PRRSV Induces HMGB1 Phosphorylation at Threonine-51 Residue to Enhance Its Secretion
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Viruses, and Chemicals
2.2. Plasmids and siRNA
2.3. Co-Immunoprecipitation (co-IP)
2.4. Immunoblotting
2.5. Immunofluorescence Assay (IFA)
2.6. RNA Isolation and Real-Time PCR
2.7. Subcellular Fractionation
2.8. Statistical Analysis
3. Results
3.1. PRRSV Infection Induces HMGB1 Phosphorylation at Threonine Residues
3.2. HMGB1 Phosphorylation at Threonine-51 Is Associated with the HMGB1 Secretion Induced by PRRSV Infection
3.3. PRRSV Enhances HMGB1 Interaction with RPS3
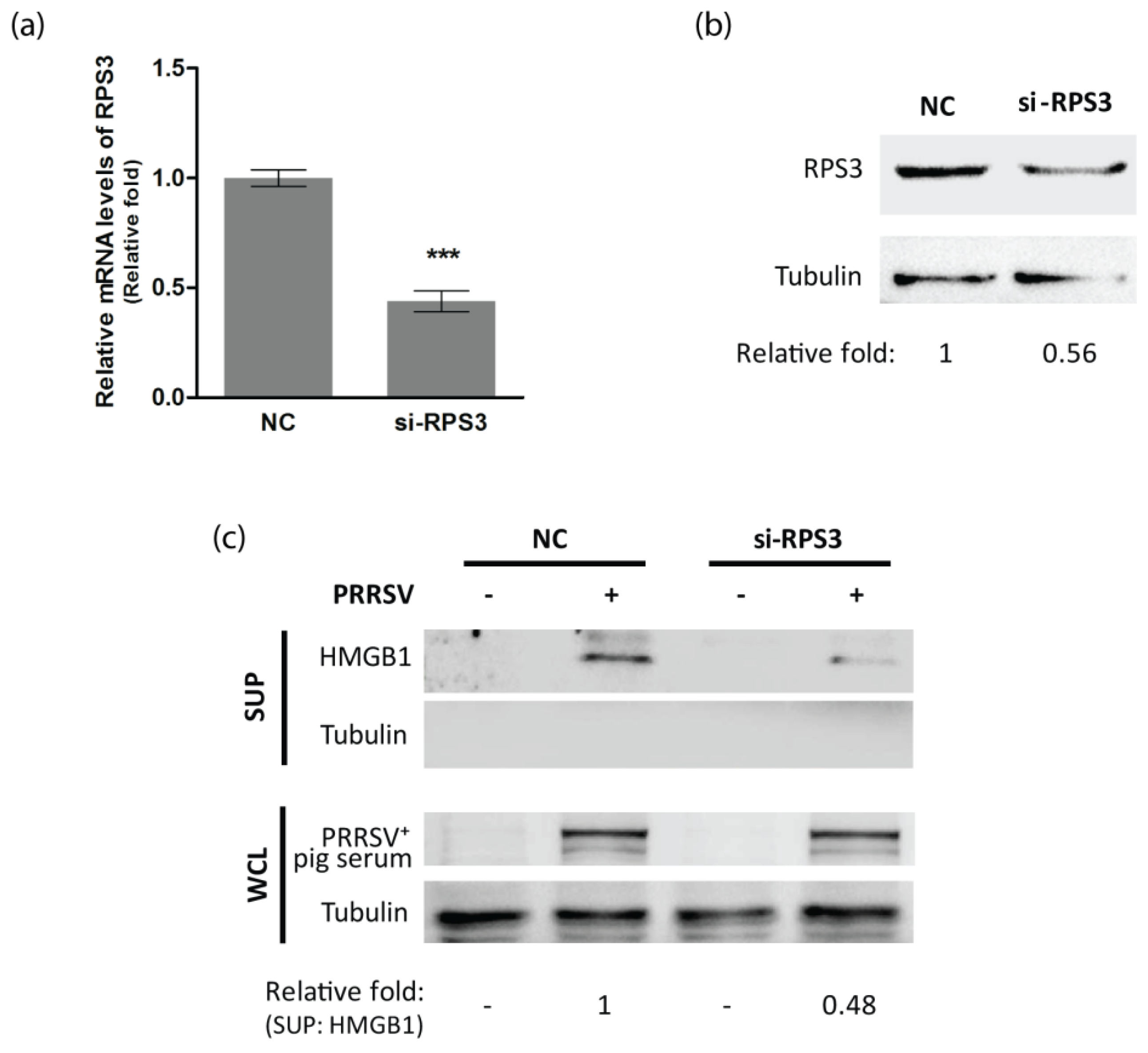
3.4. RPS3 Is Involved in HMGB1 Secretion in PRRSV-Infected Cells
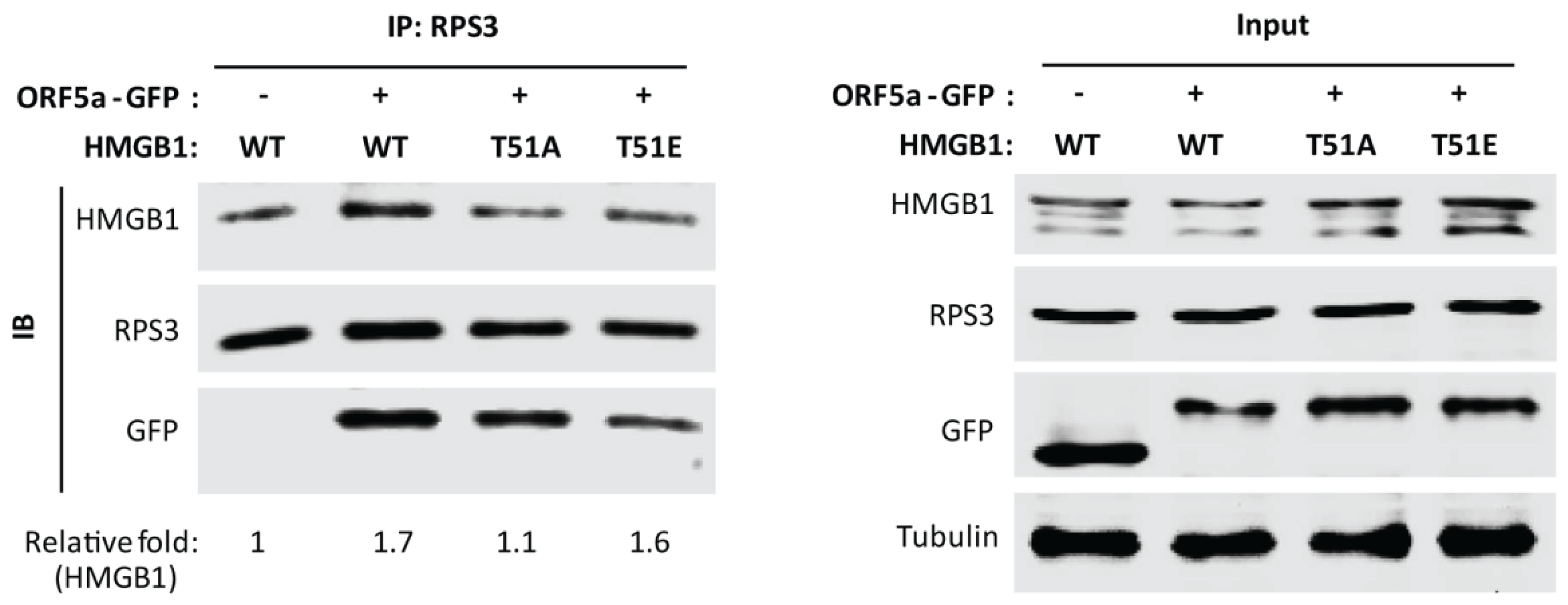
3.5. HMGB1 Phosphorylation at Threonine-51 Is Involved in HMGB1 and RPS3 Interaction
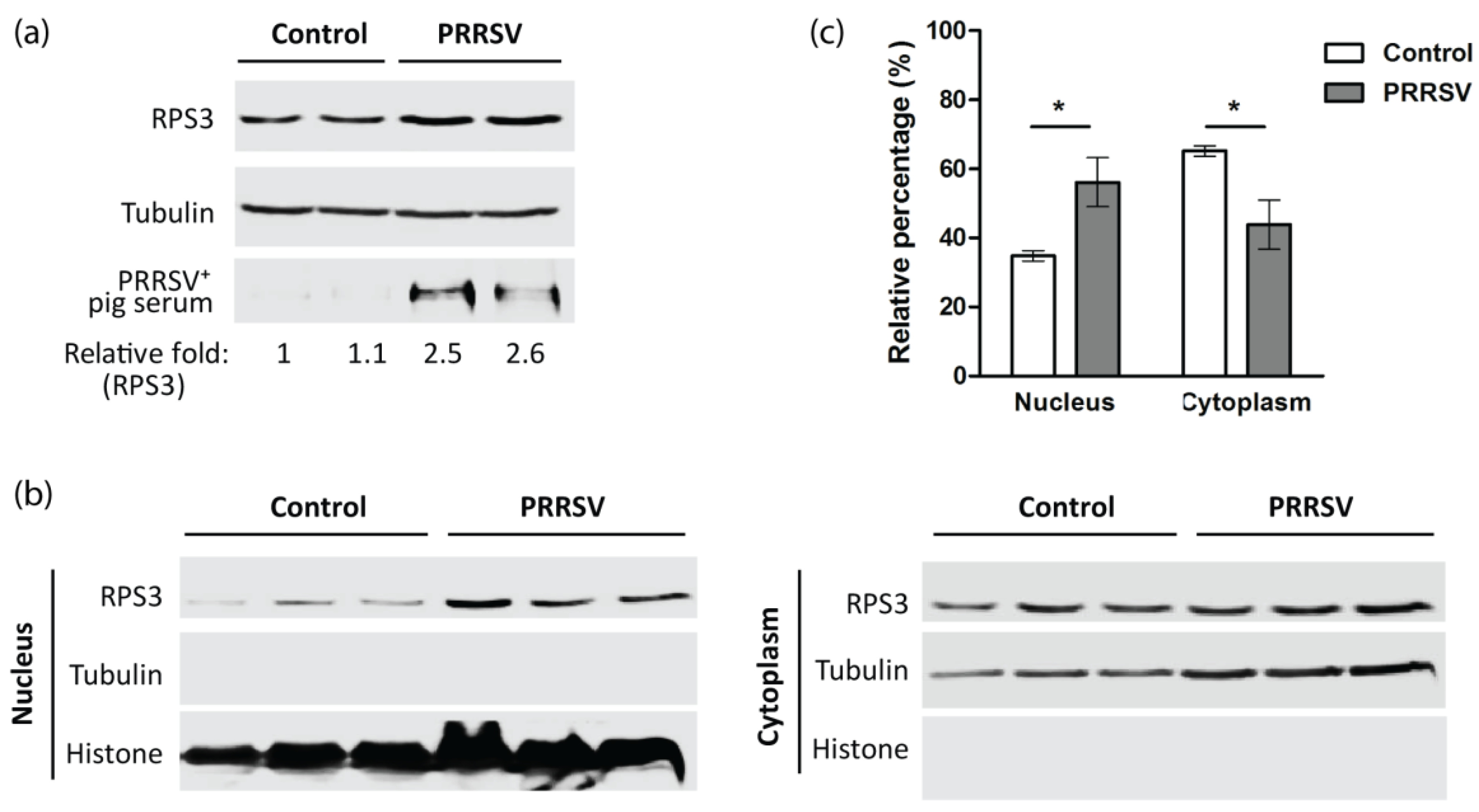
3.6. PRRSV Increases RPS3 Expression and Nuclear Accumulation In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rossow, K.D.; Collins, J.E.; Goyal, S.M.; Nelson, E.A.; Christopherhennings, J.; Benfield, D.A. Pathogenesis of porcine reproductive and respiratory syndrome virus-infection in gnotobiotic pigs. Vet. Pathol. 1995, 32, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.H.; Lauck, M.; Bailey, A.L.; Shchetinin, A.M.; Vishnevskaya, T.V.; Bao, Y.M.; Ng, T.F.F.; LeBreton, M.; Schneider, B.S.; Gillis, A.; et al. Reorganization and expansion of the Nidoviral family Arteriviridae. Arch. Virol. 2016, 161, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Conzelmann, K.K.; Visser, N.; Van Woensel, P.; Thiel, H.J. Molecular characterization of porcine reproductive and respiratory syndrome virus, a member of the Arterivirus group. Virology 1993, 193, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dokland, T. The structural biology of PRRSV. Virus Res. 2010, 154, 86–97. [Google Scholar] [CrossRef]
