mRNA- and Adenovirus-Based Vaccines against SARS-CoV-2 in HIV-Positive People
Abstract
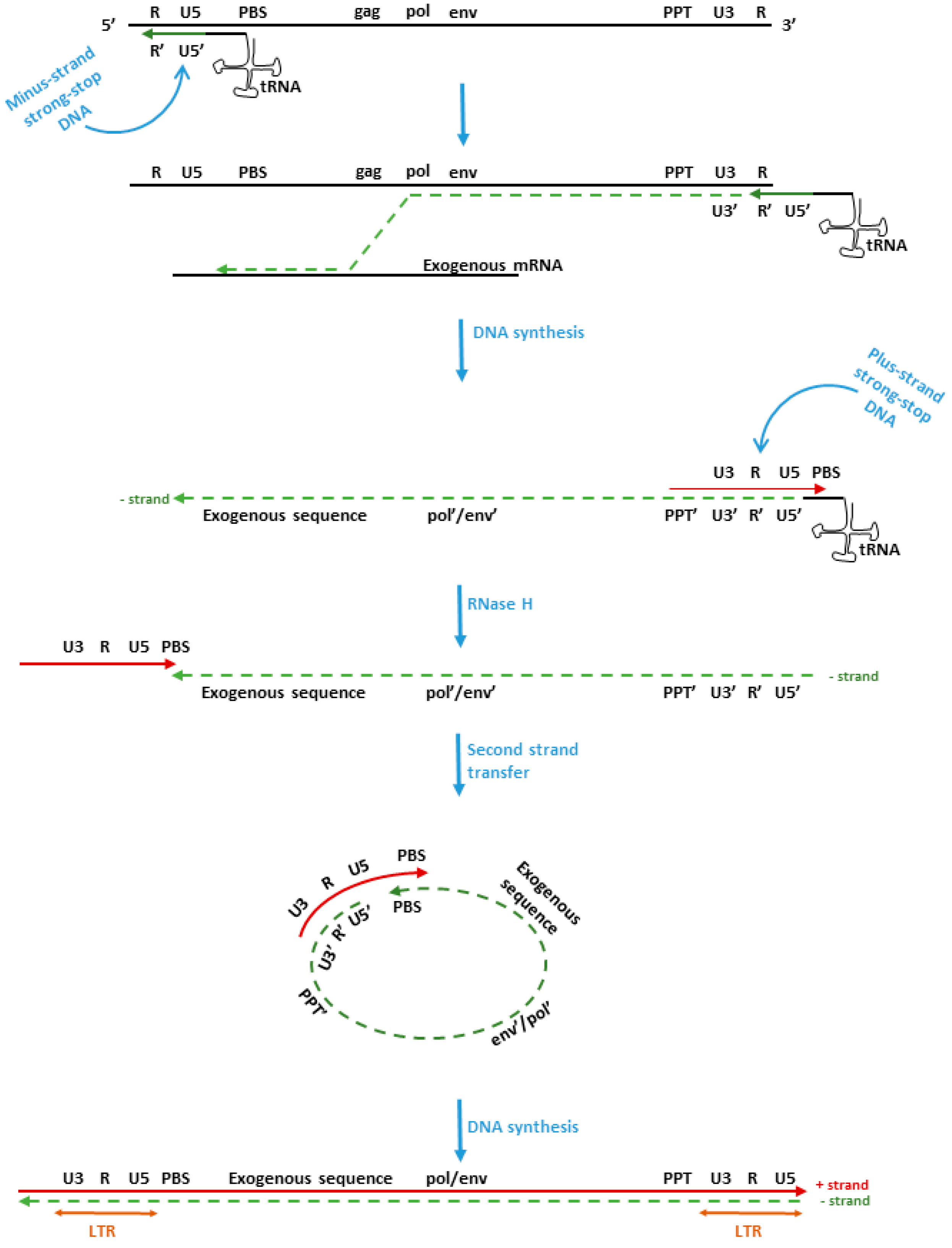
:1. Introduction
2. Adenovirus Vaccines
Adenovirus Vectors
3. mRNA Vaccines
- The speed of their development. In fact, it usually takes >10 years to develop a conventional vaccine. Conversely, the SARS-CoV-2 vaccine Moderna mRNA-1273 was prepared in only 42 days following the online availability of the SARS-CoV-2 spike-protein coding sequence in GenBank, and after 10 months, the vaccine was authorized for emergency use by the FDA [41];
- They are not infectious;
- The mRNA is rapidly delivered into the host cell cytoplasm by lipid nanoparticles (LNP). Shortly after protein translation, it is degraded by cellular enzymes;
- mRNA vaccines are able to induce both humoral and cell-mediated immunity, stimulating potent MHC-class-I- and MHC-class-II-restricted T-cell responses;
- The mRNA vaccine does not stimulate adaptive immune responses, thus no pre-existing immunity can interfere with the efficacy of the vaccine after the booster doses;
- Both mRNA and LNP have adjuvant properties.
