Aptamer-Based High-Throughput Screening Model for Efficient Selection and Evaluation of Natural Ingredients against SGIV Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Virus, and Reagents
2.2. Aptamer Q2 to Monitor SGIV Infection
2.3. Confocal Fluorescence Imaging Analysis
2.4. Gene Expression Detection by RT-qPCR
2.5. Percentage Inhibition of SGIV Infection by Q2-AHTS or RT-qPCR
2.6. Statistical Analysis
3. Results
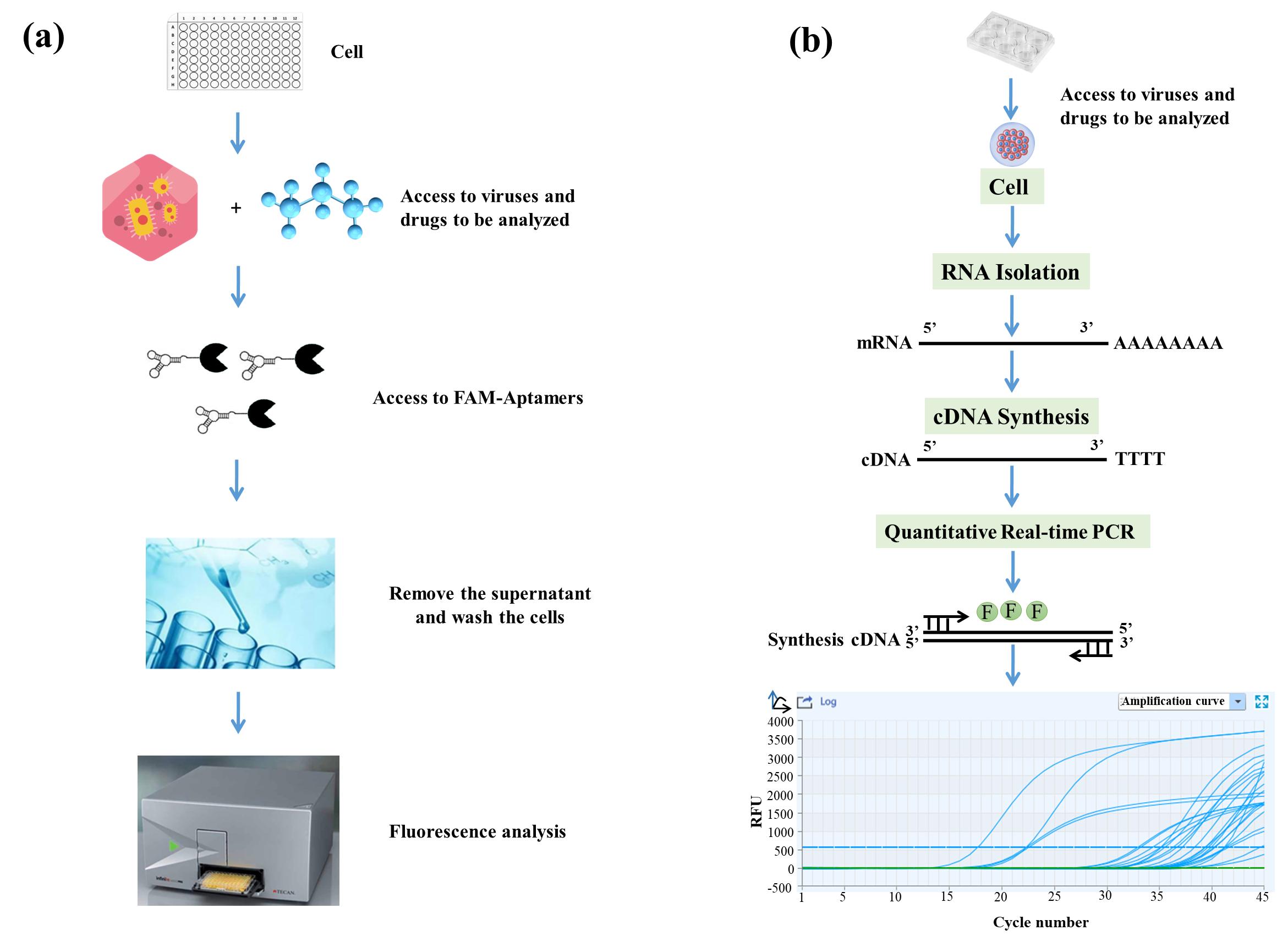
3.1. Flow Chart of Q2-AHTS and RT-qPCR Screening Technology
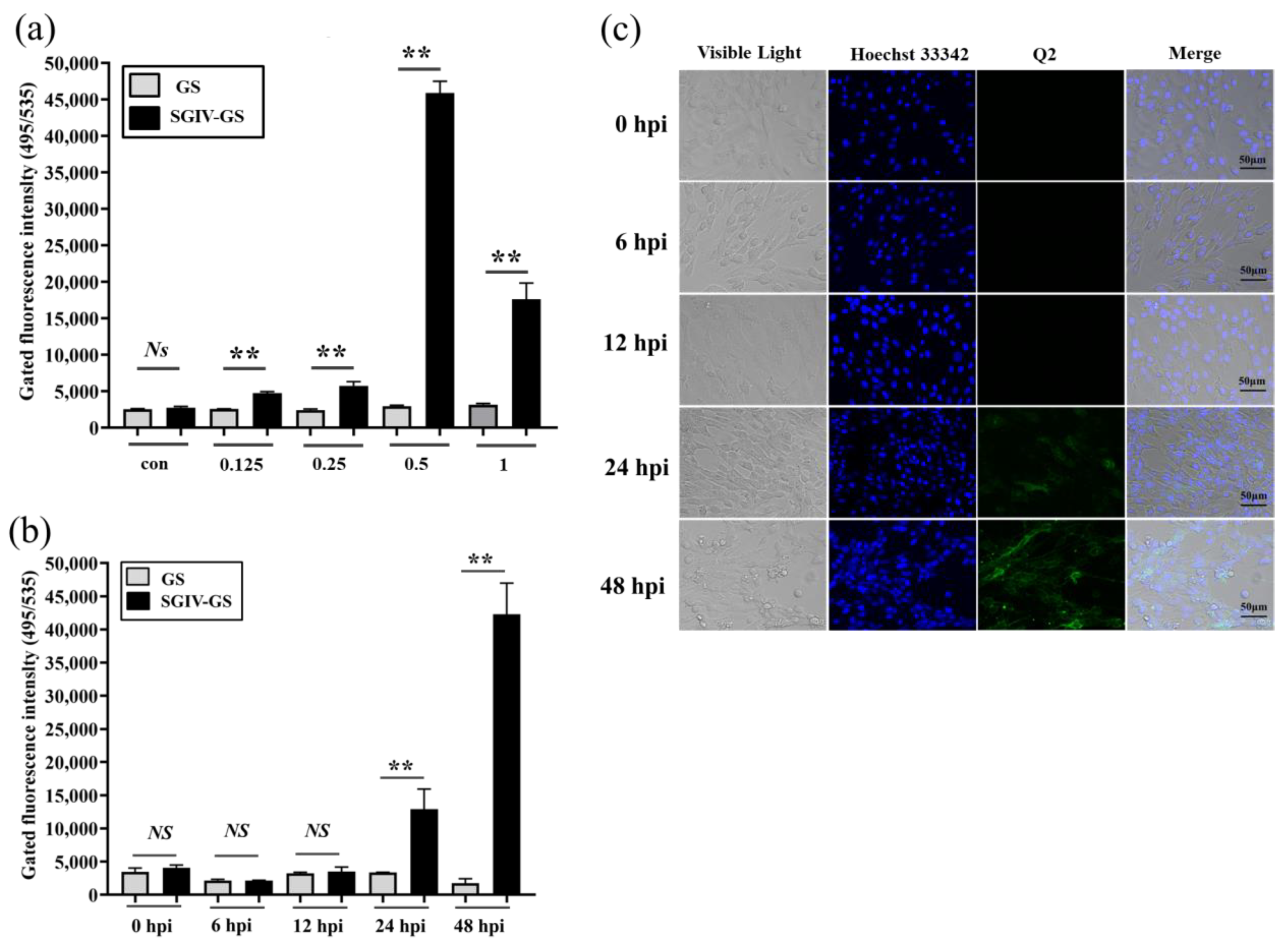
3.2. Fluorescent-Labeled Aptamer Probe for Specific Detection of SGIV Infection
3.3. RT-qPCR for Specific Detection Analysis of SGIV Infection
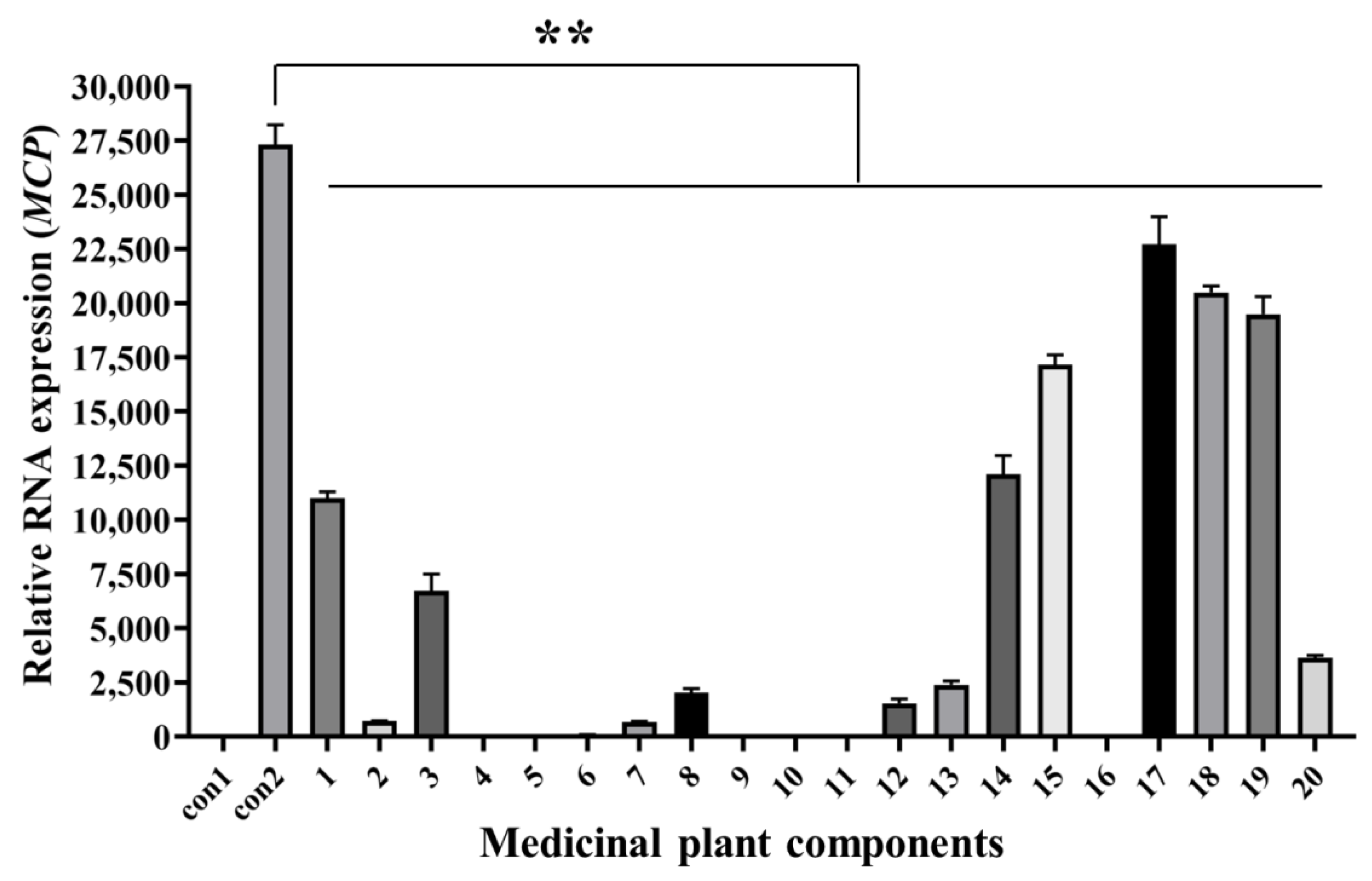
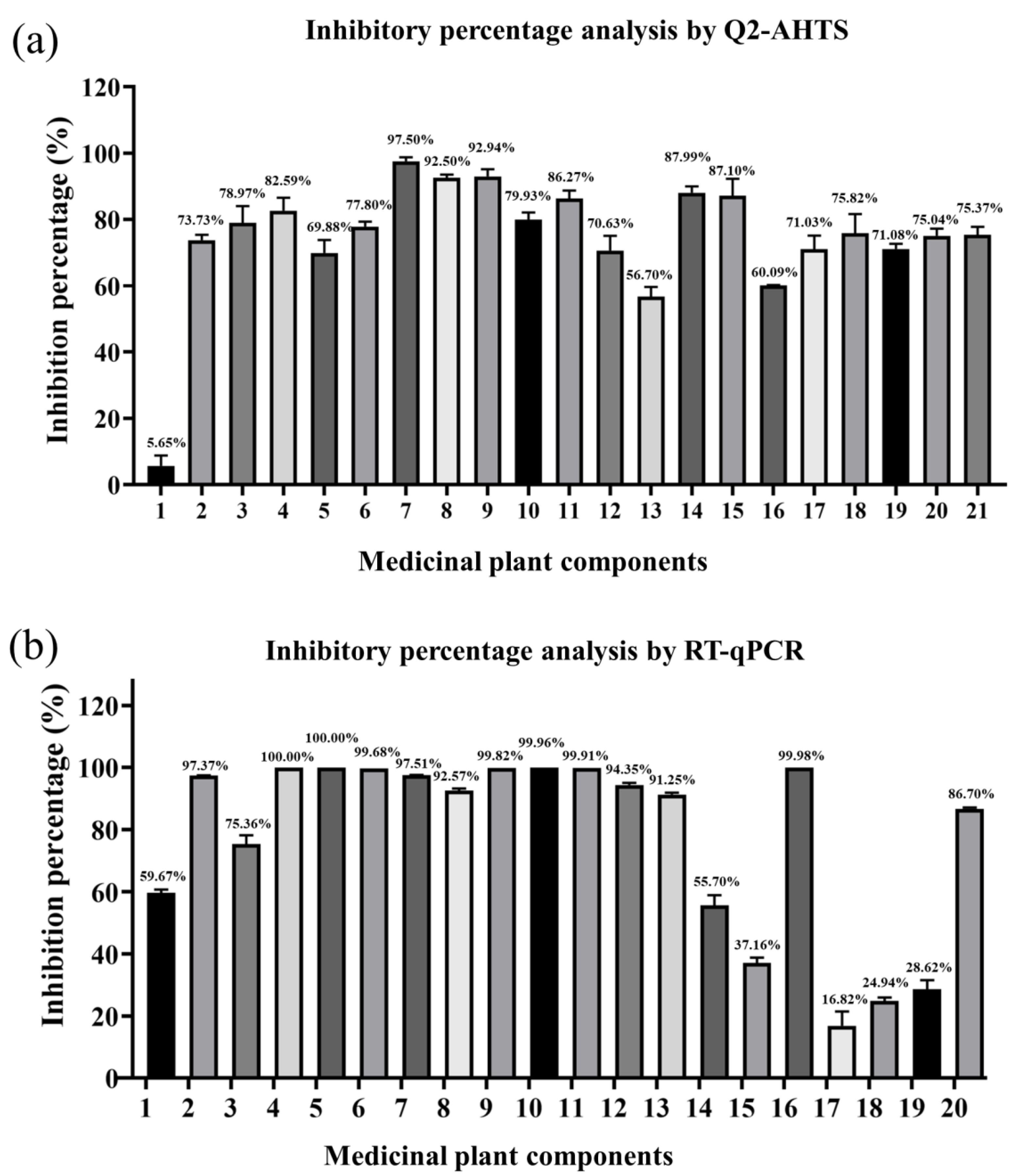
3.4. Antiviral Activity Analysis of Medicinal Plants Components with Q2-AHTS
3.5. Antiviral Activity Analysis of Medicinal Plants Components with RT-qPCR
3.6. Comparison of Inhibition Rates between Q2-AHTS and RT-qPCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, X.; Cui, L.; Li, S.; Han, x.; Jiang, K.; Yuan, X.; Yu, X. China Fisheries Statistical Yearbook 2021; Chinese Agriculture Express: Beijing, China, 2021. [Google Scholar]
- Xiao, H.; Liu, M.; Li, S.; Shi, D.; Zhu, D.; Ke, K.; Xu, Y.; Dong, D.; Zhu, L.; Yu, Q.; et al. Isolation and Characterization of a Ranavirus Associated with Disease Outbreaks in Cultured Hybrid Grouper (♀ Tiger Grouper Epinephelus fuscoguttatus × ♂ Giant Grouper E. lanceolatus) in Guangxi, China. J. Aquat. Anim. Health 2019, 31, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Whittington, R.J.; Becker, J.A.; Dennis, M.M. Iridovirus infections in finfish-critical review with emphasis on ranaviruses. J. Fish Dis. 2010, 33, 95–122. [Google Scholar] [CrossRef]
