Global Extension and Predominance of Human Metapneumovirus A2 Genotype with Partial G Gene Duplication
Abstract
:1. Introduction
2. Materials and Methods
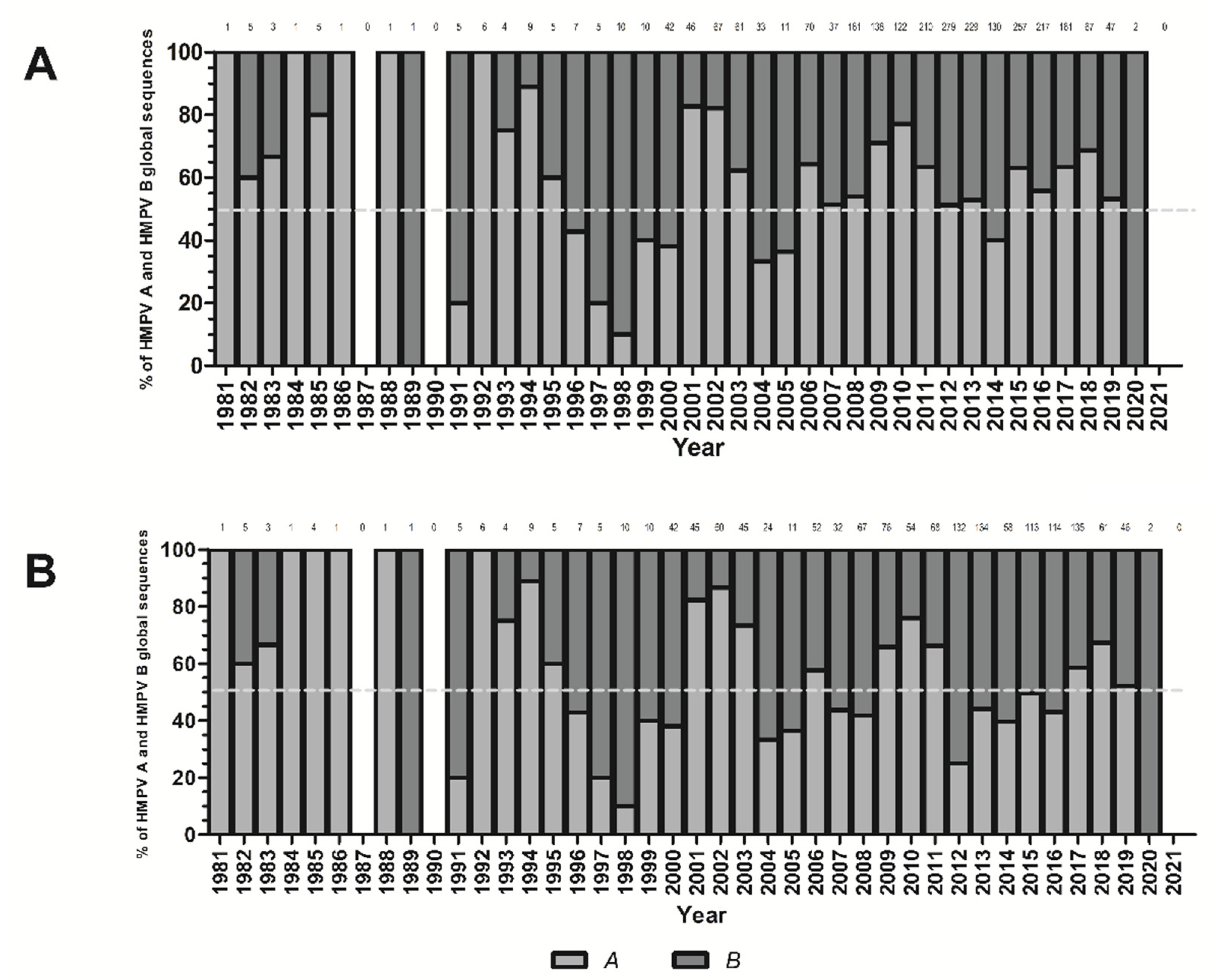
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Van Den Hoogen, B.G.; De Jong, J.C.; Groen, J.; Kuiken, T.; De Groot, R.; Fouchier, R.A.; Osterhaus, A.D. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat. Med. 2001, 7, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Shafagati, N.; Williams, J. Human metapneumovirus—What we know now. F1000Research 2018, 7, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nao, N.; Saikusa, M.; Sato, K.; Sekizuka, T.; Usuku, S.; Tanaka, N.; Nishimura, H.; Takeda, M. Recent Molecular Evolution of Human Metapneumovirus (HMPV): Subdivision of HMPV A2b Strains. Microorganisms 2020, 8, 1280. [Google Scholar] [CrossRef]
- Huck, B.; Scharf, G.; Neumann-Haefelin, D.; Puppe, W.; Weigl, J.; Falcone, V. Novel human metapneumovirus sublineage. Emerg. Infect. Dis. 2006, 12, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Nidaira, M.; Taira, K.; Hamabata, H.; Kawaki, T.; Gushi, K.; Mahoe, Y.; Maeshiro, N.; Azama, Y.; Okano, S.; Kyan, H.; et al. Molecular Epidemiology of Human Metapneumovirus from 2009 to 2011 in Okinawa, Japan. Jpn. J. Infect. Dis. 2012, 65, 337–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oketch, J.W.; Kamau, E.; Otieno, G.P.; Otieno, J.R.; Agoti, C.N.; Nokes, D.J. Human metapneumovirus prevalence and patterns of subgroup persistence identified through surveillance of pediatric pneumonia hospital admissions in coastal Kenya, 2007–2016. BMC Infect. Dis. 2019, 19, 757. [Google Scholar] [CrossRef] [Green Version]
- Piñana, M.; Vila, J.; Maldonado, C.; Galano-Frutos, J.J.; Valls, M.; Sancho, J.; Nuvials, F.X.; Andrés, C.; Martín-Gómez, M.T.; Esperalba, J.; et al. Insights into immune evasion of human metapneumovirus: Novel 180- and 111-nucleotide duplications within viral G gene throughout 2014–2017 seasons in Barcelona, Spain. J. Clin. Virol. 2020, 132, 104590. [Google Scholar] [CrossRef]
- Xie, Z.; Xu, J.; Ren, Y.; Cui, A.; Wang, H.; Song, J.; Zhang, Q.; Hu, M.; Xu, W.; Zhang, Y. Emerging Human Metapneumovirus Gene Duplication Variants in Patients with Severe Acute Respiratory Infection, China, 2017–2019. Emerg. Infect. Dis. 2021, 27, 275–277. [Google Scholar] [CrossRef]
- Jagusic, M.; Slovic, A.; Ivancic-Jelecki, J.; Ljubin-Sternak, S.; Vilibić-Čavlek, T.; Tabain, I.; Forcic, D. Molecular epidemiology of human respiratory syncytial virus and human metapneumovirus in hospitalized children with acute respiratory infections in Croatia, 2014–2017. Infect. Genet. Evol. 2019, 76, 104039. [Google Scholar] [CrossRef]
