Epidemic and Evolutionary Characteristics of Swine Enteric Viruses in South-Central China from 2018 to 2021
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection and Pre-Treatment of Clinical Samples
2.2. Reverse Transcription PCR (RT-PCR)
2.3. Sanger Sequencing
2.4. Phylogenetic and Evolution Analysis
2.5. Cell Culture and Virus Isolation
2.6. Viral Growth Kinetics
2.7. Swine Enteric Virus Investigation
2.8. Statistical Analysis
3. Results
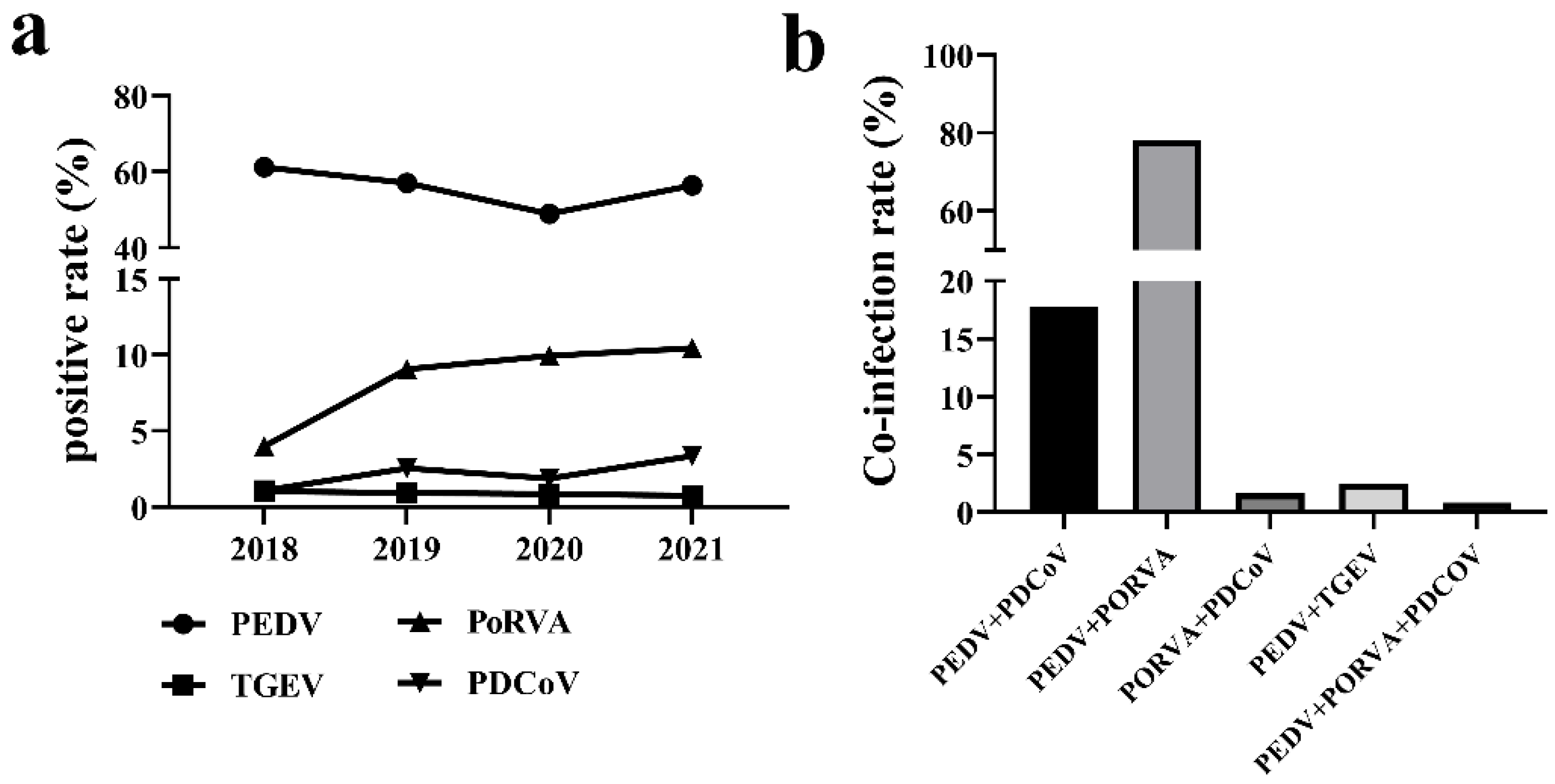
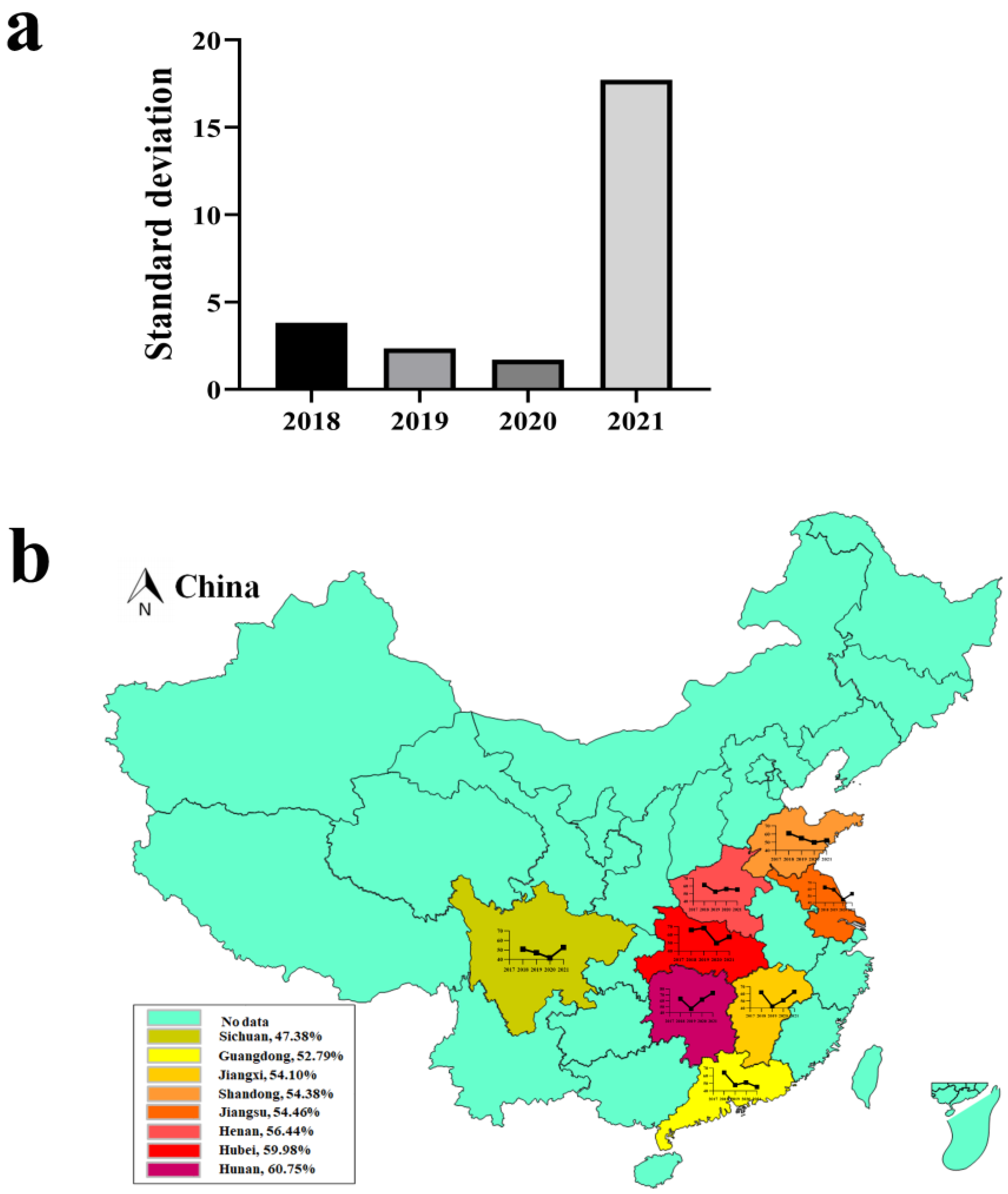
3.1. Positive Rate of Swine Enteric Viruses from 2018 to 2021
3.2. Seasonal Characteristics and Distribution of PEDV
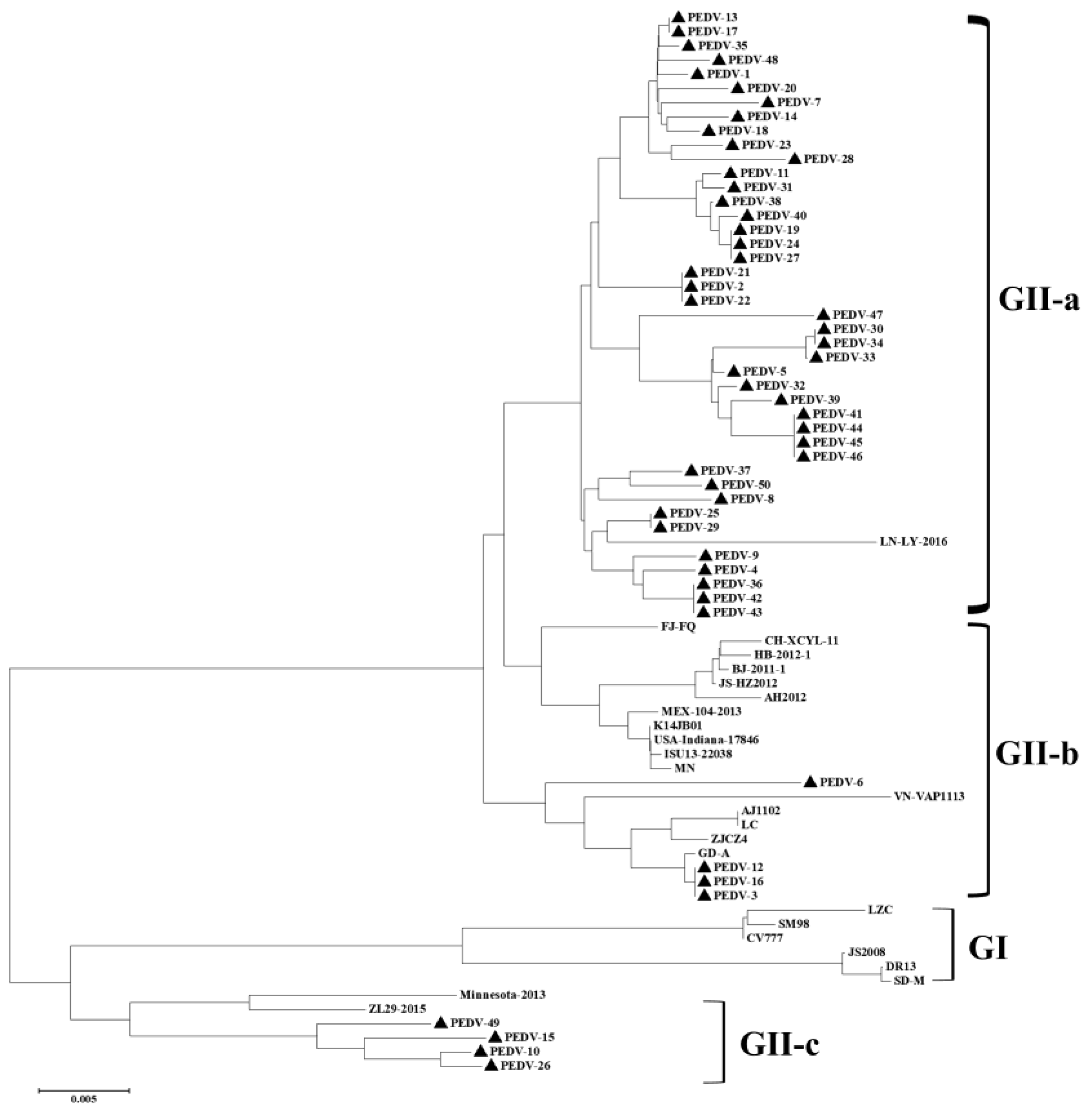
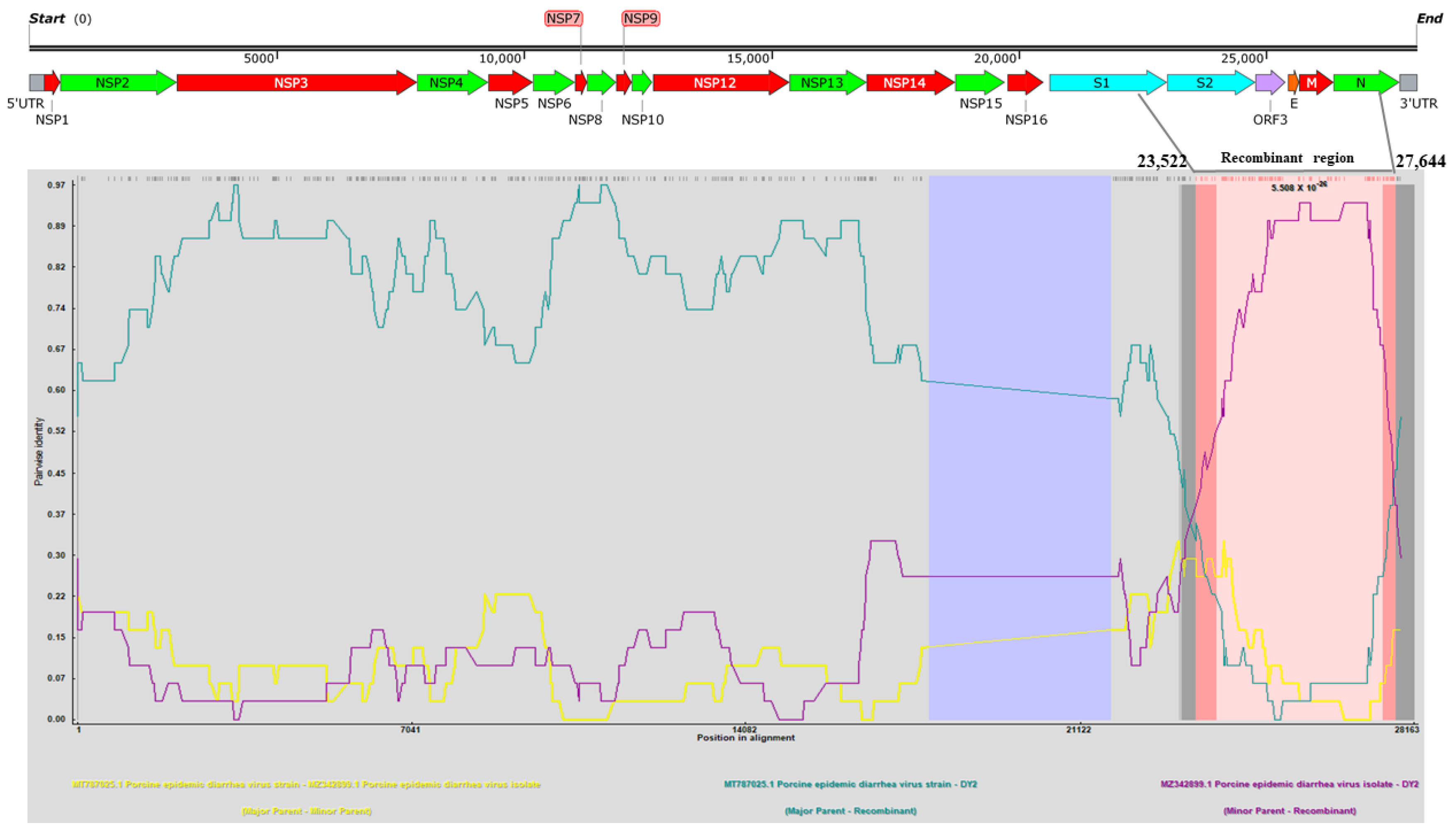
3.3. Phylogenetic and Recombination Analysis of PEDV
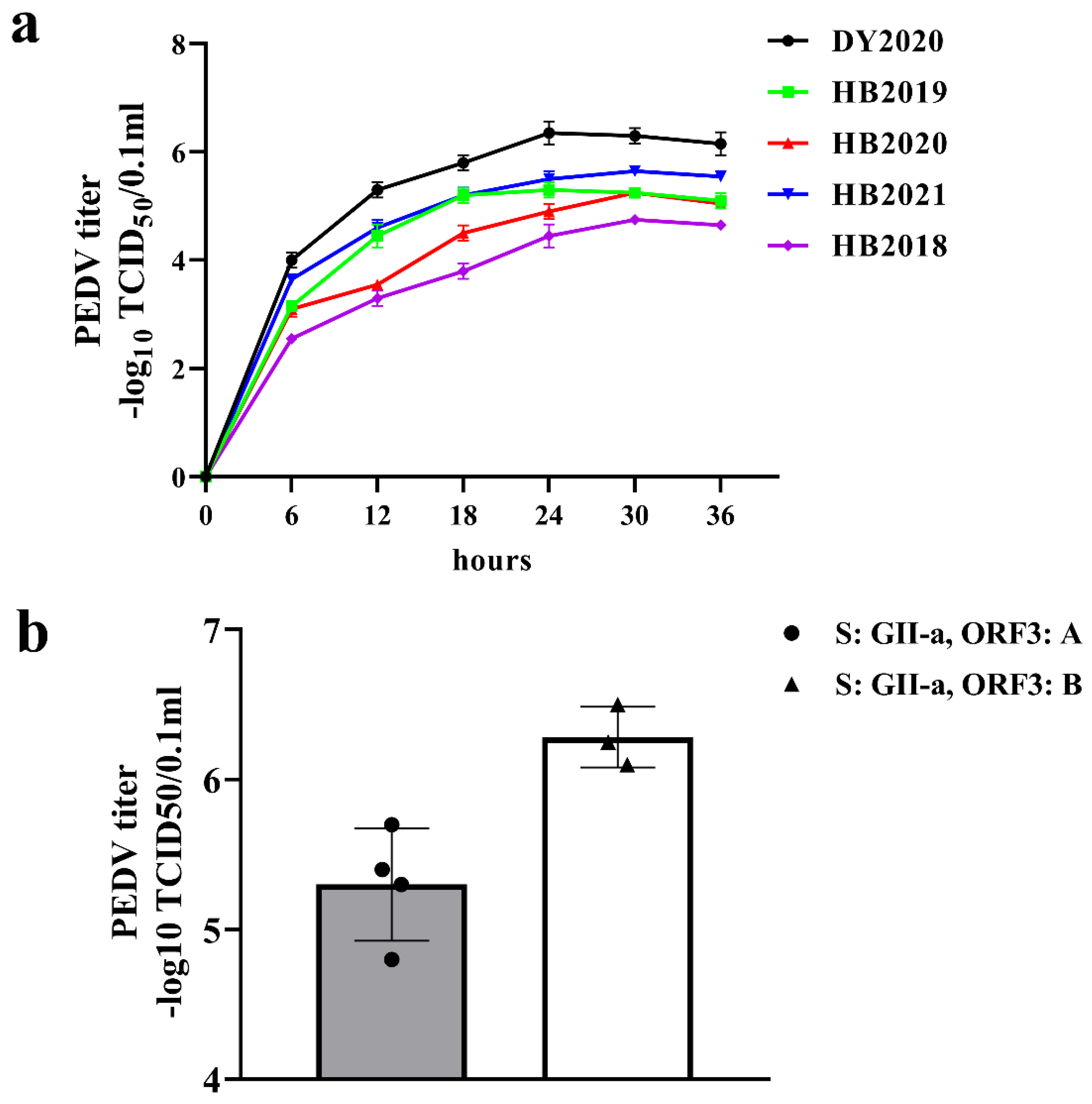
3.4. Isolation and Titer Analysis of PEDV