- Kim, H.S.; Kwang, J.; Yoon, I.J.; Joo, H.S.; Frey, M.L. Enhanced replication of porcine reproductive and respiratory syndrome (Prrs) virus in a homogeneous subpopulation of Ma-104 cell-line. Arch. Virol. 1993, 133, 477–483. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H.C.; Chavan, S.S.; Andersson, U. High Mobility Group Box Protein 1 (HMGB1): The prototypical endogenous danger molecule. Mol. Med. 2015, 21, S6–S12. [Google Scholar] [CrossRef]
- Harris, H.E.; Andersson, U.; Pisetsky, D.S. HMGB1: A multifunctional alarmin driving autoimmune and inflammatory disease. Nat. Rev. Rheumatol. 2012, 8, 195–202. [Google Scholar] [CrossRef]
- Andersson, U.; Wang, H.C.; Palmblad, K.; Aveberger, A.C.; Bloom, O.; Erlandsson-Harris, H.; Janson, A.; Kokkola, R.; Zhang, M.H.; Yang, H.; et al. High mobility group 1 protein (HMG-1) stimulates proinflammatory cytokine synthesis in human monocytes. J. Exp. Med. 2000, 192, 565–570. [Google Scholar] [CrossRef]
- Klune, J.R.; Dhupar, R.; Cardinal, J.; Billiar, T.R.; Tsung, A. HMGB1: Endogenous danger signaling. Mol. Med. 2008, 14, 476–484. [Google Scholar] [CrossRef]
- Wang, R.; Xiao, Y.Q.; Zhang, Q.; Bai, L.; Wang, W.R.; Zhao, S.H.; Liu, E.Q. Upregulation of HMGB1 secretion in lungs of pigs infected by highly pathogenic porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2021, 252, 108922. [Google Scholar] [CrossRef]
- Senthilkumar, D.; Rajukumar, K.; Kumar, M.; Kalaiyarasu, S.; Shrivastava, D.; Katare, M.; Kulkarni, D.D.; Singh, V.P. Porcine reproductive and respiratory syndrome virus induces concurrent elevation of High Mobility Group Box-1 protein and pro-inflammatory cytokines in experimentally infected piglets. Cytokine 2019, 113, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Yang, L.P.; Zhang, Y.L.; Li, J.Y.; Xu, L.R.; Xiao, Y.Q.; Zhang, Q.; Bai, L.; Zhao, S.H.; Liu, E.Q.; et al. Porcine reproductive and respiratory syndrome virus induces HMGB1 secretion via activating PKC-delta to trigger inflammatory response. Virology 2018, 518, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Duan, E.Z.; Wang, D.; Luo, R.; Luo, J.Y.; Gao, L.; Chen, H.C.; Fang, L.R.; Xiao, S.B. Porcine