- Jancovich, J.K.; Michel, B.; Touchman, J.W.; Jacobs, B.L. Evidence for multiple recent host species shifts among the ranaviruses (family Iridoviridae). J. Virol. 2010, 84, 2636–2647. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.T.; Ather, A.; Thompson, K.D.; Gambari, R. Extracts and molecules from medicinal plants against herpes simplex viruses. Antivir. Res. 2005, 67, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Pan, H.; Li, M.; Miao, X.; Ding, H. Lonicera japonica Thunb.: Ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. J. Ethnopharmacol. 2011, 138, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, Q.; Xiao, H.; Yi, Y.; Cheng, H.; Putra, D.F.; Huang, Y.; Zhang, Q.; Li, P. Antiviral activity of Illicium verum Hook. f. extracts against grouper iridovirus infection. J. Fish Dis. 2020, 43, 531–540. [Google Scholar] [CrossRef]
- Liu, M.; Yu, Q.; Yi, Y.; Xiao, H.; Putra, D.; Ke, K.; Zhang, Q.; Li, P. Antiviral activities of Lonicera japonica Thunb. Components against grouper iridovirus in vitro and in vivo. Aquaculture 2020, 519, 734882. [Google Scholar] [CrossRef]
- Huang, L.; Li, Z.; Fu, Q.; Liang, C.; Liu, Z.; Liu, Q.; Pu, G.; Li, J. Genome-Wide Identification of CBL-CIPK Gene Family in Honeysuckle (Lonicera japonica Thunb.) and Their Regulated Expression Under Salt Stress. Front. Genet. 2021, 12, 751040. [Google Scholar] [CrossRef]