- Generally, a linear plasmid or amplicon is used as a template for RNA synthesis. The most commonly used RNA-polymerases are T3, T7 or SP6. The translational efficiency is improved by codon optimization and nucleoside modification (generally pseudouridine replaces the uridine). The activation of TLR-3, -7 and -8 is abrogated by introducing pseudouridine in the mRNA or m5C, m6-A, m5U or s2U [42,43]. Furthermore, the presence of pseudouridine m6A and s2U in the mRNA vaccine molecules hampers the degradation of RNA by cellular RNAse [44]. In all COVID-19 mRNA vaccines, uridine has been replaced by pseudouridine (m6A and s2U);
- A 7-methylguanosine (m7G)5′ trisphosphate cap is added to the 5′ end to allow the recognition of the mRNA vaccine by cytoplasmic factors involved in the translation process [45]. This cap is able to eliminate free phosphate groups in the mRNA sequence, thus enhancing mRNA stability [45]. The 5′ cap represents a determining factor through which the host can discriminate between self- vs. non-self-mRNA. Moreover, anti-reverse cap analogs (ARCA) have been introduced to prevent the reverse incorporation of the 5′ cap [46,47]. ARCA is modified at the C2 or C3 positions to ensure that the methyl groups react with hydroxyl groups at the correct site during transcription, enhancing the translational efficiency [46,48]. Innovative protocols set up the addition of the 5′cap to a given start sequence during in vitro transcription [49,50]. Gene expression is enhanced by the untranslated (UTR) sequence addition to mRNA [51,52]. The 5′UTR shows a direct influence on translation of the downstream (sequence) open reading frame (ORF). Furthermore, some specific sequences can be added to the 5′UTR to strengthen the accuracy of translation and the stability of the mRNA [53,54]. The stability and the extension of the mRNA half-life are markedly increased by the 3′UTR sequence in the mRNA vaccine [55,56];
- Polyadenylation tail poly (a) reduces the degradation of mRNA mediated by the RNA exonuclease, guaranteeing a great efficiency in translation [57]. The length of poly (a) is not absolute, and it depends on the cellular milieu where the mRNA is translated. A poly (a) sequence of over 300 nucleotides in length may be more effective in ensuring mRNA expression in primary T cells, while small poly (a) (120–150 nt in length) sequences were found to be optimal for mRNA expression in dendritic cells [58]. A poly (a) sequence which is shorter than 20 nt reduces mRNA translational efficiency in all human cell types [59]. However, the information on the 5′ and 3′UTR and poly (a) composition of the SARS-CoV-2 mRNA vaccines remains undisclosed and the intellectual property of pharmaceutical companies;
- mRNA purification: the mRNA produced by in vitro transcription (IVT) should be purified before being incorporated into the LNP. In fact, the double-stranded RNA–RNA and DNA–RNA hybrid molecules can stimulate the innate immune response, which can weaken the effectiveness of the vaccine since the innate immune activation could provoke mRNA degradation and reduce the production of the immunogenic protein [60,61]. The main method employed for mRNA purification is high performance liquid chromatography (HPLC) [50,57]. In an alternative and cheaper method, the RNA–RNA hybrid molecules are adsorbed onto cellulose polysaccharide [61]. Nevertheless, the mRNA of COVID-19 vaccines is purified by HPLC.
Delivery System
4. Immunogenicity of mRNA and Adenoviral-Vectored Vaccines against SARS-CoV-2 in People without HIV/Healthy Individuals
4.1. Adenoviral-Vector-Based Vaccines
4.2. mRNA Vaccines
4.3. Safety
5. HIV and SARS-CoV-2
- The age of the patients. For example, in the United States, many HIV-positive patients are over 50 years of age with cardiovascular diseases;
- Obesity. This factor is certainly not negligible and it can worsen the course of the COVID-19 disease;
- Cardiovascular problems;
6. SARS-CoV-2 Vaccines in HIV-Positive People (PLWH)
6.1. AdV-Vector-Based Vaccines
6.2. mRNA Vaccines
7. Specific/Potential Side Effects of SARS-CoV-2 Vaccines in People Living with HIV (PLWH)
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard. Available online: https://covid19.who.int (accessed on 22 February 2022).
- HAdV Working Group. Available online: http://hadvwg.gmu.edu/ (accessed on 18 September 2021).
- Gao, J.; Mese, K.; Bunz, O.; Ehrhardt, A. State-of-the-art Human Adenovirus Vectorology for Therapeutic Approaches. FEBS Lett. 2019, 593, 3609–3622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crenshaw, B.J.; Jones, L.B.; Bell, C.R.; Kumar, S.; Matthews, Q.L. Perspective on Adenoviruses: Epidemiology, Pathogenicity, and Gene Therapy. Biomedicines 2019, 7, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B. New Vaccine Technologies to Combat Outbreak Situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zheng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-Mediated Gene Delivery: Potential Applications for Gene and Cell-Based Therapies in the New Era of Personalized Medicine. Genes Dis. 2017, 4, 43–63. [Google Scholar] [CrossRef]
- Zhang, Y.; Bergelson, J.M. Adenovirus Receptors. J. Virol. 2005, 79, 12125–12131. [Google Scholar] [CrossRef] [Green Version]
- Baker, A.T.; Greenshields-Watson, A.; Coughlan, L.; Davies, J.A.; Uusi-Kerttula, H.; Cole, D.K.; Rizkallah, P.J.; Parker, A.L. Diversity within the Adenovirus Fiber Knob Hypervariable Loops Influences Primary Receptor Interactions. Nat. Commun. 2019, 10, 741. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Tian, X. Vaccine Development for Human Mastadenovirus. J. Thorac. Dis. 2018, 10, S2280–S2294. [Google Scholar] [CrossRef]
- Jobran, S.; Kattan, R.; Shamaa, J.; Marzouqa, H.; Hindiyeh, M. Adenovirus Respiratory Tract Infections in Infants: A Retrospective Chart-Review Study. Lancet 2018, 391 (Suppl. 2), S43. [Google Scholar] [CrossRef]
- Radke, J.R.; Cook, J.L. Human Adenovirus Infections: Update and Consideration of Mechanisms of Viral Persistence. Curr. Opin. Infect. Dis. 2018, 31, 251–256. [Google Scholar] [CrossRef]
- Cook, J.; Radke, J. Mechanisms of Pathogenesis of Emerging Adenoviruses. F1000Research 2017, 6, 90. [Google Scholar] [CrossRef] [Green Version]
- Lamson, D.M.; Kajon, A.; Shudt, M.; Girouard, G.; St. George, K. Detection and Genetic Characterization of Adenovirus Type 14 Strain in Students with Influenza-Like Illness, New York, USA, 2014–2015. Emerg. Infect. Dis. 2017, 23, 1194–1197. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/mm5645a1.htm (accessed on 20 December 2021).