reproductive and respiratory syndrome virus infection triggers HMGB1 release to promote inflammatory cytokine production. Virology 2014, 468–470, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, Y.T.; Zhao, X.; Antoine, D.; Xiao, X.Z.; Wang, H.C.; Andersson, U.; Billiar, T.R.; Tracey, K.J.; Lu, B. Regulation of posttranslational modifications of HMGB1 during immune responses. Antioxid. Redox Signal. 2016, 24, 620–634. [Google Scholar] [CrossRef]
- Andersson, U.; Antoine, D.J.; Tracey, K.J. The functions of HMGB1 depend on molecular localization and post-translational modifications Introduction. J. Intern. Med. 2014, 276, 420–424. [Google Scholar] [CrossRef] [Green Version]
- Bonaldi, T.; Talamo, F.; Scaffidi, P.; Ferrera, D.; Porto, A.; Bachi, A.; Rubartelli, A.; Agresti, A.; Bianchi, M.E. Monocytic cells hyperacetylate chromatin protein HMGB1 to redirect it towards secretion. EMBO J. 2003, 22, 5551–5560. [Google Scholar] [CrossRef] [Green Version]
- Youn, J.H.; Shin, J.S. Nucleocytoplasmic shuttling of HMGB1 is regulated by phosphorylation that redirects it toward secretion. J. Immunol. 2006, 177, 7889–7897. [Google Scholar] [CrossRef] [Green Version]
- Lu, B.; Antoine, D.J.; Kwan, K.; Lundback, P.; Wahamaa, H.; Schierbeck, H.; Robinson, M.; Van Zoelen, M.A.D.; Yang, H.; Li, J.H.; et al. JAK/STAT1 signaling promotes HMGB1 hyperacetylation and nuclear translocation. Proc. Natl. Acad. Sci. USA 2014, 111, 3068–3073. [Google Scholar] [CrossRef] [Green Version]
- Oh, Y.J.; Youn, J.H.; Ji, Y.; Lee, S.E.; Lim, K.J.; Choi, J.E.; Shin, J.S. HMGB1 is phosphorylated by classical protein kinase C and is secreted by a Calcium-dependent mechanism. J. Immunol. 2009, 182, 5800–5809. [Google Scholar] [CrossRef] [Green Version]
