Bao, H., Pang, M., Zhou, Y., Wang, R., & Zhang, H. (2022). The Broad Host Range Phage vB_CpeS_BG3P Is Able to Inhibit Clostridium perfringens Growth. Viruses, 14(4), 676. https://doi.org/10.3390/v14040676