- Louie, J.K.; Kajon, A.E.; Holodniy, M.; Guardia-LaBar, L.; Lee, B.; Petru, A.M.; Hacker, J.K.; Schnurr, D.P. Severe Pneumonia Due to Adenovirus Serotype 14: A New Respiratory Threat? Clin. Infect. Dis. 2008, 46, 421–425. [Google Scholar] [CrossRef] [PubMed]
- de Mezerville, M.H.; Tellier, R.; Richardson, S.; Hébert, D.; Doyle, J.; Allen, U. Adenoviral Infections in Pediatric Transplant Recipients: A Hospital-Based Study. Pediatr. Infect. Dis. J. 2006, 25, 815–818. [Google Scholar] [CrossRef] [PubMed]
- Garnett, C.T.; Erdman, D.; Xu, W.; Gooding, L.R. Prevalence and Quantitation of Species C Adenovirus DNA in Human Mucosal Lymphocytes. J. Virol. 2002, 76, 10608–10616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.; Calcedo, R.; Medina-Jaszek, A.; Keough, M.; Peng, H.; Wilson, J.M. Adenoviruses in Lymphocytes of the Human Gastro-Intestinal Tract. PLoS ONE 2011, 6, e24859. [Google Scholar] [CrossRef] [PubMed]
- Kosulin, K.; Geiger, E.; Vécsei, A.; Huber, W.-D.; Rauch, M.; Brenner, E.; Wrba, F.; Hammer, K.; Innerhofer, A.; Pötschger, U.; et al. Persistence and Reactivation of Human Adenoviruses in the Gastrointestinal Tract. Clin. Microbiol. Infect. 2016, 22, e1–e381. [Google Scholar] [CrossRef] [Green Version]
- Tebruegge, M.; Curtis, N. Adenovirus: An Overview for Pediatric Infectious Diseases Specialists. Pediatr. Infect. Dis. J. 2012, 31, 626–627. [Google Scholar] [CrossRef]
- Adeyemi, O.A.; Yeldandi, A.V.; Ison, M.G. Fatal Adenovirus Pneumonia in a Person with AIDS and Burkitt Lymphoma: A Case Report and Review of the Literature. AIDS Read. 2008, 18, 196–198, 201–202, 206–207. [Google Scholar]
- Nebbia, G.; Chawla, A.; Schutten, M.; Atkinson, C.; Raza, M.; Johnson, M.; Geretti, A. Adenovirus Viraemia and Dissemination Unresponsive to Antiviral Therapy in Advanced HIV-1 Infection. AIDS 2005, 19, 1339–1340. [Google Scholar] [CrossRef]
- Heemskerk, B.; van Vreeswijk, T.; Veltrop-Duits, L.A.; Sombroek, C.C.; Franken, K.; Verhoosel, R.M.; Hiemstra, P.S.; van Leeuwen, D.; Ressing, M.E.; Toes, R.E.M.; et al. Adenovirus-Specific CD4 + T Cell Clones Recognizing Endogenous Antigen Inhibit Viral Replication In Vitro through Cognate Interaction. J. Immunol. 2006, 177, 8851–8859. [Google Scholar] [CrossRef] [Green Version]
- Shirali, G.S.; Ni, J.; Chinnock, R.E.; Johnston, J.K.; Rosenthal, G.L.; Bowles, N.E.; Towbin, J.A. Association of Viral Genome with Graft Loss in Children after Cardiac Transplantation. N. Engl. J. Med. 2001, 344, 1498–1503. [Google Scholar] [CrossRef] [PubMed]
- McGrath, D.; Falagas, M.E.; Freeman, R.; Rohrer, R.; Fairchild, R.; Colbach, C.; Snydman, D.R. Adenovirus Infection in Adult Orthotopic Liver Transplant Recipients: Incidence and Clinical Significance. J. Infect. Dis. 1998, 177, 459–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Öhrmalm, L.; Smedman, C.; Wong, M.; Broliden, K.; Tolfvenstam, T.; Norbeck, O. Decreased functional T lymphocyte-mediated cytokine responses in patients with chemotherapy-induced neutropenia. J. Intern. Med. 2013, 274, 363–370. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.; Foster, R.E.; Horgan, K.; Mounsey, K.; Nixon, H.; Smalle, N.; Hughes, T.A.; Carter, C.R.D. Lymphocyte Depletion and Repopulation after Chemotherapy for Primary Breast Cancer. Breast Cancer Res. 2016, 18, 10. [Google Scholar] [CrossRef] [Green Version]
- Peng, B.; Wang, L.R.; Gómez-Román, V.R.; Davis-Warren, A.; Montefiori, D.C.; Kalyanaraman, V.S.; Venzon, D.; Zhao, J.; Kan, E.; Rowell, T.J.; et al. Replicating Rather than Nonreplicating Adenovirus-Human Immunodeficiency Virus Recombinant Vaccines Are Better at Eliciting Potent Cellular Immunity and Priming High-Titer Antibodies. J. Virol. 2005, 79, 10200–10209. [Google Scholar] [CrossRef] [Green Version]
- Wold, W.; Toth, K. Adenovirus Vectors for Gene Therapy, Vaccination and Cancer Gene Therapy. Curr. Gene Ther. 2014, 13, 421–433. [Google Scholar] [CrossRef]
- Tan, W.G.; Jin, H.-T.; West, E.E.; Penaloza-MacMaster, P.; Wieland, A.; Zilliox, M.J.; McElrath, M.J.; Barouch, D.H.; Ahmed, R. Comparative Analysis of Simian Immunodeficiency Virus Gag-Specific Effector and Memory CD8 + T Cells Induced by Different Adenovirus Vectors. J. Virol. 2013, 87, 1359–1372. [Google Scholar] [CrossRef] [Green Version]
- Humphreys, I.R.; Sebastian, S. Novel Viral Vectors in Infectious Diseases. Immunology 2018, 153, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Mennechet, F.J.D.; Paris, O.; Ouoba, A.R.; Salazar Arenas, S.; Sirima, S.B.; Takoudjou Dzomo, G.R.; Diarra, A.; Traore, I.T.; Kania, D.; Eichholz, K.; et al. A Review of 65 Years of Human Adenovirus Seroprevalence. Expert Rev. Vaccines 2019, 18, 597–613. [Google Scholar] [CrossRef]
- Kostense, S.; Koudstaal, W.; Sprangers, M.; Weverling, G.J.; Penders, G.; Helmus, N.; Vogels, R.; Bakker, M.; Berkhout, B.; Havenga, M.; et al. Adenovirus Types 5 and 35 Seroprevalence in AIDS Risk Groups Supports Type 35 as a Vaccine Vector. AIDS 2004, 18, 1213–1216. [Google Scholar] [CrossRef]