- Kang, H.J.; Lee, H.; Choi, H.J.; Youn, J.H.; Shin, J.S.; Ahn, Y.H.; Yoo, J.S.; Paik, Y.K.; Kim, H. Non-histone nuclear factor HMGB1 is phosphorylated and secreted in colon cancers. Lab. Investig. 2009, 89, 948–959. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Park, M.; Shin, N.; Kim, G.; Kim, Y.G.; Shin, J.S.; Kim, H. High mobility group box-1 is phosphorylated by protein kinase C zeta and secreted in colon cancer cells. Biochem. Biophys. Res. Commun. 2012, 424, 321–326. [Google Scholar] [CrossRef]
- Graifer, D.; Malygin, A.; Zharkov, D.O.; Karpova, G. Eukaryotic ribosomal protein S3: A constituent of translational machinery and an extraribosomal player in various cellular processes. Biochimie 2014, 99, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.F.; Hardwidge, P.R. Ribosomal protein S3: A multifunctional target of attaching/effacing bacterial pathogens. Front. Microbiol. 2011, 2, 137. [Google Scholar] [CrossRef] [Green Version]
- Tong, G.Z.; Zhou, Y.J.; Hao, X.F.; Tian, Z.J.; An, T.Q.; Qiu, H.J. Highly pathogenic porcine reproductive and respiratory syndrome, china. Emerg. Infect. Dis. 2007, 13, 1434–1436. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Nan, Y.C.; Yu, Y.; Zhang, Y.J. Porcine reproductive and respiratory syndrome virus Nsp1 beta inhibits interferon-activated JAK/STAT signal transduction by inducing Karyopherin-alpha 1 degradation. J. Virol. 2013, 87, 5219–5228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Nan, Y.C.; Yu, Y.; Yang, Z.Q.; Zhang, Y.J. Variable interference with interferon signal transduction by different strains of porcine reproductive and respiratory syndrome virus. Vet. Microbiol. 2013, 166, 493–503. [Google Scholar] [CrossRef]
- Wan, F.Y.; Anderson, D.E.; Barnitz, R.A.; Snow, A.; Bidere, N.; Zheng, L.X.; Hegde, V.; Lam, L.T.; Staudt, L.M.; Levens, D.; et al. Ribosomal protein S3: A KH domain subunit in NF-kappa B complexes that mediates selective gene regulation. Cell 2007, 131, 927–939. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Andersson, U.; Tracey, K.J. HMGB1 is a therapeutic target for sterile inflammation and infection. Annu. Rev. Immunol. 2011, 29, 139–162. [Google Scholar] [CrossRef] [Green Version]