- Regev, L.; Meningher, T.; Hindiyeh, M.; Mendelson, E.; Mandelboim, M. Increase Human Metapneumovirus Mediated Morbidity following Pandemic Influenza Infection. PLoS ONE 2012, 7, e34750. [Google Scholar] [CrossRef] [Green Version]
- Neemuchwala, A.; Duvvuri, V.R.; Marchand-Austin, A.; Li, A.; Gubbay, J.B. Human metapneumovirus prevalence and molecular epidemiology in respiratory outbreaks in Ontario, Canada: HMPV Outbreaks in Long-Term Care Facilities, Ontario. J. Med. Virol. 2015, 87, 269–274. [Google Scholar] [CrossRef] [PubMed]
- Chow, W.Z.; Chan, Y.F.; Oong, X.Y.; Ng, L.J.; Nor’E, S.S.; Ng, K.T.; Chan, K.G.; Hanafi, N.S.; Pang, Y.K.; Kamarulzaman, A.; et al. Genetic diversity, seasonality and transmission network of human metapneumovirus: Identification of a unique sub-lineage of the fusion and attachment genes. Sci. Rep. 2016, 6, 27730. [Google Scholar] [CrossRef] [PubMed]
- Oong, X.Y.; Chook, J.B.; Ng, K.T.; Chow, W.Z.; Chan, K.G.; Hanafi, N.S.; Pang, Y.K.; Chan, Y.F.; Kamarulzaman, A.; Tee, K.K. The role of human Metapneumovirus genetic diversity and nasopharyngeal viral load on symptom severity in adults. Virol. J. 2018, 15, 91. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Lewandowski, K.; Jeffery, K.; Downs, L.O.; Foster, D.; Sanderson, N.D.; Kavanagh, J.; Vaughan, A.; Salvagno, C.; Vipond, R.; et al. Nanopore metagenomic sequencing to investigate nosocomial transmission of human metapneumovirus from a unique genetic group among haematology patients in the United Kingdom. J. Infect. 2020, 80, 571–577. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oketch, J.W.; Kamau, E.; Otieno, J.R.; Mwema, A.; Lewa, C.; Isoe, E.; Nokes, D.J.; Agoti, C.N. Comparative analysis of spatial-temporal patterns of human metapneumovirus and respiratory syncytial virus in Africa using genetic data, 2011–2014. Virol. J. 2021, 18, 104. [Google Scholar] [CrossRef]
- Saikusa, M.; Kawakami, C.; Nao, N.; Takeda, M.; Usuku, S.; Sasao, T.; Nishimoto, K.; Toyozawa, T. 180-Nucleotide Duplication in the G Gene of Human metapneumovirus A2b Subgroup Strains Circulating in Yokohama City, Japan, since 2014. Front. Microbiol. 2017, 8, 402. [Google Scholar] [CrossRef] [Green Version]
- Saikusa, M.; Nao, N.; Kawakami, C.; Usuku, S.; Sasao, T.; Toyozawa, T.; Takeda, M.; Okubo, I. A novel 111-nucleotide duplication in the G gene of human metapneumovirus. Microbiol. Immunol. 2017, 61, 507–512. [Google Scholar] [CrossRef]
- Trento, A.; Galiano, M.; Videla, C.; Carballal, G.; García-Barreno, B.; Melero, J.A.; Palomo, C. Major changes in the G protein of human respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J. Gen. Virol. 2003, 84, 3115–3120. [Google Scholar] [CrossRef]