- Feng, L.; Wang, Q.; Shan, C.; Yang, C.; Feng, Y.; Wu, J.; Liu, X.; Zhou, Y.; Jiang, R.; Hu, P.; et al. An Adenovirus-Vectored COVID-19 Vaccine Confers Protection from SARS-COV-2 Challenge in Rhesus Macaques. Nat. Commun. 2020, 11, 4207. [Google Scholar] [CrossRef]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit Vaccines Against Emerging Pathogenic Human Coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.-C.; Li, Y.-H.; Guan, X.-H.; Hou, L.-H.; Wang, W.-J.; Li, J.-X.; Wu, S.-P.; Wang, B.-S.; Wang, Z.; Wang, L.; et al. Safety, Tolerability, and Immunogenicity of a Recombinant Adenovirus Type-5 Vectored COVID-19 Vaccine: A Dose-Escalation, Open-Label, Non-Randomised, First-in-Human Trial. Lancet 2020, 395, 1845–1854. [Google Scholar] [CrossRef]
- Zhu, F.-C.; Guan, X.-H.; Li, Y.-H.; Huang, J.-Y.; Jiang, T.; Hou, L.-H.; Li, J.-X.; Yang, B.-F.; Wang, L.; Wang, W.-J.; et al. Immunogenicity and Safety of a Recombinant Adenovirus Type-5-Vectored COVID-19 Vaccine in Healthy Adults Aged 18 Years or Older: A Randomised, Double-Blind, Placebo-Controlled, Phase 2 Trial. Lancet 2020, 396, 479–488. [Google Scholar] [CrossRef]
- Sadoff, J.; Le Gars, M.; Shukarev, G.; Heerwegh, D.; Truyers, C.; de Groot, A.M.; Stoop, J.; Tete, S.; Van Damme, W.; Leroux-Roels, I.; et al. Interim Results of a Phase 1-2a Trial of Ad26.COV2.S Covid-19 Vaccine. N. Engl. J. Med. 2021, 384, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Logunov, D.Y.; Dolzhikova, I.V.; Zubkova, O.V.; Tukhvatulin, A.I.; Shcheblyakov, D.V.; Dzharullaeva, A.S.; Grousova, D.M.; Erokhova, A.S.; Kovyrshina, A.V.; Botikov, A.G.; et al. Safety and Immunogenicity of an RAd26 and RAd5 Vector-Based Heterologous Prime-Boost COVID-19 Vaccine in Two Formulations: Two Open, Non-Randomised Phase 1/2 Studies from Russia. Lancet 2020, 396, 887–897. [Google Scholar] [CrossRef]
- Voysey, M.; Clemens, S.A.C.; Madhi, S.A.; Weckx, L.Y.; Folegatti, P.M.; Aley, P.K.; Angus, B.; Baillie, V.L.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and Efficacy of the ChAdOx1 NCoV-19 Vaccine (AZD1222) against SARS-CoV-2: An Interim Analysis of Four Randomised Controlled Trials in Brazil, South Africa, and the UK. Lancet 2021, 397, 99–111. [Google Scholar] [CrossRef]
- Hodgson, J. The Pandemic Pipeline. Nat. Biotechnol. 2020, 38, 523–532. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA Recognition by Toll-like Receptors: The Impact of Nucleoside Modification and the Evolutionary Origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [Green Version]
- Karikó, K.; Muramatsu, H.; Welsh, F.A.; Ludwig, J.; Kato, H.; Akira, S.; Weissman, D. Incorporation of Pseudouridine Into MRNA Yields Superior Nonimmunogenic Vector With Increased Translational Capacity and Biological Stability. Mol. Ther. 2008, 16, 1833–1840. [Google Scholar] [CrossRef]
- Anderson, B.R.; Muramatsu, H.; Jha, B.K.; Silverman, R.H.; Weissman, D.; Kariko, K. Nucleoside Modifications in RNA Limit Activation of 2’-5’-Oligoadenylate Synthetase and Increase Resistance to Cleavage by RNase L. Nucleic Acids Res. 2011, 39, 9329–9338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarkar, S.C.; Wang, C.; Miller, M.T.; Ramanathan, A.; Jiang, F.; Khan, A.G.; Patel, S.S.; Marcotrigiano, J. Structural Basis for M7G Recognition and 2′-O-Methyl Discrimination in Capped RNAs by the Innate Immune Receptor RIG-I. Proc. Natl. Acad. Sci. USA 2016, 113, 596–601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stepinski, J.; Waddell, C.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Synthesis and Properties of MRNAs Containing the Novel “Anti-Reverse” Cap Analogs 7-Methyl(3′-O-Methyl)GpppG and 7-Methyl (3′-Deoxy)GpppG. RNA 2001, 7, 1486–1495. [Google Scholar] [PubMed]
- Jemielity, J.; Fowler, T.; Zuberek, J.; Stepinski, J.; Lewdorowicz, M.; Niedzwiecka, A.; Stolarski, R.; Darzynkiewicz, E.; Rhoads, R.E. Novel “Anti-Reverse” Cap Analogs with Superior Translational Properties. RNA 2003, 9, 1108–1122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grudzien-Nogalska, E.; Stepinski, J.; Jemielity, J.; Zuberek, J.; Stolarski, R.; Rhoads, R.E.; Darzynkiewicz, E. Synthesis of Anti-Reverse Cap Analogs (ARCAs) and Their Applications in MRNA Translation and Stability. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2007; Volume 431, pp. 203–227. ISBN 978-0-12-373964-3. [Google Scholar]
- Vaidyanathan, S.; Azizian, K.T.; Haque, A.K.M.A.; Henderson, J.M.; Hendel, A.; Shore, S.; Antony, J.S.; Hogrefe, R.I.; Kormann, M.S.D.; Porteus, M.H.; et al. Uridine Depletion and Chemical Modification Increase Cas9 MRNA Activity and Reduce Immunogenicity without HPLC Purification. Mol. Ther.—Nucleic Acids 2018, 12, 530–542. [Google Scholar] [CrossRef]
- Pardi, N.; Hogan, M.J.; Weissman, D. Recent Advances in MRNA Vaccine Technology. Curr. Opin. Immunol. 2020, 65, 14–20. [Google Scholar] [CrossRef]
- Leppek, K.; Das, R.; Barna, M. Functional 5’ UTR MRNA Structures in Eukaryotic Translation Regulation and How to Find Them. Nat. Rev. Mol. Cell Biol. 2018, 19, 158–174. [Google Scholar] [CrossRef]
- Tanguay, R.L.; Gallie, D.R. Translational Efficiency Is Regulated by the Length of the 3′ Untranslated Region. Mol. Cell. Biol. 1996, 16, 146–156. [Google Scholar] [CrossRef] [Green Version]
- Gray, N.K.; Wickens, M. Control of translation initiation in animals. Annu. Rev. Cell Dev. Biol. 1998, 14, 399–458. [Google Scholar] [CrossRef] [Green Version]