- Magna, M.; Pisetsky, D.S. The role of HMGB1 in the pathogenesis of inflammatory and autoimmune diseases. Mol. Med. 2014, 20, 138–146. [Google Scholar] [CrossRef]
- Muller, S.; Scaffidi, P.; Degryse, B.; Bonaldi, T.; Ronfani, L.; Agresti, A.; Beltrame, M.; Bianchi, M.E. New EMBO members’ review: The double life of HMGB1 chromatin protein: Architectural factor and extracellular signal. EMBO J. 2001, 20, 4337–4340. [Google Scholar] [CrossRef] [PubMed]
- Chikhirzhina, E.; Starkova, T.; Beljajev, A.; Polyanichko, A.; Tomilin, A. Functional diversity of non-histone chromosomal protein HmgB1. Int. J. Mol. Sci. 2020, 21, 7948. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.W.; Gong, Z.J. Regulation of acetylation in high mobility group protein B1 cytosol translocation. DNA Cell Biol. 2019, 38, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Taira, J.; Kida, Y.; Kuwano, K.; Higashimoto, Y. Protein phosphatase 2A dephosphorylates phosphoserines in nucleocytoplasmic shuttling and secretion of high mobility group box 1. J. Biochem. 2013, 154, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Taira, J.; Higashimoto, Y. Evaluation of in vitro properties of predicted kinases that phosphorylate serine residues within nuclear localization signal 1 of high mobility group box 1. J. Pept. Sci. 2014, 20, 613–617. [Google Scholar] [CrossRef]
- Shirai, Y.; Saito, N. Activation mechanisms of protein kinase C: Maturation, catalytic activation, and targeting. J. Biochem. 2002, 132, 663–668. [Google Scholar] [CrossRef] [Green Version]
- Hegde, V.; Wang, M.; Deutsch, W.A. Characterization of human ribosomal protein S3 binding to 7,8-dihydro-8-oxoguanine and abasic sites by surface plasmon resonance. DNA Repair 2004, 3, 121–126. [Google Scholar] [CrossRef]
- Kim, J.; Chubatsu, L.S.; Admon, A.; Stahl, J.; Fellous, R.; Linn, S. Implication of mammalian ribosomal protein S3 in the processing of DNA damage. J. Biol. Chem. 1995, 270, 13620–13629. [Google Scholar] [CrossRef] [Green Version]