- Pu, H.; Li, X.; Du, Q.; Cui, H.; Xu, Y. Research progress in the application of Chinese herbal medicines in aquaculture: A review. Engineering 2017, 3, 731–737. [Google Scholar] [CrossRef]
- Akram, M.; Tahir, I.M.; Shah, S.M.A.; Mahmood, Z.; Altaf, A.; Ahmad, K.; Munir, N.; Daniyal, M.; Nasir, S.; Mehboob, H. Antiviral potential of medicinal plants against HIV, HSV, influenza, hepatitis, and coxsackievirus: A systematic review. Phytother. Res. 2018, 32, 811–822. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, X.; Xu, S.; Hou, C.; Xu, J.; Zhao, D.; Chen, Y. Antiviral Activities of a Medicinal Plant Extract Against Sacbrood Virus in Honeybees. Virol. J. 2021, 18, 83. [Google Scholar] [CrossRef]
- Cui, Q.; Du, R.; Liu, M.; Rong, L. Lignans and Their Derivatives from Plants as Antivirals. Molecules 2020, 25, 183. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.C.; Du, J.; But, P.P.; Deng, X.L.; Zhang, Y.W.; Ooi, V.E.; Xu, H.X.; Lee, S.H.; Lee, S.F. Antiviral Chinese medicinal herbs against respiratory syncytial virus. J. Ethnopharmacol. 2002, 79, 205–211. [Google Scholar] [CrossRef]
- Wang, X.; Jia, W.; Zhao, A.; Wang, X. Anti-influenza agents from plants and traditional Chinese medicine. Phytother. Res. 2006, 20, 335–341. [Google Scholar] [CrossRef]
- Li, P.; Zhou, L.; Wei, J.; Yu, Y.; Yang, M.; Wei, S.; Qin, Q. Development and characterization of aptamer-based enzyme-linked apta-sorbent assay for the detection of Singapore grouper iridovirus infection. J. Appl. Microbiol. 2016, 121, 634–643. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Lee, Y.B.; Lee, J.H.; Lee, D.H.; Cho, E.J.; Yu, S.J.; Kim, Y.J.; Kim, J.I.; Im, J.H.; Lee, J.H.; et al. Modified AS1411 Aptamer Suppresses Hepatocellular Carcinoma by Up-Regulating Galectin-14. PLoS ONE 2016, 11, e0160822. