Development of DNA Vaccine Candidate against SARS-CoV-2
Abstract
:1. Introduction
2. Materials and Methods
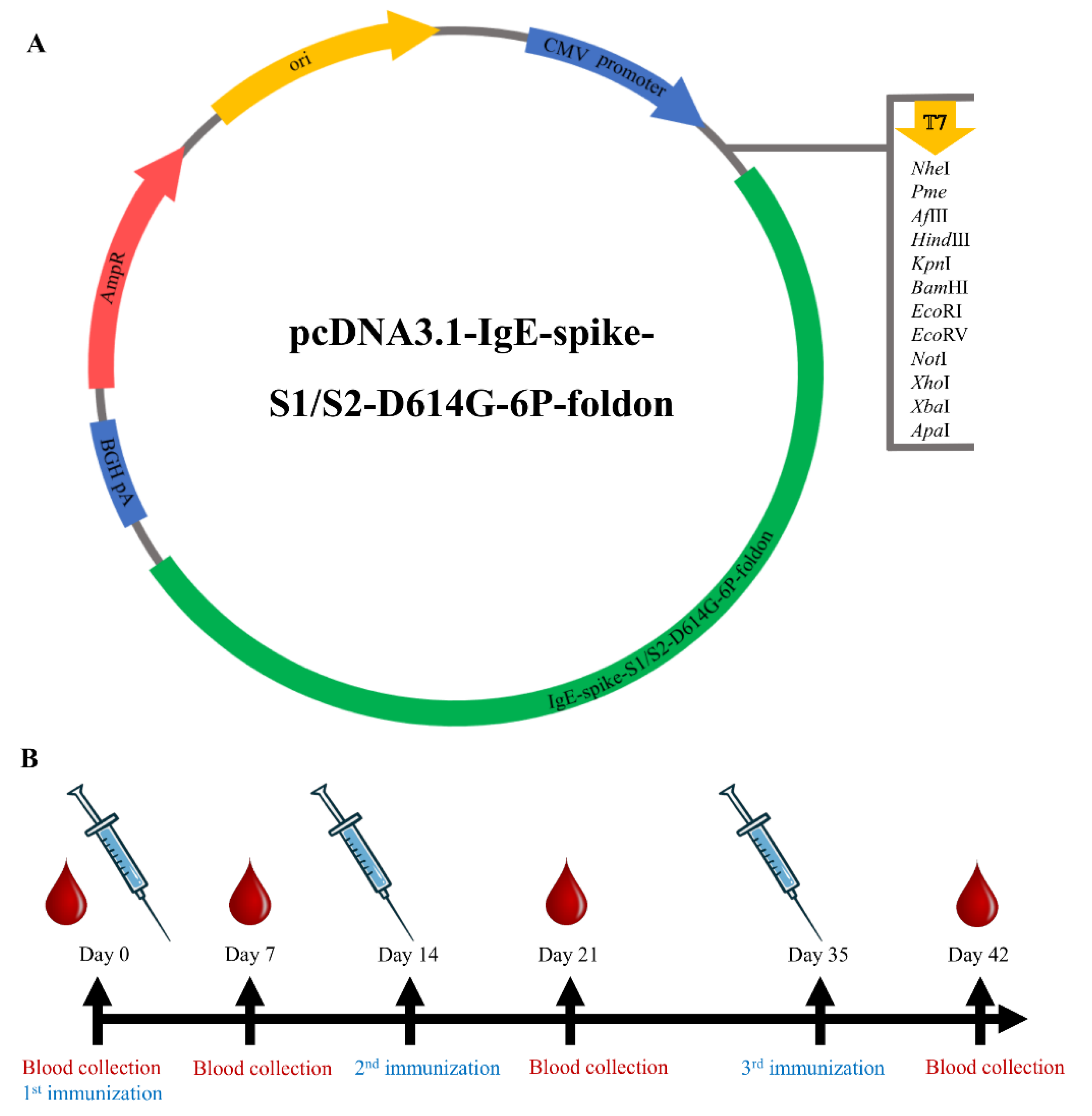
2.1. Design and Synthesis of SARS-CoV-2 DNA Vaccine Construct
2.2. Amplification of the Target Gene
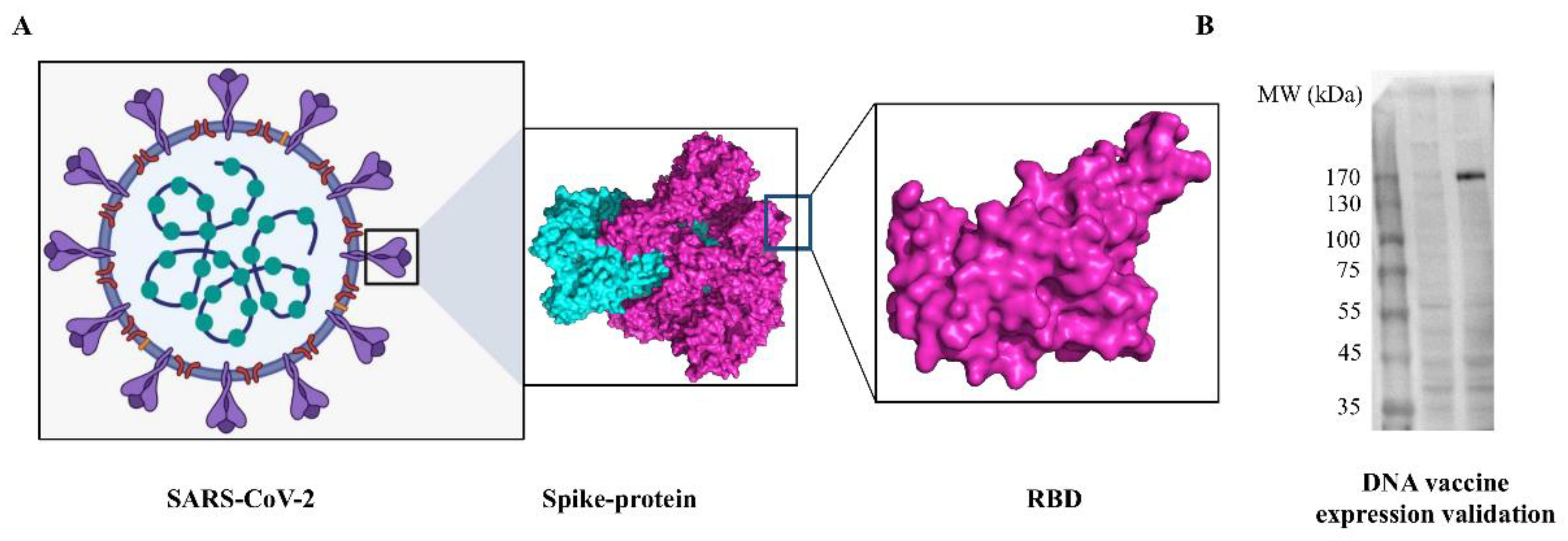
2.3. In Vitro Validation of DNA Vaccine Expression
2.4. Mouse Immunization Experiment
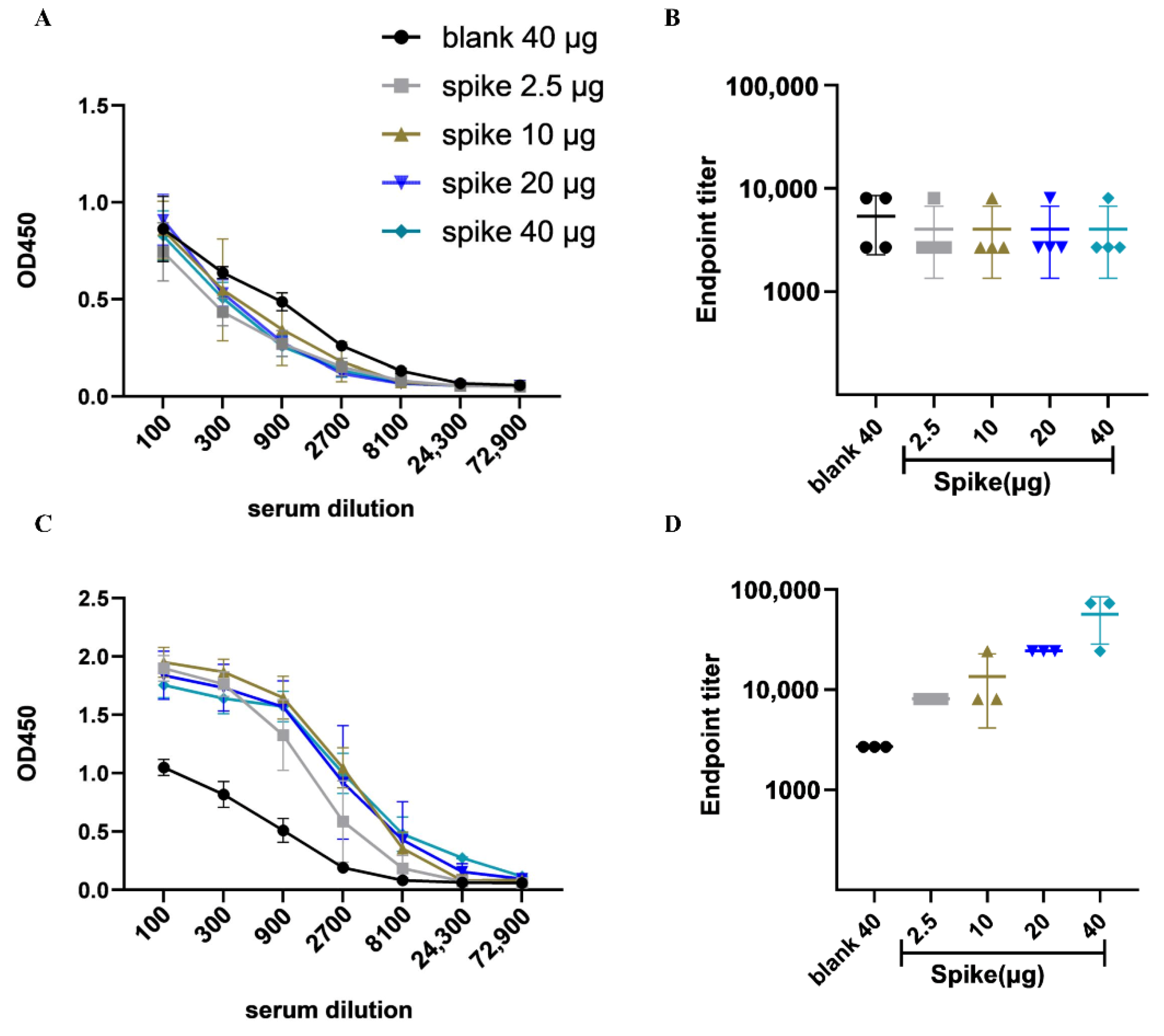
2.5. Detection of Plasma IgG and Antibody Titers by ELISA
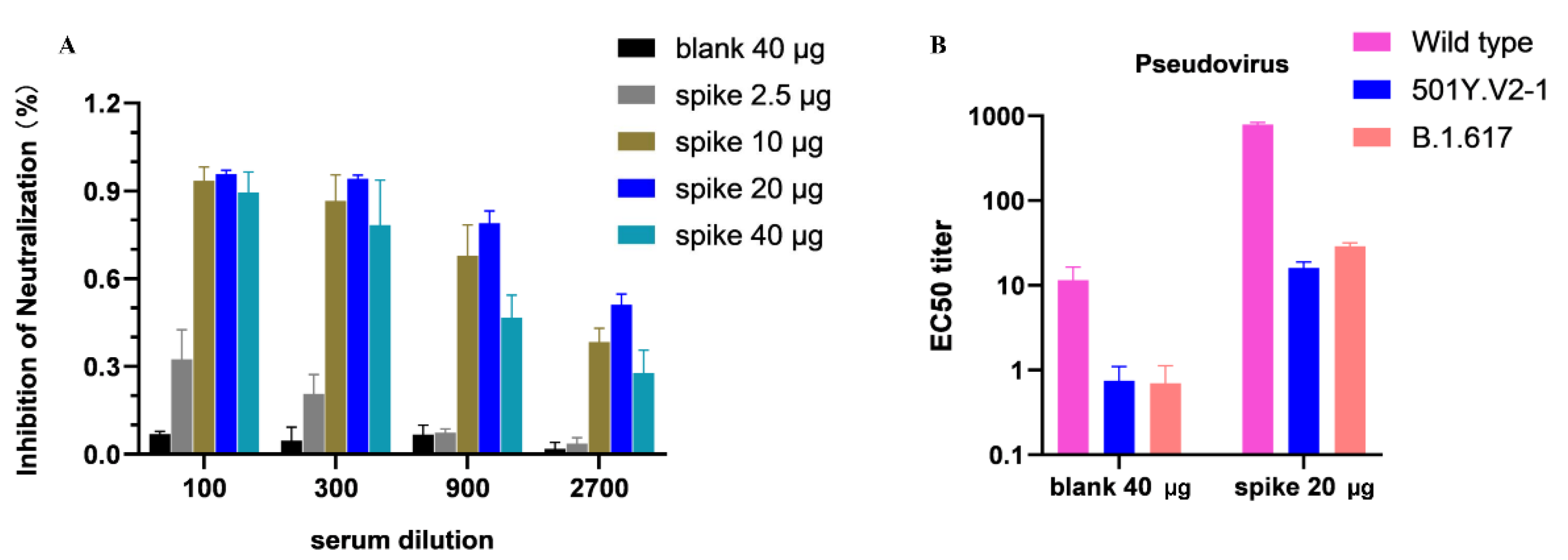