- Stanborough, T.; Niederhauser, J.; Koch, B.; Bergler, H.; Pertschy, B. Ribosomal protein S3 interacts with the NF-kappa B inhibitor I kappa B alpha. FEBS Lett. 2014, 588, 659–664. [Google Scholar] [CrossRef]
- Wier, E.M.; Neighoff, J.; Sun, X.; Fu, K.; Wan, F.Y. Identification of an N-terminal truncation of the NF-kappa B p65 subunit that specifically modulates ribosomal protein S3-dependent NF-kappa B gene expression. J. Biol. Chem. 2012, 287, 43019–43029. [Google Scholar] [CrossRef] [Green Version]
- Jang, C.Y.; Kim, H.D.; Kim, J. Ribosomal protein S3 interacts with TRADD to induce apoptosis through caspase dependent JNK activation. Biochem. Biophys. Res. Commun. 2012, 421, 474–478. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Jiang, H.W.; Chen, H.; Zhang, H.N.; Liu, Y.; Xu, Z.W.; Wu, F.L.; Guo, S.J.; Hou, J.L.; Yang, M.K.; et al. Systematic Identification of Mycobacterium tuberculosis Effectors Reveals that BfrB Suppresses Innate Immunity. Mol. Cell. Proteom. 2017, 16, 2243–2253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, F.; Weaver, A.; Gao, X.; Bern, M.; Hardwidge, P.R.; Lenardo, M.J. IKKbeta phosphorylation regulates RPS3 nuclear translocation and NF-kappaB function during infection with Escherichia coli strain O157:H7. Nat. Immunol. 2011, 12, 335–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Wan, F.; Mateo, K.; Callegari, E.; Wang, D.; Deng, W.; Puente, J.; Li, F.; Chaussee, M.S.; Finlay, B.B.; et al. Bacterial effector binding to ribosomal protein s3 subverts NF-kappaB function. PLoS Pathog. 2009, 5, e1000708. [Google Scholar] [CrossRef]
- Pham, T.H.; Gao, X.; Singh, G.; Hardwidge, P.R. Escherichia coli virulence protein NleH1 interaction with the v-Crk sarcoma virus CT10 oncogene-like protein (CRKL) governs NleH1 inhibition of the ribosomal protein S3 (RPS3)/nuclear factor kappaB (NF-kappaB) pathway. J. Biol. Chem. 2013, 288, 34567–34574. [Google Scholar] [CrossRef] [Green Version]