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Wei, S.; Zhou, L.; Yang, M.; Yu, Y.; Wei, J.; Jiang, G.; Qin, Q. Selection and characterization of novel DNA aptamers specifically recognized by Singapore grouper iridovirus-infected fish cells. J. Gen. Virol. 2015, 96, 3348–3359. [Google Scholar] [CrossRef]
- Syed, M.A.; Pervaiz, S. Advances in aptamers. Oligonucleotides 2010, 20, 215–224. [Google Scholar] [CrossRef] [PubMed]
- Kurt, H.; Eyüpoğlu, A.E.; Sütlü, T.; Budak, H.; Yüce, M. Plasmonic Selection of ssDNA Aptamers against Fibroblast Growth Factor Receptor. ACS Comb. Sci. 2019, 21, 578–587. [Google Scholar] [CrossRef] [PubMed]
- Yüce, M.; Kurt, H.; Hussain, B.; Ow-Yang, C.W.; Budak, H. Exploiting Stokes and anti-Stokes type emission profiles of aptamer-functionalized luminescent nanoprobes for multiplex sensing applications. Chem. Select. 2018, 3, 5814–5823. [Google Scholar] [CrossRef]
- Bai, C.; Lu, Z.; Jiang, H.; Yang, Z.; Liu, X.; Ding, H.; Li, H.; Dong, J.; Huang, A.; Fang, T.; et al. Aptamer selection and application in multivalent binding-based electrical impedance detection of inactivated H1N1 virus. Biosens. Bioelectron. 2018, 110, 162–167. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, Y.; Di, Y.; Xiu, C.; He, L.; Liao, S.; Li, D.; Huang, B. DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer. RSC Adv. 2019, 9, 950–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zamay, G.S.; Ivanchenko, T.I.; Zamay, T.N.; Grigorieva, V.L.; Glazyrin, Y.E.; Kolovskaya, O.S.; Garanzha, I.V.; Barinov, A.A.; Krat, A.V.; Mironov, G.G.; et