2.6. SARS-CoV-2 Surrogate Virus Neutralization ELISA
2.7. SARS-CoV-2 Pseudovirus Neutralization Assay
2.8. Statistics
3. Results
3.1. Characterization of SARS-CoV-2 DNA Vaccine Candidate and Validation In Vitro
3.2. DNA Vaccine Candidate Induced Humoral Immune Response In Vivo
3.3. Inhibition of SARS-CoV-2 RBD Binding to a Host Receptor ACE2 via Competitive ELISA
3.4. Neutralization Ability DNA Vaccine-Induced Antibodies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Liu, S.M.; Yu, X.H.; Tang, S.L.; Tang, C.K. Coronavirus disease 2019 (COVID-19): Current status and future perspectives. Int. J. Antimicrob. Agents 2020, 55, 105951. [Google Scholar] [CrossRef] [PubMed]
- Van Bavel, J.J.; Cichocka, A.; Capraro, V.; Sjåstad, H.; Nezlek, J.B.; Pavlović, T.; Alfano, M.; Gelfand, M.J.; Azevedo, F.; Birtel, M.D.; et al. National identity predicts public health support during a global pandemic. Nat. Commun. 2022, 13, 517. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422. [Google Scholar] [CrossRef]
- Da Silva Junior, F.J.G.; Sales, J.C.E.S.; Monteiro, C.F.D.S.; Costa, A.P.C.; Campos, L.R.B.; Miranda, P.I.G.; Monteiro, T.A.D.S.; Lima, R.A.G.; Lopes-Junior, L.C. Impact of COVID-19 pandemic on mental health of young people and adults: A systematic review protocol of observational studies. BMJ Open 2020, 10, e039426. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization (WHO). COVID-19 Explorer. Available online: https://covid19.who.int/ (accessed on 9 April 2022).