- Eshaghi, A.; Duvvuri, V.R.; Lai, R.; Nadarajah, J.T.; Li, A.; Patel, S.N.; Low, D.E.; Gubbay, J.B. Genetic Variability of Human Respiratory Syncytial Virus A Strains Circulating in Ontario: A Novel Genotype with a 72 Nucleotide G Gene Duplication. PLoS ONE 2012, 7, e32807. [Google Scholar] [CrossRef] [Green Version]
- Cantú-Flores, K.; Rivera-Alfaro, G.; Muñoz-Escalante, J.C.; Noyola, D.E. Global distribution of respiratory syncytial virus A and B infections: A systematic review. Pathog. Glob. Health 2022, 1–12. [Google Scholar] [CrossRef]
- Kenmoe, S.; Vernet, M.A.; Penlap Beng, V.; Vabret, A.; Njouom, R. Phylogenetic variability of Human Metapneumovirus in patients with acute respiratory infections in Cameroon, 2011–2014. J. Infect. Public Health. 2020, 13, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Du, L.N.; Zhang, Z.Y.; Qin, X.; Yang, X.; Liu, P.; Chen, X.; Zhao, Y.; Liu, E.M.; Zhao, X.D. Detection and genetic diversity of human metapneumovirus in hospitalized children with acute respiratory infections in Southwest China. J. Clin. Microbiol. 2012, 50, 2714–2719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, A.S.; Roy, T.; Ghosh, S.; Chawla-Sarkar, M. Genetic variability of attachment (G) and fusion (F) protein genes of human metapneumovirus strains circulating during 2006–2009 in Kolkata, Eastern India. Virol. J. 2011, 8, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, R.; Guo, C.; Zhao, L.; Deng, J.; Wang, F.; Sun, Y.; Qian, Y. Epidemiological and genetic characteristics of human metapneumovirus in pediatric patients across six consecutive seasons in Beijing, China. Int. J. Infect. Dis. 2020, 91, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Perchetti, G.A.; Wilcox, N.; Chu, H.Y.; Katz, J.; Khatry, S.K.; LeClerq, S.C.; Tielsch, J.M.; Jerome, K.R.; Englund, J.A.; Kuypers, J. Human Metapneumovirus Infection and Genotyping of Infants in Rural Nepal. J. Pediatric. Infect. Dis. Soc. 2021, 10, 408–416. [Google Scholar] [CrossRef]
- Al-Turab, M.; Chehadeh, W.; Al-Nakib, W. Phylogenetic analysis of human metapneumovirus detected in hospitalized patients in Kuwait during the years 2009–2011. J. Infect. Public Health 2015, 8, 448–457. [Google Scholar] [CrossRef] [Green Version]
- Rahman, M.Z.; Sumiya, M.; Sahabuddin, M.; Pell, L.G.; Gubbay, J.B.; Rahman, R.; Momtaz, F.; Azmuda, N.; Shanta, S.S.; Jahan, I.; et al. Genetic characterization of human metapneumovirus identified through community and facility-based surveillance of infants in Dhaka, Bangladesh. J. Med. Virol. 2019, 91, 549–554. [Google Scholar] [CrossRef]