al. DNA Aptamers for the Characterization of Histological Structure of Lung Adenocarcinoma. Mol. Ther. Nucleic Acids 2017, 6, 150–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, K.S. Nucleic acid aptamer-based methods for diagnosis of infections. Biosens. Bioelectron. 2018, 102, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Balogh, Z.; Lautner, G.; Bardoczy, V.; Komorowska, B.; Gyurcsanyi, R.E.; Meszaros, T. Selection and versatile application of virus-specific aptamers. FASEB J. 2010, 24, 4187–4195. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.J.; Platonova, O.; Stockley, P.G. Development of aptamer therapeutics. Curr. Opin. Pharmacol. 2010, 10, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Farokhzad, O.C. Aptamer-functionalized nanoparticles for medical applications: Challenges and opportunities. ACS Nano 2012, 6, 3670–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Q.; Liu, M.Z.; Wei, S.; Qin, X.; Qin, Q.; Li, P. Research progress and prospects for the use of aptamers in aquaculture biosecurity. Aquaculture 2021, 534, 736257. [Google Scholar] [CrossRef]
- Liu, M.; Yu, Q.; Xiao, H.; Li, M.; Huang, Y.; Zhang, Q.; Li, P. The inhibitory activities and antiviral mechanism of medicinal plant ingredient quercetin against grouper iridovirus infection. Front. Microbiol. 2020, 11, 586331. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–428. [Google Scholar] [CrossRef] [PubMed]