- Wood, J.; Frederickson, R.M.; Fields, S.; Patel, A.H. Hepatitis C virus 3 ′X region interacts with human ribosomal proteins. J. Virol. 2001, 75, 1348–1358. [Google Scholar] [CrossRef] [Green Version]
- Boon, A.C.M.; de Beauchamp, J.; Hollmann, A.; Luke, J.; Kotb, M.; Rowe, S.; Finkelstein, D.; Neale, G.; Lu, L.; Williams, R.W.; et al. Host genetic variation affects resistance to infection with a highly pathogenic H5N1 influenza A virus in mice. J. Virol. 2009, 83, 10417–10426. [Google Scholar] [CrossRef] [Green Version]
- Petrova, E.; Gracias, S.; Beauclair, G.; Tangy, F.; Jouvenet, N. Uncovering Flavivirus host dependency factors through a genome-wide gain-of-function screen. Viruses 2019, 11, 68. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.S.; Kim, H.D.; Kim, J. PKC delta-dependent functional switch of rpS3 between translation and DNA repair. Biochim. Biophys. Acta-Mol. Cell Res. 2009, 1793, 395–405. [Google Scholar] [CrossRef] [Green Version]
- Kim, T.S.; Kim, H.D.; Shin, H.S.; Kim, J. Phosphorylation status of nuclear ribosomal protein S3 is reciprocally regulated by protein kinase C delta and protein phosphatase 2A. J. Biol. Chem. 2009, 284, 21201–21208. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.J.; Youn, H.; Seong, K.M.; Jin, Y.W.; Kim, J.; Youn, B. Phosphorylation of ribosomal protein S3 and antiapoptotic TRAF2 protein mediates radioresistance in non-small cell lung cancer cells. J. Biol. Chem. 2013, 288, 2965–2975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.D.; Lee, J.Y.; Kim, J. Erk phosphorylates threonine 42 residue of ribosomal protein S3. Biochem. Biophys. Res. Commun. 2005, 333, 110–115. [Google Scholar] [CrossRef] [PubMed]




| Primer a | Sequences (5′-3′) | Target Gene |
|---|---|---|
| T51A-F | GTGGAAGGCCATGTCTGC | HMGB1-T51A |
| T51A-R | GCAGACATGGCCTTCCAC | HMGB1-T51A |
| T51E-F | GGTGGAAGGAGATGTCTG | HMGB1-T51E |
| T51E-R | CAGACATCTCCTTCCACC | HMGB1-T51E |
| RPL32-F | CAACTGGCCATCAGAGTCAC | RPL32 |
| RPL32-R | GTGCACATGAGCTGCCTACT | RPL32 |
| RPS3-F | GCTGAAGATGGCTACTCTGGAG | RPS3 |
| RPS3-R | ACAGCAGTCAGTTCCCGAATCC | RPS3 |