- Kozak, M. At Least Six Nucleotides Preceding the AUG Initiator Codon Enhance Translation in Mammalian Cells. J. Mol. Biol. 1987, 196, 947–950. [Google Scholar] [CrossRef]
- Ferizi, M.; Leonhardt, C.; Meggle, C.; Aneja, M.K.; Rudolph, C.; Plank, C.; Rädler, J.O. Stability Analysis of Chemically Modified MRNA Using Micropattern-Based Single-Cell Arrays. Lab Chip 2015, 15, 3561–3571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orlandini von Niessen, A.G.; Poleganov, M.A.; Rechner, C.; Plaschke, A.; Kranz, L.M.; Fesser, S.; Diken, M.; Löwer, M.; Vallazza, B.; Beissert, T.; et al. Improving MRNA-Based Therapeutic Gene Delivery by Expression-Augmenting 3′ UTRs Identified by Cellular Library Screening. Mol. Ther. 2019, 27, 824–836. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linares-Fernández, S.; Lacroix, C.; Exposito, J.-Y.; Verrier, B. Tailoring MRNA Vaccine to Balance Innate/Adaptive Immune Response. Trends Mol. Med. 2020, 26, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Grier, A.E.; Burleigh, S.; Sahni, J.; Clough, C.A.; Cardot, V.; Choe, D.C.; Krutein, M.C.; Rawlings, D.J.; Jensen, M.C.; Scharenberg, A.M.; et al. PEVL: A Linear Plasmid for Generating MRNA IVT Templates With Extended Encoded Poly(A) Sequences. Mol. Ther.—Nucleic Acids 2016, 5, e306. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Yi, H.; Kim, Y.; Chang, H.; Kim, V.N. Regulation of Poly(A) Tail and Translation during the Somatic Cell Cycle. Mol. Cell 2016, 62, 462–471. [Google Scholar] [CrossRef] [Green Version]
- Karikó, K.; Muramatsu, H.; Ludwig, J.; Weissman, D. Generating the Optimal MRNA for Therapy: HPLC Purification Eliminates Immune Activation and Improves Translation of Nucleoside-Modified, Protein-Encoding MRNA. Nucleic Acids Res. 2011, 39, e142. [Google Scholar] [CrossRef] [Green Version]
- Baiersdörfer, M.; Boros, G.; Muramatsu, H.; Mahiny, A.; Vlatkovic, I.; Sahin, U.; Karikó, K. A Facile Method for the Removal of DsRNA Contaminant from In Vitro-Transcribed MRNA. Mol. Ther.—Nucleic Acids 2019, 15, 26–35. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Tuyishime, S.; Muramatsu, H.; Kariko, K.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; Hope, M.J.; Weissman, D. Expression Kinetics of Nucleoside-Modified MRNA Delivered in Lipid Nanoparticles to Mice by Various Routes. J. Control. Release Off. J. Control. Release Soc. 2015, 217, 345–351. [Google Scholar] [CrossRef] [Green Version]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. MRNA Vaccines—A New Era in Vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Semple, S.C.; Akinc, A.; Chen, J.; Sandhu, A.P.; Mui, B.L.; Cho, C.K.; Sah, D.W.Y.; Stebbing, D.; Crosley, E.J.; Yaworski, E.; et al. Rational Design of Cationic Lipids for SiRNA Delivery. Nat. Biotechnol. 2010, 28, 172–176. [Google Scholar] [CrossRef]
- Kowalski, P.S.; Rudra, A.; Miao, L.; Anderson, D.G. Delivering the Messenger: Advances in Technologies for Therapeutic MRNA Delivery. Mol. Ther. 2019, 27, 710–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lonez, C.; Vandenbranden, M.; Ruysschaert, J.-M. Cationic Lipids Activate Intracellular Signaling Pathways. Adv. Drug Deliv. Rev. 2012, 64, 1749–1758. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Folegatti, P.M.; Ewer, K.J.; Aley, P.K.; Angus, B.; Becker, S.; Belij-Rammerstorfer, S.; Bellamy, D.; Bibi, S.; Bittaye, M.; Clutterbuck, E.A.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 Vaccine against SARS-CoV-2: A Preliminary Report of a Phase 1/2, Single-Blind, Randomised Controlled Trial. Lancet 2020, 396, 467–478. [Google Scholar] [CrossRef]
- Ewer, K.J.; Barrett, J.R.; Belij-Rammerstorfer, S.; Sharpe, H.; Makinson, R.; Morter, R.; Flaxman, A.; Wright, D.; Bellamy, D.; Bittaye, M.; et al. T Cell and Antibody Responses Induced by a Single Dose of ChAdOx1 NCoV-19 (AZD1222) Vaccine in a Phase 1/2 Clinical Trial. Nat. Med. 2021, 27, 270–278. [Google Scholar] [CrossRef]
- Barrett, J.R.; Belij-Rammerstorfer, S.; Dold, C.; Ewer, K.J.; Folegatti, P.M.; Gilbride, C.; Halkerston, R.; Hill, J.; Jenkin, D.; Stockdale, L.; et al. Phase 1/2 Trial of SARS-CoV-2 Vaccine ChAdOx1 NCoV-19 with a Booster Dose Induces Multifunctional Antibody Responses. Nat. Med. 2021, 27, 279–288. [Google Scholar] [CrossRef]
- Ramasamy, M.N.; Minassian, A.M.; Ewer, K.J.; Flaxman, A.L.; Folegatti, P.M.; Owens, D.R.; Voysey, M.; Aley, P.K.; Angus, B.; Babbage, G.; et al. Safety and Immunogenicity of ChAdOx1 NCoV-19 Vaccine Administered in a Prime-Boost Regimen in Young and Old Adults (COV002): A Single-Blind, Randomised, Controlled, Phase 2/3 Trial. Lancet 2021, 396, 1979–1993. [Google Scholar] [CrossRef]
- Walsh, E.E.; Frenck, R.W.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020, 383, 2439–2450. [Google Scholar] [CrossRef]
- Sahin, U.; Muik, A.; Vogler, I.; Derhovanessian, E.; Kranz, L.M.; Vormehr, M.; Quandt, J.; Bidmon, N.; Ulges, A.; Baum, A.; et al. BNT162b2 Vaccine Induces Neutralizing Antibodies and Poly-Specific T Cells in Humans. Nature 2021, 595, 572–577. [Google Scholar] [CrossRef]