- Rehman, M.F.; Fariha, C.; Anwar, A.; Shahzad, N.; Ahmad, M.; Mukhtar, S.; Farhan Ul Haque, M. Novel coronavirus disease (COVID-19) pandemic: A recent mini review. Comput. Struct. Biotechnol. J. 2021, 19, 612–623. [Google Scholar] [CrossRef] [PubMed]
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615. [Google Scholar] [CrossRef] [PubMed]
- Haas, E.J.; Angulo, F.J.; McLaughlin, J.M.; Anis, E.; Singer, S.R.; Khan, F.; Brooks, N.; Smaja, M.; Mircus, G.; Pan, K.; et al. Impact and effectiveness of mRNA BNT162b2 vaccine against SARS-CoV-2 infections and COVID-19 cases, hospitalisations, and deaths following a nationwide vaccination campaign in Israel: An observational study using national surveillance data. Lancet 2021, 397, 1819–1829. [Google Scholar] [CrossRef]
- Mascellino, M.T.; Di Timoteo, F.; De Angelis, M.; Oliva, A. Overview of the main anti-Sars-CoV-2 vaccines: Mechanism of action, efficacy and safety. Infect. Drug Resist. 2021, 14, 3459–3476. [Google Scholar] [CrossRef]
- Tavilani, A.; Abbasi, E.; Kian Ara, F.; Darini, A.; Asefy, Z. COVID-19 vaccines: Current evidence and considerations. Metab. Open 2021, 12, 100124. [Google Scholar] [CrossRef]
- Fang, E.; Liu, X.; Li, M.; Zhang, Z.; Song, L.; Zhu, B.; Wu, X.; Liu, J.; Zhao, D.; Li, Y. Advances in COVID-19 mRNA vaccine development. Signal Transduct. Target Ther. 2022, 23, 94. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines-a new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flingai, S.; Czerwonko, M.; Goodman, J.; Kudchodkar, S.B.; Muthumani, K.; Weiner, D.B. Synthetic DNA vaccines: Improved vaccine potency by electroporation and co-delivered genetic adjuvants. Front. Immunol. 2013, 4, 354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, J. Vector Design for Improved DNA Vaccine Efficacy, Safety and Production. Vaccines 2013, 1, 225–249. [Google Scholar] [CrossRef] [PubMed]
- Xia, D.; Jin, R.; Byagathvalli, G.; Yu, H.; Ye, L.; Lu, C.Y.; Saad Bhamla, M.; Yang, C.; Prausnitz, M.R. An ultra-low-cost electroporator with microneedle electrodes (ePatch) for SARS-CoV-2 vaccination. Proc. Natl. Acad. Sci. USA 2021, 118, e2110817118. [Google Scholar] [CrossRef] [PubMed]
- Chavda, V.P.; Pandya, R.; Apostolopoulos, V. DNA vaccines for SARS-CoV-2: Toward third-generation vaccination era. Expert Rev. Vaccines 2021, 20, 1549–1560. [Google Scholar] [CrossRef]
- Ye, G.; Liu, B.; Li, F. Cryo-EM structure of a SARS-CoV-2 omicron spike protein ectodomain. Nat. Commun. 2022, 13, 1214. [Google Scholar] [CrossRef]
- Hsieh, C.; Goldsmith, J.A.; Schaub, J.M.; Divenere, A.M.; Kuo, H.; Javanmardi, K.; Le, K.C.; Wrapp, D.; Lee, A.G.; Liu, Y.; et al. Structure-based design of prefusion-stabilized SARS-CoV-2 spikes. Science 2020, 369, 1501–1505. [Google Scholar] [CrossRef]
- Yilmaz, I.C.; Ipekoglu, E.M.; Bulbul, A.; Turay, N.; Yildirim, M.; Evcili, I.; Yilmaz, N.S.; Guvencli, N.; Aydin, Y.; Gungor, B.; et al. Development and preclinical evaluation of virus-like particle vaccine against COVID-19 infection. Allergy Eur. J. Allergy Clin. Immunol. 2022, 77, 258–270. [Google Scholar] [CrossRef]