- Phan, N.; Hong, J.J.; Tofig, B.; Mapua, M.; Elashoff, D.; Moatamed, N.A.; Huang, J.; Memarzadeh, S.; Damoiseaux, R.; Soragni, A. A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids. Commun. Biol. 2019, 25, 78. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.; Liu, M.; Wei, S.; Xiao, H.; Wu, S.; Ke, K.; Huang, X.; Qin, Q.; Li, P. Identification of Major Capsid Protein as a Potential Biomarker of Grouper Iridovirus-Infected Cells Using Aptamers Selected by SELEX. Front. Microbiol. 2019, 10, 2684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, Q.W.; Chang, S.F.; Ngoh-Lim, G.H.; Gibson-Kueh, S.; Shi, C.; Lam, T.J. Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Dis. Aquat. Org. 2003, 53, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, R.; Yuan, B.; Liu, Y.; Liu, C. The antiviral and antimicrobial activities of licorice, a widely-used Chinese herb. Acta Pharm. Sin. 2015, 5, 310–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.F.; Hou, Y.L.; Huang, J.C.; Pan, W.Q.; Ma, Q.H.; Shi, Y.X.; Li, C.F.; Zhao, J.; Jia, Z.H.; Jiang, H.M.; et al. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2). Pharmacol. Res. 2020, 156, 104761. [Google Scholar]
- Takashita, E.; Kinoshita, N.; Yamayoshi, S.; Sakai-Tagawa, Y.; Fujisaki, S.; Ito, M.; Iwatsuki-Horimoto, K.; Chiba, S.; Halfmann, P.; Nagai, H.; et al. Efficacy of Antibodies and Antiviral Drugs against Covid-19 Omicron Variant. N. Engl. J. Med. 2022, 386, 995–998. [Google Scholar] [CrossRef]
- Bulfon, C.; Volpatti, D.; Galeotti, M. Current research on the use of plant-derived products in farmed fish. Aquac. Res. 2015, 46, 513–551. [Google Scholar] [CrossRef]
- Liu, M.; Xiao, H.; Wu, S.; Yu, Q.; Li, P. Aptamer-based high-throughput screening model for medicinal plant drugs against SGIV. J. Fish Dis. 2020, 43, 1479–1482. [Google Scholar] [CrossRef]