- Frenck, R.W.; Klein, N.P.; Kitchin, N.; Gurtman, A.; Absalon, J.; Lockhart, S.; Perez, J.L.; Walter, E.B.; Senders, S.; Bailey, R.; et al. Safety, Immunogenicity, and Efficacy of the BNT162b2 Covid-19 Vaccine in Adolescents. N. Engl. J. Med. 2021, 385, 239–250. [Google Scholar] [CrossRef]
- Jackson, L.A.; Anderson, E.J.; Rouphael, N.G.; Roberts, P.C.; Makhene, M.; Coler, R.N.; McCullough, M.P.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; et al. An MRNA Vaccine against SARS-CoV-2—Preliminary Report. N. Engl. J. Med. 2020, 383, 1920–1931. [Google Scholar] [CrossRef] [PubMed]
- Anderson, E.J.; Rouphael, N.G.; Widge, A.T.; Jackson, L.A.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Safety and Immunogenicity of SARS-CoV-2 MRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020, 383, 2427–2438. [Google Scholar] [CrossRef] [PubMed]
- Widge, A.T.; Rouphael, N.G.; Jackson, L.A.; Anderson, E.J.; Roberts, P.C.; Makhene, M.; Chappell, J.D.; Denison, M.R.; Stevens, L.J.; Pruijssers, A.J.; et al. Durability of Responses after SARS-CoV-2 MRNA-1273 Vaccination. N. Engl. J. Med. 2021, 384, 80–82. [Google Scholar] [CrossRef]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 MRNA Covid-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef]
- Kircheis, R. Coagulopathies after Vaccination against SARS-CoV-2 May Be Derived from a Combined Effect of SARS-CoV-2 Spike Protein and Adenovirus Vector-Triggered Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 10791. [Google Scholar] [CrossRef] [PubMed]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 NCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.H.; Sørvoll, I.H.; Michelsen, A.E.; Munthe, L.A.; Lund-Johansen, F.; Ahlen, M.T.; Wiedmann, M.; Aamodt, A.-H.; Skattør, T.H.; Tjønnfjord, G.E.; et al. Thrombosis and Thrombocytopenia after ChAdOx1 NCoV-19 Vaccination. N. Engl. J. Med. 2021, 384, 2124–2130. [Google Scholar] [CrossRef]
- Scully, M.; Singh, D.; Lown, R.; Poles, A.; Solomon, T.; Levi, M.; Goldblatt, D.; Kotoucek, P.; Thomas, W.; Lester, W. Pathologic Antibodies to Platelet Factor 4 after ChAdOx1 NCoV-19 Vaccination. N. Engl. J. Med. 2021, 384, 2202–2211. [Google Scholar] [CrossRef]
- Anand, P.; Stahel, V.P. The Safety of Covid-19 MRNA Vaccines: A Review. Patient Saf. Surg. 2021, 15, 20. [Google Scholar] [CrossRef]
- Blanco, J.L.; Ambrosioni, J.; Garcia, F.; Martínez, E.; Soriano, A.; Mallolas, J.; Miro, J.M. COVID-19 in Patients with HIV: Clinical Case Series. Lancet HIV 2020, 7, e314–e316. [Google Scholar] [CrossRef]
- Okoh, A.K.; Sossou, C.; Dangayach, N.S.; Meledathu, S.; Phillips, O.; Raczek, C.; Patti, M.; Kang, N.; Hirji, S.A.; Cathcart, C.; et al. Coronavirus Disease 19 in Minority Populations of Newark, New Jersey. Int. J. Equity Health 2020, 19, 93. [Google Scholar] [CrossRef] [PubMed]
- Cooper, T.; Woodward, B.; Alom, S.; Harky, A. Coronavirus Disease 2019 (COVID-19) Outcomes in HIV/AIDS Patients: A Systematic Review. HIV Med. 2020, 21, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Bhaskaran, K.; Rentsch, C.T.; MacKenna, B.; Schultze, A.; Mehrkar, A.; Bates, C.J.; Eggo, R.M.; Morton, C.E.; Bacon, S.C.J.; Inglesby, P.; et al. HIV infection and COVID-19 death: A population-based cohort analysis of UK primary care data and linked national death registrations within the OpenSAFELY platform. Lancet HIV 2021, 8, e24–e32. [Google Scholar] [CrossRef]
- Vizcarra, P.; Pérez-Elías, M.J.; Quereda, C.; Moreno, A.; Vivancos, M.J.; Dronda, F.; Casado, J.L.; Moreno, S.; Pérez-Elías, M.J.; Fortún, J.; et al. Description of COVID-19 in HIV-Infected Individuals: A Single-Centre, Prospective Cohort. Lancet HIV 2020, 7, e554–e564. [Google Scholar] [CrossRef]
- Dandachi, D.; Geiger, G.; Montgomery, M.W.; Karmen-Tuohy, S.; Golzy, M.; Antar, A.A.R.; Llibre, J.M.; Camazine, M.; Díaz-De Santiago, A.; Carlucci, P.M.; et al. Characteristics, Comorbidities, and Outcomes in a Multicenter Registry of Patients With Human Immunodeficiency Virus and Coronavirus Disease 2019. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, e1964–e1972. [Google Scholar] [CrossRef]
- Nomah, D.K.; Reyes-Urueña, J.; Díaz, Y.; Moreno, S.; Aceiton, J.; Bruguera, A.; Vivanco-Hidalgo, R.M.; Llibre, J.M.; Domingo, P.; Falcó, V.; et al. Sociodemographic, Clinical, and Immunological Factors Associated with SARS-CoV-2 Diagnosis and Severe COVID-19 Outcomes in People Living with HIV: A Retrospective Cohort Study. Lancet HIV 2021, 8, e701–e710. [Google Scholar] [CrossRef]
- Vaughn, V.M.; Gandhi, T.N.; Petty, L.A.; Patel, P.K.; Prescott, H.C.; Malani, A.N.; Ratz, D.; McLaughlin, E.; Chopra, V.; Flanders, S.A. Empiric Antibacterial Therapy and Community-Onset Bacterial Coinfection in Patients Hospitalized With Coronavirus Disease 2019 (COVID-19): A Multi-Hospital Cohort Study. Clin. Infect. Dis. 2021, 72, e533–e541. [Google Scholar] [CrossRef]
- Nagarakanti, S.R.; Okoh, A.K.; Grinberg, S.; Bishburg, E. Clinical Outcomes of Patients with COVID-19 and HIV Coinfection. J. Med. Virol. 2021, 93, 1687–1693. [Google Scholar] [CrossRef]
- Wang, M.; Luo, L.; Bu, H.; Xia, H. One Case of Coronavirus Disease 2019 (COVID-19) in a Patient Co-Infected by HIV with a Low CD4+ T-Cell Count. Int. J. Infect. Dis. 2020, 96, 148–150. [Google Scholar] [CrossRef]