- Weissman, D.; Alameh, M.G.; de Silva, T.; Collini, P.; Hornsby, H.; Brown, R.; LaBranche, C.C.; Edwards, R.J.; Sutherland, L.; Santra, S.; et al. D614G Spike Mutation Increases SARS-CoV-2 Susceptibility to Neutralization. Cell Host Microbe 2021, 29, 23–31.e4. [Google Scholar] [CrossRef]
- Flanagan, K.L.; Best, E.; Crawford, N.W.; Giles, M.; Koirala, A.; Macartney, K.; Russell, F.; Teh, B.W.; Wen, S.C.H. Progress and Pitfalls in the Quest for Effective SARS-CoV-2 (COVID-19) Vaccines. Front. Immunol. 2020, 11, 579250. [Google Scholar] [CrossRef] [PubMed]
- Dai, L.; Gao, G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021, 21, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Li, Y.; Guo, Y.; Wang, L.; Yang, J.; Zhao, G.; Zhou, Y.; Du, L.; Jiang, S. Intranasal vaccination of recombinant H5N1 HA1 proteins fused with foldon and Fc induces strong mucosal immune responses with neutralizing activity: Implication for developing novel mucosal influenza vaccines. Hum. Vaccines Immunother. 2015, 11, 2831–2838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Cai, Y.; Xiao, T.; Lu, J.; Peng, H.; Sterling, S.M.; Jr, R.M.W.; Rits-Volloch, S.; Zhu, H.; Woosley, A.N.; et al. Structural impact on SARS-CoV-2 spike protein by D614G substitution. Science 2021, 372, 525–530. [Google Scholar] [CrossRef]
- Yu, J.; Tostanosk, L.H.; Peter, L.; Mercad, N.B.; McMahan, K.; Mahrokhia, S.H.; Nkolol, J.P.; Liu, J.; Li, Z.; Chandrashekar, A.; et al. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science 2020, 369, 806–811. [Google Scholar] [CrossRef]
- Nie, J.; Li, Q.; Wu, J.; Zhao, C.; Hao, H.; Liu, H.; Zhang, L.; Nie, L.; Qin, H.; Wang, M.; et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020, 15, 3699–3715. [Google Scholar] [CrossRef]
- Smith, T.R.F.; Patel, A.; Ramos, S.; Elwood, D.; Zhu, X.; Yan, J.; Gary, E.N.; Walker, S.N.; Schultheis, K.; Purwar, M.; et al. Immunogenicity of a DNA vaccine candidate. Nat. Commun. 2020, 11, 2601. [Google Scholar] [CrossRef]
- Fiolet, T.; Kherabi, Y.; Macdonald, C.; Ghosn, J.; Peiffer-smadja, N. Comparing COVID-19 vaccines for their characteristics, efficacy and effectiveness against SARS-CoV-2 and variants of concern: A narrative review. Clin. Microbiol. Infect. 2022, 28, 202–221. [Google Scholar] [CrossRef]
- Zheng, C.; Shao, W.; Chen, X.; Zhang, B.; Wang, G.; Zhang, W. Real-world effectiveness of COVID-19 vaccines: A literature review and meta-analysis. Int. J. Infect. Dis. 2022, 114, 52–260. [Google Scholar] [CrossRef]
- Kim, J.H.; Marks, F.; Clemens, J.D. Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med. 2021, 27, 205–211. [Google Scholar] [CrossRef]
- Kantarcioglu, B.; Iqbal, O.; Lewis, J.; Carter, C.A.; Singh, M.; Lievano, F.; Ligocki, M.; Jeske, W.; Adiguzel, C.; Gerotziafas, G.T.; et al. An Update on the Status of Vaccine Development for SARS-CoV-2 Including Variants. Practical Considerations for COVID-19 Special Populations. Clin. Appl. Thromb Hemost. 2022, 28, 10760296211056648. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Qin, C.; Liu, M.; Liu, J. Effectiveness and safety of SARS-CoV-2 vaccine in real-world studies: A systematic review and meta-analysis. Infect. Dis. Poverty 2021, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, N.; Weissman, D.; Whitehead, K.A. mRNA vaccines for infectious diseases: Principles, delivery and clinical translation. Nat. Rev. Drug Discov. 2021, 20, 817–838. [Google Scholar] [CrossRef] [PubMed]