- Shi, K.Q.; Fan, Y.C.; Liu, W.Y.; Li, L.F.; Chen, Y.P.; Zheng, M.H. Traditional Chinese medicines benefit to nonalcoholic fatty liver disease: A systematic review and meta-analysis. Mol. Biol. Rep. 2012, 39, 9715–9722. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequences |
|---|---|
| qMCP-F | 5′-GCACGCTTCTCTCACCTTCA-3′ |
| qMCP-R | 5′-AACGGCAACGGGAGCACTA-3′ |
| β-actin-F | 5′-TACGAGCTGCCTGACGGACA-3 |
| β-actin-R | 5′-GGCTGTGATCTCCTTCTGCA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wei, H.; Guo, Z.; Long, Y.; Liu, M.; Xiao, J.; Huang, L.; Yu, Q.; Li, P. Aptamer-Based High-Throughput Screening Model for Efficient Selection and Evaluation of Natural Ingredients against SGIV Infection. Viruses 2022, 14, 1242. https://doi.org/10.3390/v14061242
Wei H, Guo Z, Long Y, Liu M, Xiao J, Huang L, Yu Q, Li P. Aptamer-Based High-Throughput Screening Model for Efficient Selection and Evaluation of Natural Ingredients against SGIV Infection. Viruses. 2022; 14(6):1242. https://doi.org/10.3390/v14061242
Chicago/Turabian StyleWei, Hongling, Zhongbao Guo, Yu Long, Mingzhu Liu, Jun Xiao, Lin Huang, Qing Yu, and Pengfei Li. 2022. "Aptamer-Based High-Throughput Screening Model for Efficient Selection and Evaluation of Natural Ingredients against SGIV Infection" Viruses 14, no. 6: 1242. https://doi.org/10.3390/v14061242
APA StyleWei, H., Guo, Z., Long, Y., Liu, M., Xiao, J., Huang, L., Yu, Q., & Li, P. (2022). Aptamer-Based High-Throughput Screening Model for Efficient Selection and Evaluation of Natural Ingredients against SGIV Infection. Viruses, 14(6), 1242. https://doi.org/10.3390/v14061242