- Amer, H.M. Molecular epidemiology of human metapneumovirus in Riyadh Province, Saudi Arabia. J. Mol. Microbiol. Biotechnol. 2016, 26, 414–421. [Google Scholar] [CrossRef]
- Ábrego, L.E.; Mirazo, S.; Delfraro, A.; Franco, D.; Castillo, M.; Gaitán, M.; Castillo, J.; Moreno, B.; Pascale, J.M.; Arbiza, J. Genotypes of human metapneumovirus circulating during 2010–2012 in children from Panama. J. Med. Virol. 2018, 90, 604–608. [Google Scholar] [CrossRef]
- Jallow, M.M.; Fall, A.; Kiori, D.; Sy, S.; Goudiaby, D.; Barry, M.A.; Fall, M.; Niang, M.N.; Dia, N. Epidemiological, clinical and genotypic features of human Metapneumovirus in patients with influenza-like illness in Senegal, 2012 to 2016. BMC Infect. Dis. 2019, 19, 457. [Google Scholar] [CrossRef] [Green Version]
- Saikusa, M.; Nao, N.; Kawakami, C.; Usuku, S.; Tanaka, N.; Tahara, M.; Takeda, M.; Okubo, I. Predominant detection of the subgroup a2b human metapneumovirus strain with a 111-nucleotide duplication in the g gene in Yokohama city, Japan in 2018. Jpn. J. Infect. Dis. 2019, 72, 350–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, L.; Chen, L.; Xu, D.; Wu, X. Molecular typing and epidemiologic profiles of human metapneumovirus infection among children with severe acute respiratory infection in Huzhou, China. Mol. Biol. Rep. 2021, 48, 7697–7702. [Google Scholar] [CrossRef] [PubMed]
- Galiano, M.; Trento, A.; Ver, L.; Carballal, G.; Videla, C. Genetic heterogeneity of G and F protein genes from Argentinean human metapneumovirus strains. J. Med. Virolo. 2006, 78, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Van den Hoogen, B.G.; Herfst, S.; Sprong, L.; Cane, P.A.; Forleo-Neto, E.; De Swart, R.L.; Osterhaus, A.D.; Fouchier, R.A. Antigenic and Genetic Variability of Human Metapneumoviruses. Emerg. Infect. Dis. 2004, 10, 658–666. [Google Scholar] [CrossRef] [PubMed]
- Yi, L.; Zou, L.; Peng, J.; Yu, J.; Song, Y.; Liang, L.; Guo, Q.; Kang, M.; Ke, C.; Song, T.; et al. Epidemiology, evolution and transmission of human metapneumovirus in Guangzhou China, 2013–2017. Sci. Rep. 2019, 9, 14022. [Google Scholar] [CrossRef] [Green Version]
- Reiche, J.; Jacobsen, S.; Neubauer, K.; Hafemann, S.; Nitsche, A.; Milde, J.; Wolff, T.; Schweiger, B. Human Metapneumovirus: Insights from a Ten-Year Molecular and Epidemiological Analysis in Germany. PLoS ONE 2014, 9, e88342. [Google Scholar] [CrossRef] [Green Version]
- Lo Presti, A.; Cammarota, R.; Apostoli, P.; Cella, E.; Fiorentini, S.; Babakir-Mina, M.; Ciotti, M.; Ciccozzi, M. Genetic Variability and Circulation Pattern of Human Metapneumovirus Isolated in Italy over Five Epidemic Seasons. New Microbiol. 2011, 34, 337–344. [Google Scholar]