- Dolgin, E. The tangled history of mRNA vaccines. Nature 2021, 597, 318–324. [Google Scholar] [CrossRef]
- Qin, F.; Xia, F.; Chen, H.; Cui, B.; Feng, Y.; Zhang, P.; Chen, J.; Luo, M.; Tornesello, M.L.; Nazionale, I.; et al. A Guide to Nucleic Acid Vaccines in the Prevention and Treatment of Infectious Diseases and Cancers: From Basic Principles to Current Applications. Front. Cell Dev. Biol. 2021, 9, 633776. [Google Scholar] [CrossRef]
- Chavda, V.P.; Hossain, K.; Beladiya, J.; Apostolopoulos, V. Nucleic Acid Vaccines for COVID-19: A Paradigm Shift in the Vaccine Development Arena. Biologics 2021, 1, 337–356. [Google Scholar] [CrossRef]
- Liu, M.A. A Comparison of Plasmid DNA and mRNA as Vaccine Technologies. Vaccines 2019, 7, 37. [Google Scholar] [CrossRef] [Green Version]
- Kyriakidis, N.C.; López-cortés, A.; González, E.V.; Grimaldos, A.B.; Prado, E.O. SARS-CoV-2 vaccines strategies: A comprehensive review of phase 3 candidates. NPJ Vaccines 2021, 6, 28. [Google Scholar] [CrossRef]
- Liang, Y.; Huang, L.; Liu, T. Development and Delivery Systems of mRNA Vaccines. Front. Bioeng. Biotechnol. 2021, 9, 718753. [Google Scholar] [CrossRef]
- Shafaati, M.; Mirzaei, R.; Saidijam, M.; Amirheidari, B.; Soleimani, M.; Mahaki, H.; Hazrati, F.; Safaei, M.; Tanzadehpanah, H.; Kazemi, S.; et al. A brief review on DNA vaccines in the era of. Future Virol. 2022, 17, 49–66. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines—How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Kumar, S.A.; Jhan, Y.Y.; Bishop, C.J. Engineering DNA vaccines against infectious diseases. Acta Biomater. 2018, 15, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.A.; Farmer, E.; Huang, C.; Lin, J.; Hung, C.F.; Wu, T.C. Therapeutic DNA Vaccines for Human Papillomavirus and Associated Diseases. Hum. Gene Ther. 2018, 29, 971–996. [Google Scholar] [CrossRef] [PubMed]
- Kutzler, M.A.; Weiner, D.B. DNA vaccines: Ready for prime time? Nature Reviews. Genetics 2008, 9, 776–788. [Google Scholar] [CrossRef]
- Rauch, S.; Jasny, E.; Schmidt, K.E.; Petsch, B.; Ag, C. New vaccine technologies to combat outbreak situations. Front. Immunol. 2018, 9, 1963. [Google Scholar] [CrossRef] [Green Version]
- Sundaram, A.K.; Ewing, D.; Liang, Z.; Jani, V.; Cheng, Y.; Sun, P.; Raviprakash, K.; Wu, S.; Petrovsky, N.; Defang, G.; et al. Immunogenicity of Adjuvanted Psoralen-Inactivated SARS-CoV-2 Vaccines and SARS-CoV-2 Spike Protein DNA Vaccines in BALB/c Mice. Pathogens 2021, 10, 626. [Google Scholar] [CrossRef]
- Alluhaybi, K.A.; Alharbi, R.H.; Alhabbab, R.Y.; Aljehani, N.D.; Alamri, S.S.; Basabrain, M.; Alharbi, R.; Abdulaal, W.H.; Alfaleh, M.A.; Tamming, L.; et al. Cellular and humoral immunogenicity of a candidate dna vaccine expressing SARS-CoV-2 spike subunit 1. Vaccines 2021, 9, 852. [Google Scholar] [CrossRef]