| Accession | Description | Gene Symbol |
|---|---|---|
| A0A0D9S9M0 | Uncharacterized protein | LOC103219323 |
| A0A0D9SB94 | Uncharacterized protein | LOC103237177 |
| A0A0D9R6U8 | 60S ribosomal protein L29 | RPL29 |
| A0A0D9SC49 | Uncharacterized protein | LOC103238803 |
| A0A0D9SCL1 | Histone cluster 1 H1 family member d | LOC103222006 |
| A0A0D9R3U8 | NUFIP2, FMR1 interacting protein 2 | NUFIP2 |
| A0A0D9RHW7 | Polyadenylate-binding protein | PABPC1 |
| A0A0D9S9I8 | 60S ribosomal protein L13 | RPL13 |
| A0A0D9S7U2 | Splicing factor proline and glutamine rich | SFPQ |
| A0A0D9S7K7 | Y-box binding protein 1 | YBX1 |
| A0A0D9QV93 | Ribosomal protein S3 | RPS3 |
| A0A0D9SBN8 | Uncharacterized protein | |
| A0A0D9S125 | LSM12 homolog | LSM12 |
| A0A0D9QXD5 | G3BP stress granule assembly factor 2 | G3BP2 |
| A0A0D9SD09 | Histone H4 | LOC103247796; LOC103221971; LOC103222007 |
| LOC103221993; LOC103222008 | ||
| A0A0D9RXA3 | Uncharacterized protein | |
| A0A0D9REQ6 | G3BP stress granule assembly factor 1 | G3BP1 |
| A0A0D9RDI7 | Ribosomal protein L26 | RPL26 |
| A0A0D9QZA0 | Cell cycle associated protein 1 | CAPRIN1 |
| A0A0D9RGJ5 | Ribosomal protein S14 | RPS14 |
| A0A0D9RBS5 | EWS RNA binding protein 1 | EWSR1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, R.; Zhang, J.; Fu, Y.; Jia, L.; Zhang, Y.; Bai, L.; Wang, W.; Cheng, D.; Liu, E. PRRSV Induces HMGB1 Phosphorylation at Threonine-51 Residue to Enhance Its Secretion. Viruses 2022, 14, 1002. https://doi.org/10.3390/v14051002
Wang R, Zhang J, Fu Y, Jia L, Zhang Y, Bai L, Wang W, Cheng D, Liu E. PRRSV Induces HMGB1 Phosphorylation at Threonine-51 Residue to Enhance Its Secretion. Viruses. 2022; 14(5):1002. https://doi.org/10.3390/v14051002
Chicago/Turabian StyleWang, Rong, Jingyi Zhang, Yu Fu, Linying Jia, Yali Zhang, Liang Bai, Weirong Wang, Daxin Cheng, and Enqi Liu. 2022. "PRRSV Induces HMGB1 Phosphorylation at Threonine-51 Residue to Enhance Its Secretion" Viruses 14, no. 5: 1002. https://doi.org/10.3390/v14051002