- Kong, W.; Wang, Y.; Zhu, H.; Lin, X.; Yu, B.; Hu, Q.; Yang, X.; Guo, D.; Peng, J.; Zhou, D. Circulation of human metapneumovirus among children with influenza-like illness in Wuhan, China. J. Med. Virol. 2016, 88, 774–781. [Google Scholar] [CrossRef]
- Duvvuri, V.R.; Granados, A.; Rosenfeld, P.; Bahl, J.; Eshaghi, A.; Gubbay, J.B. Genetic diversity and evolutionary insights of respiratory syncytial virus A ON1 genotype: Global and local transmission dynamics. Sci. Rep. 2015, 5, srep14268. [Google Scholar] [CrossRef] [Green Version]
- Korsun, N.S.; Angelova, S.G.; Trifonova, I.T.; Voleva, S.E.; Grigorova, I.G.; Tzotcheva, I.S.; Mileva, S.D.; Perenovska, P.I. The Prevalence and Genetic Characterization of Human Metapneumovirus in Bulgaria, 2016–2019. Intervirology 2021, 64, 194–202. [Google Scholar] [CrossRef]
- Rodriguez, P.E.; Frutos, M.C.; Adamo, M.P.; Cuffini, C.; Cámara, J.A.; Paglini, M.G.; Moreno, L.; Cámara, A. Human Metapneumovirus: Epidemiology and genotype diversity in children and adult patients with respiratory infection in Córdoba, Argentina. PLoS ONE 2020, 15, e0244093. [Google Scholar] [CrossRef] [PubMed]
- Van den Hoogen, B.G.; Osterhaus, D.M.E.; Fouchier, R.A.M. Clinical impact and diagnosis of human metapneumovirus infection. Pediatr. Infect. Dis. J. 2004, 23, S25–S32. [Google Scholar] [CrossRef] [PubMed]
- Maertzdorf, J.; Wang, C.K.; Brown, J.B.; Quinto, J.D.; Chu, M.; de Graaf, M.; van den Hoogen, B.G.; Spaete, R.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Real-Time Reverse Transcriptase PCR Assay for Detection of Human Metapneumoviruses from All Known Genetic Lineages. J. Clin. Microbiol. 2004, 42, 981–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Muñoz-Escalante, J.C.; Mata-Moreno, G.; Rivera-Alfaro, G.; Noyola, D.E. Global Extension and Predominance of Human Metapneumovirus A2 Genotype with Partial G Gene Duplication. Viruses 2022, 14, 1058. https://doi.org/10.3390/v14051058
Muñoz-Escalante JC, Mata-Moreno G, Rivera-Alfaro G, Noyola DE. Global Extension and Predominance of Human Metapneumovirus A2 Genotype with Partial G Gene Duplication. Viruses. 2022; 14(5):1058. https://doi.org/10.3390/v14051058
Chicago/Turabian StyleMuñoz-Escalante, Juan Carlos, Gabriel Mata-Moreno, Gerardo Rivera-Alfaro, and Daniel E. Noyola. 2022. "Global Extension and Predominance of Human Metapneumovirus A2 Genotype with Partial G Gene Duplication" Viruses 14, no. 5: 1058. https://doi.org/10.3390/v14051058
APA StyleMuñoz-Escalante, J. C., Mata-Moreno, G., Rivera-Alfaro, G., & Noyola, D. E. (2022). Global Extension and Predominance of Human Metapneumovirus A2 Genotype with Partial G Gene Duplication. Viruses, 14(5), 1058. https://doi.org/10.3390/v14051058