- Jazayeri, S.D.; Poh, C.L. Recent advances in delivery of veterinary DNA vaccines against avian pathogens. Vet. Res. 2019, 50, 78. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.A. DNA vaccines: An historical perspective and view to the future. Immunol. Rev. 2011, 239, 62–84. [Google Scholar] [CrossRef]
- Sheridan, C. First COVID-19 DNA vaccine approved, others in hot pursuit. Nat. Biotechnol. 2021, 39, 1479–1482. [Google Scholar] [CrossRef]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- Conforti, A.; Marra, E.; Palombo, F.; Roscilli, G.; Ravà, M.; Fumagalli, V.; Muzi, A.; Maffei, M.; Luberto, L.; Lione, L.; et al. COVID-eVax, an electroporated DNA vaccine candidate encoding the SARS-CoV-2 RBD, elicits protective responses in animal models. Mol. Ther. 2022, 30, 311–326. [Google Scholar] [CrossRef] [PubMed]
- Borgoyakova, M.B.; Karpenko, L.I.; Rudometov, A.P.; Volosnikova, E.A.; Merkuleva, I.A.; Starostina, E.V.; Zadorozhny, A.M.; Isaeva, A.A.; Nesmeyanova, V.S.; Shanshin, D.V.; et al. Self-Assembled Particles Combining SARS-CoV-2 RBD Protein and RBD DNA Vaccine Induce Synergistic Enhancement of the Humoral Response in Mice. Int. J. Mol. Sci. 2022, 23, 2188. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, R.; Gao, L.J.; Gao, X.F.; Wang, D.P.; Cao, J.M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Chi, X.; Yan, R.; Zhang, J.; Zhang, G.; Zhang, Y.; Hao, M.; Zhang, Z.; Fan, P.; Dong, Y.; Yang, Y.; et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 2020, 369, 650–655. [Google Scholar] [CrossRef]
- Lan, J.; Ge, J.; Yu, J.; Shan, S.; Zhou, H.; Fan, S.; Zhang, Q. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 2020, 581, 215–220. [Google Scholar] [CrossRef] [Green Version]
- Ju, B.; Zhang, Q.; Ge, J.; Wang, R.; Sun, J.; Ge, X.; Yu, J.; Shan, S.; Zhou, B.; Song, S.; et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 2020, 584, 115–119. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Wang, X.; Pascal, K.E.; Tomkins-Tinch, C.; Nyalile, T.P.; Wang, Y.; Baum, A.; Diehl, W.E.; Dauphin, A.; Carbone, C.; et al. Structural and Functional Analysis of the D614G SARS-CoV-2 Spike Protein Variant. Cell 2020, 183, 739–751.e8. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, X.; Rcheulishvili, N.; Cai, J.; Liu, C.; Xie, F.; Hu, X.; Yang, N.; Hou, M.; Papukashvili, D.; He, Y.; et al. Development of DNA Vaccine Candidate against SARS-CoV-2. Viruses 2022, 14, 1049. https://doi.org/10.3390/v14051049
Wang X, Rcheulishvili N, Cai J, Liu C, Xie F, Hu X, Yang N, Hou M, Papukashvili D, He Y, et al. Development of DNA Vaccine Candidate against SARS-CoV-2. Viruses. 2022; 14(5):1049. https://doi.org/10.3390/v14051049
Chicago/Turabian StyleWang, Xingyun, Nino Rcheulishvili, Jie Cai, Cong Liu, Fengfei Xie, Xing Hu, Nuo Yang, Mengqi Hou, Dimitri Papukashvili, Yunjiao He, and et al. 2022. "Development of DNA Vaccine Candidate against SARS-CoV-2" Viruses 14, no. 5: 1049. https://doi.org/10.3390/v14051049
APA StyleWang, X., Rcheulishvili, N., Cai, J., Liu, C., Xie, F., Hu, X., Yang, N., Hou, M., Papukashvili, D., He, Y., & Wang, P. G. (2022). Development of DNA Vaccine Candidate against SARS-CoV-2. Viruses, 14(5), 1049. https://doi.org/10.3390/v14051049