- Western Cape Department of Health in Collaboration with the National Institute for Communicable Diseases, South Africa. Risk Factors for Coronavirus Disease 2019 (COVID-19) Death in a Population Cohort Study from the Western Cape Province, South Africa. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, e2005–e2015. [Google Scholar] [CrossRef]
- Jacobs, G.P.; Bhat, P.; Owiti, P.; Edwards, J.K.; Tweya, H.; Najjemba, R. Did the 2014 Ebola Outbreak in Liberia Affect HIV Testing, Linkage to Care and ART Initiation? Public Health Action 2017, 7, S70–S75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Triant, V.A. Cardiovascular Disease and HIV Infection. Curr. HIV/AIDS Rep. 2013, 10, 199–206. [Google Scholar] [CrossRef] [PubMed]
- Gervasoni, C.; Meraviglia, P.; Riva, A.; Giacomelli, A.; Oreni, L.; Minisci, D.; Atzori, C.; Ridolfo, A.; Cattaneo, D. Clinical Features and Outcomes of Patients With Human Immunodeficiency Virus With COVID-19. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2020, 71, 2276–2278. [Google Scholar] [CrossRef] [PubMed]
- Byrd, K.M.; Beckwith, C.G.; Garland, J.M.; Johnson, J.E.; Aung, S.; Cu-Uvin, S.; Farmakiotis, D.; Flanigan, T.; Gillani, F.S.; Macias-Gil, R.; et al. SARS-CoV-2 and HIV Coinfection: Clinical Experience from Rhode Island, United States. J. Int. AIDS Soc. 2020, 23, e25573. [Google Scholar] [CrossRef]
- Calza, L.; Colangeli, V.; Borderi, M.; Bon, I.; Borioni, A.; Volpato, F.; Re, M.C.; Viale, P. Weight Gain in Antiretroviral Therapy-Naive HIV-1-Infected Patients Starting a Regimen Including an Integrase Strand Transfer Inhibitor or Darunavir/Ritonavir. Infection 2020, 48, 213–221. [Google Scholar] [CrossRef]
- Guo, W.; Ming, F.; Feng, Y.; Zhang, Q.; Mo, P.; Liu, L.; Gao, M.; Tang, W.; Liang, K. Patterns of HIV and SARS-CoV-2 Co-infection in Wuhan, China. J. Int. AIDS Soc. 2020, 23, e25568. [Google Scholar] [CrossRef]
- Gudipati, S.; Brar, I.; Murray, S.; McKinnon, J.E.; Yared, N.; Markowitz, N. Descriptive Analysis of Patients Living With HIV Affected by COVID-19. J. Acquir. Immune Defic. Syndr. 2020, 85, 123–126. [Google Scholar] [CrossRef]
- Meyerowitz, E.A.; Kim, A.Y.; Ard, K.L.; Basgoz, N.; Chu, J.T.; Hurtado, R.M.; Lee, C.K.; He, W.; Minukas, T.; Nelson, S.; et al. Disproportionate Burden of Coronavirus Disease 2019 among Racial Minorities and Those in Congregate Settings among a Large Cohort of People with HIV. AIDS 2020, 34, 1781–1787. [Google Scholar] [CrossRef]
- Yang, X.; Sun, J.; Patel, R.C.; Zhang, J.; Guo, S.; Zheng, Q.; Olex, A.L.; Olatosi, B.; Weissman, S.B.; Islam, J.Y.; et al. Associations between HIV Infection and Clinical Spectrum of COVID-19: A Population Level Analysis Based on US National COVID Cohort Collaborative (N3C) Data. Lancet HIV 2021, 8, e690–e700. [Google Scholar] [CrossRef]
- Use of COVID-19 Vaccines in the United States. Available online: https://www.cdc.gov/vaccines/covid-19/clinical-considerations/immunocompromised.html (accessed on 16 November 2021).
- Frater, J.; Ewer, K.J.; Ogbe, A.; Pace, M.; Adele, S.; Adland, E.; Alagaratnam, J.; Aley, P.K.; Ali, M.; Ansari, M.A.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 (AZD1222) Vaccine against SARS-CoV-2 in HIV Infection: A Single-Arm Substudy of a Phase 2/3 Clinical Trial. Lancet HIV 2021, 8, e474–e485. [Google Scholar] [CrossRef]
- Access to Antiretroviral Therapy in Africa. Available online: https://www.unaids.org/sites/default/files/media_asset/20131219_AccessARTAfricaStatusReportProgresstowards2015Targets_en_0.pdf (accessed on 12 December 2021).
- Madhi, S.A.; Koen, A.L.; Izu, A.; Fairlie, L.; Cutland, C.L.; Baillie, V.; Padayachee, S.D.; Dheda, K.; Barnabas, S.L.; Bhorat, Q.E.; et al. Safety and Immunogenicity of the ChAdOx1 NCoV-19 (AZD1222) Vaccine against SARS-CoV-2 in People Living with and without HIV in South Africa: An Interim Analysis of a Randomised, Double-Blind, Placebo-Controlled, Phase 1B/2A Trial. Lancet HIV 2021, 8, e568–e580. [Google Scholar] [CrossRef]
- Ruddy, J.A.; Boyarsky, B.J.; Werbel, W.A.; Bailey, J.R.; Karaba, A.H.; Garonzik-Wang, J.M.; Segev, D.L.; Durand, C.M. Safety and Antibody Response to the First Dose of Severe Acute Respiratory Syndrome Coronavirus 2 Messenger RNA Vaccine in Persons with HIV. AIDS 2021, 35, 1872–1874. [Google Scholar] [CrossRef] [PubMed]
- Ruddy, J.A.; Boyarsky, B.J.; Bailey, J.R.; Karaba, A.H.; Garonzik-Wang, J.M.; Segev, D.L.; Durand, C.M.; Werbel, W.A. Safety and Antibody Response to Two-Dose SARS-CoV-2 Messenger RNA Vaccination in Persons with HIV. AIDS 2021, 35, 2399–2401. [Google Scholar] [CrossRef] [PubMed]
- Levy, I.; Wieder-Finesod, A.; Litchevsky, V.; Biber, A.; Indenbaum, V.; Olmer, L.; Huppert, A.; Mor, O.; Goldstein, M.; Levin, E.G.; et al. Immunogenicity and Safety of the BNT162b2 MRNA COVID-19 Vaccine in People Living with HIV-1. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2021, 27, 1851–1855. [Google Scholar] [CrossRef]
- Sklar, P.A.; Ward, D.J.; Baker, R.K.; Wood, K.C.; Gafoor, Z.; Alzola, C.F.; Moorman, A.C.; Holmberg, S.D.; HIV Outpatient Study (HOPS) Investigators. Prevalence and Clinical Correlates of HIV Viremia (‘blips’) in Patients with Previous Suppression below the Limits of Quantification. AIDS 2002, 16, 2035–2041. [Google Scholar] [CrossRef]
- Jedicke, N.; Stankov, M.V.; Cossmann, A.; Dopfer-Jablonka, A.; Knuth, C.; Ahrenstorf, G.; Ramos, G.M.; Behrens, G.M.N. Humoral Immune Response Following Prime and Boost BNT162b2 Vaccination in People Living with HIV on Antiretroviral Therapy. HIV Med. 2021. [Google Scholar] [CrossRef]
- Xu, X.; Vesterbacka, J.; Aleman, S.; Nowak, P.; COVAXID Study Group. High Seroconversion Rate after Vaccination with MRNA BNT162b2 Vaccine against SARS-CoV-2 among People with HIV—but HIV Viremia Matters? AIDS 2022, 36, 479–481. [Google Scholar] [CrossRef]
- Woldemeskel, B.A.; Karaba, A.H.; Garliss, C.C.; Beck, E.J.; Wang, K.H.; Laeyendecker, O.; Cox, A.L.; Blankson, J.N. The BNT162b2 MRNA Vaccine Elicits Robust Humoral and Cellular Immune Responses in People Living with HIV. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, ciab648. [Google Scholar] [CrossRef]
- Lombardi, A.; Butta, G.M.; Donnici, L.; Bozzi, G.; Oggioni, M.; Bono, P.; Matera, M.; Consonni, D.; Ludovisi, S.; Muscatello, A.; et al. Anti-Spike Antibodies and Neutralising Antibody Activity in People Living with HIV Vaccinated with COVID-19 MRNA-1273 Vaccine: A Prospective Single-Centre Cohort Study. Lancet Reg. Health Eur. 2022, 13, 100287. [Google Scholar] [CrossRef]
- Tombácz, I.; Weissman, D.; Pardi, N. Vaccination with Messenger RNA: A Promising Alternative to DNA Vaccination. Methods Mol. Biol. 2021, 2197, 13–31. [Google Scholar] [CrossRef]
- Varmus, H.; Brown, P. Retroviruses. In Mobile DNA; Berg, D.E., Howe, M.M., Eds.; American Society for Microbiology: Washington, DC, USA, 1989; pp. 53–108. [Google Scholar]
- Hwang, C.K.; Svarovskaia, E.S.; Pathak, V.K. Dynamic Copy Choice: Steady State between Murine Leukemia Virus Polymerase and Polymerase-Dependent RNase H Activity Determines Frequency of in Vivo Template Switching. Proc. Natl. Acad. Sci. USA 2001, 98, 12209–12214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hajjar, A.M.; Linial, M.L. A Model System for Nonhomologous Recombination between Retroviral and Cellular RNA. J. Virol. 1993, 67, 3845–3853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, P.C.Y.; Lau, S.K.P.; Yip, C.C.Y.; Huang, Y.; Tsoi, H.-W.; Chan, K.-H.; Yuen, K.-Y. Comparative Analysis of 22 Coronavirus HKU1 Genomes Reveals a Novel Genotype and Evidence of Natural Recombination in Coronavirus HKU1. J. Virol. 2006, 80, 7136–7145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woo, P.C.; Lau, S.K.; Yuen, K. Infectious Diseases Emerging from Chinese Wet-Markets: Zoonotic Origins of Severe Respiratory Viral Infections. Curr. Opin. Infect. Dis. 2006, 19, 401–407. [Google Scholar] [CrossRef] [PubMed]
- Woo, P.C.Y.; Lau, S.K.P.; Huang, Y.; Yuen, K.-Y. Coronavirus Diversity, Phylogeny and Interspecies Jumping. Exp. Biol. Med. 2009, 234, 1117–1127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobay, L.-M.; O’Donnell, A.C.; Ochman, H. Recombination Events Are Concentrated in the Spike Protein Region of Betacoronaviruses. PLoS Genet. 2020, 16, e1009272. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Woo, P.C.Y.; Yip, C.C.Y.; Tse, H.; Tsoi, H.; Cheng, V.C.C.; Lee, P.; Tang, B.S.F.; Cheung, C.H.Y.; Lee, R.A.; et al. Coronavirus HKU1 and Other Coronavirus Infections in Hong Kong. J. Clin. Microbiol. 2006, 44, 2063–2071. [Google Scholar] [CrossRef] [Green Version]
- Ebner, K.; Pinsker, W.; Lion, T. Comparative Sequence Analysis of the Hexon Gene in the Entire Spectrum of Human Adenovirus Serotypes: Phylogenetic, Taxonomic, and Clinical Implications. J. Virol. 2005, 79, 12635–12642. [Google Scholar] [CrossRef] [Green Version]
- Doerfler, W. A New Concept in (Adenoviral) Oncogenesis: Integration of Foreign DNA and Its Consequences. Biochim. Biophys. Acta 1996, 1288, F79–F99. [Google Scholar] [CrossRef]
- Buchbinder, S.P.; McElrath, M.J.; Dieffenbach, C.; Corey, L. Use of Adenovirus Type-5 Vectored Vaccines: A Cautionary Tale. Lancet 2020, 396, e68–e69. [Google Scholar] [CrossRef]
- Auclair, S.; Liu, F.; Niu, Q.; Hou, W.; Churchyard, G.; Morgan, C.; Frahm, N.; Nitayaphan, S.; Pitisuthithum, P.; Rerks-Ngarm, S.; et al. Distinct Susceptibility of HIV Vaccine Vector-Induced CD4 T Cells to HIV Infection. PLoS Pathog. 2018, 14, e1006888. [Google Scholar] [CrossRef] [PubMed]
- Vallée, A.; Fourn, E.; Majerholc, C.; Touche, P.; Zucman, D. COVID-19 Vaccine Hesitancy among French People Living with HIV. Vaccines 2021, 9, 302. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garbuglia, A.R.; Minosse, C.; Del Porto, P. mRNA- and Adenovirus-Based Vaccines against SARS-CoV-2 in HIV-Positive People. Viruses 2022, 14, 748. https://doi.org/10.3390/v14040748
Garbuglia AR, Minosse C, Del Porto P. mRNA- and Adenovirus-Based Vaccines against SARS-CoV-2 in HIV-Positive People. Viruses. 2022; 14(4):748. https://doi.org/10.3390/v14040748
Chicago/Turabian StyleGarbuglia, Anna Rosa, Claudia Minosse, and Paola Del Porto. 2022. "mRNA- and Adenovirus-Based Vaccines against SARS-CoV-2 in HIV-Positive People" Viruses 14, no. 4: 748. https://doi.org/10.3390/v14040748
APA StyleGarbuglia, A. R., Minosse, C., & Del Porto, P. (2022). mRNA- and Adenovirus-Based Vaccines against SARS-CoV-2 in HIV-Positive People. Viruses, 14(4), 748. https://doi.org/10.3390/v14040748