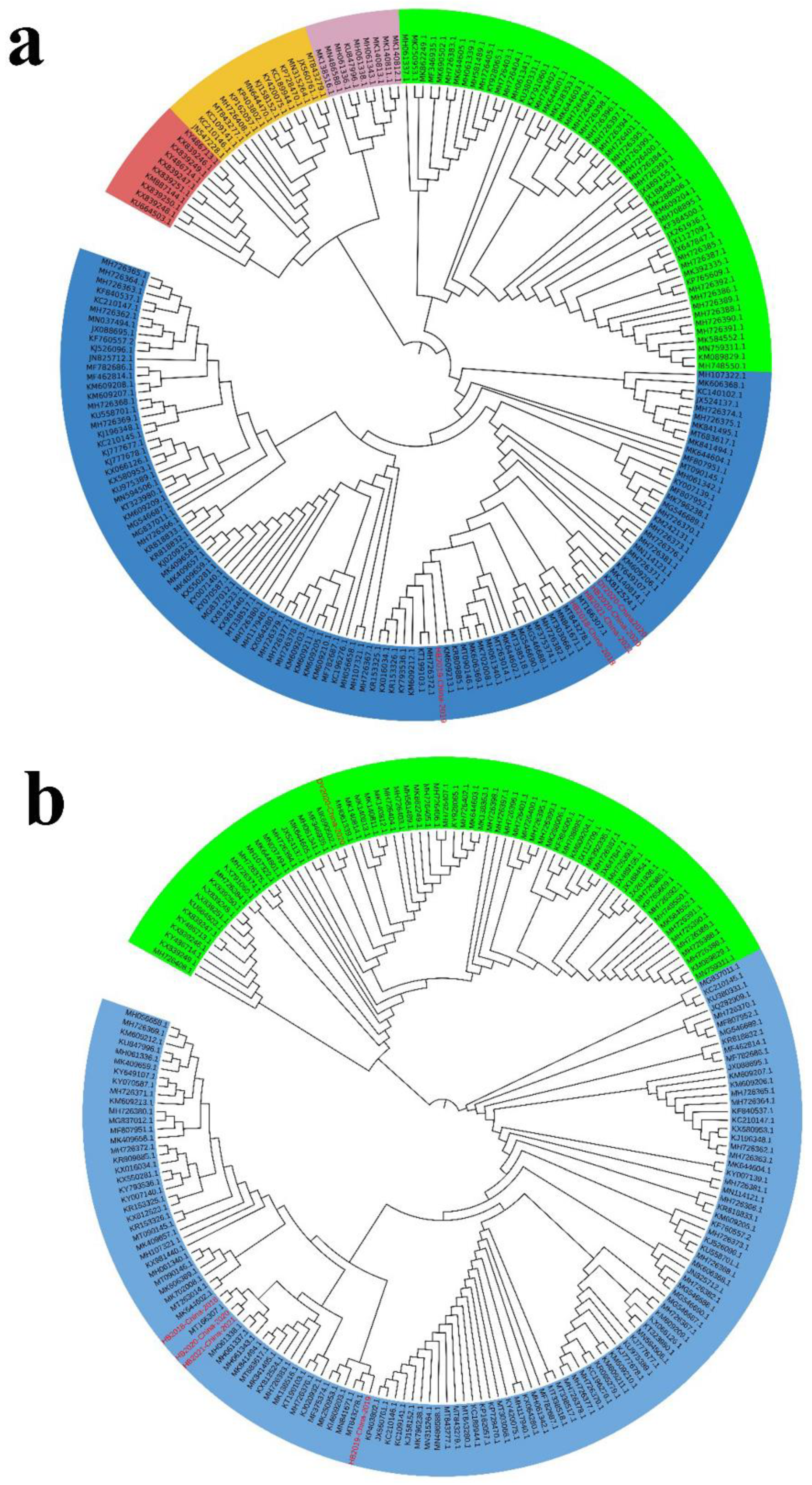
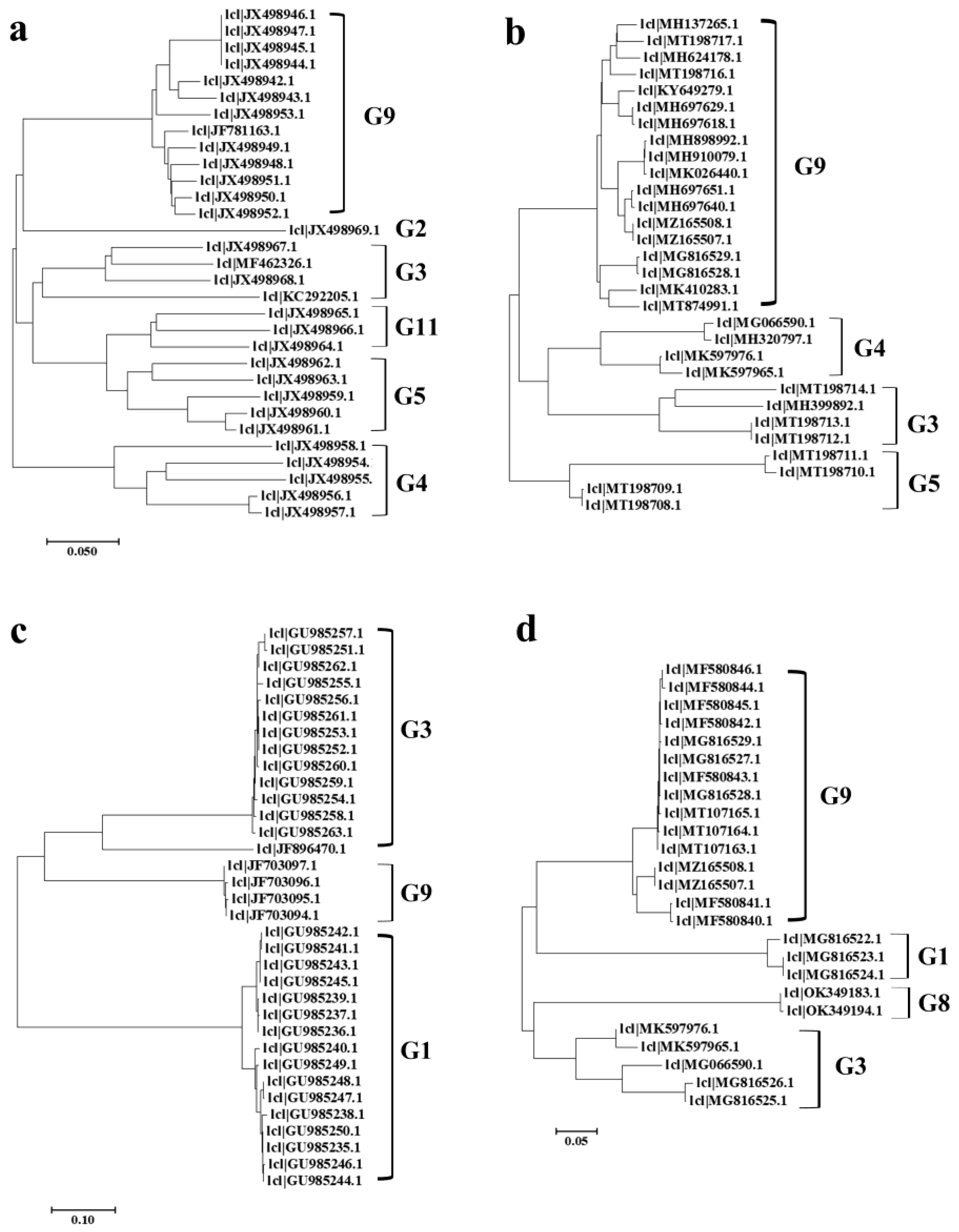
3.5. Phylogenetic Analysis of PoRVA
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pan, Z.; Lu, J.; Wang, N.; He, W.-T.; Zhang, L.; Zhao, W.; Su, S. Development of a TaqMan-probe-based multiplex real-time PCR for the simultaneous detection of emerging and reemerging swine coronaviruses. Virulence 2020, 11, 707–718. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.-L.; Wei, W.-K.; Li, X.-P.; Wen, X.-H.; Zhou, X.; Zhang, H.; Lv, D.-H.; Li, F.; Wang, D. Occurrence and sequence analysis of porcine deltacoronaviruses in southern China. Virol. J. 2016, 13, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353. [Google Scholar] [CrossRef]
- Cheng, S.; Wu, H.; Chen, Z. Evolution of Transmissible Gastroenteritis Virus (TGEV): A Codon Usage Perspective. Int. J. Mol. Sci. 2020, 21, 7898. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.-Q.; Cai, R.-J.; Chen, Y.-Q.; Liang, P.-S.; Chen, D.-K.; Song, C.-X. Outbreak of porcine epidemic diarrhea in suckling piglets, China. Emerg. Infect. Dis. 2012, 18, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Schwegmann-Wessels, C.; Herrler, G. Transmissible gastroenteritis virus infection: A vanishing specter. Dtsch. Tierarztl. Wochenschr. 2006, 113, 157–159. [Google Scholar]
- Gao, D.-M.; Yu, H.-Y.; Zhou, W.; Xia, B.-B.; Li, H.-Z.; Wang, M.-L.; Zhao, J. Inhibitory effects of recombinant porcine interferon-α on porcine transmissible gastroenteritis virus infections in TGEV-seronegative piglets. Vet. Microbiol. 2021, 252, 108930. [Google Scholar] [CrossRef]
- Jones, T.; Pritchard, G.; Paton, D. Transmissible gastroenteritis of pigs. Vet. Rec. 1997, 141, 427–428. [Google Scholar]
- Martella, V.; Bányai, K.; Matthijnssens, J.; Buonavoglia, C.; Ciarlet, M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010, 140, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Midgley, S.; Böttiger, B.; Jensen, T.G.; Friis-Møller, A.; Person, L.K.; Nielsen, L.; Barzinci, S.; Fischer, T.K. Human group A rotavirus infections in children in Denmark: Detection of reassortant G9 strains and zoonotic P[14] strains. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2014, 27, 114–120. [Google Scholar] [CrossRef]
- Midgley, S.E.; Bányai, K.; Buesa, J.; Halaihel, N.; Hjulsager, C.K.; Jakab, F.; Kaplon, J.; Larsen, L.E.; Monini, M.; Poljšak-Prijatelj, M.; et al. Diversity and zoonotic potential of rotaviruses in swine and cattle across Europe. Vet. Microbiol. 2012, 156, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Atii, D.J.; Ojeh, C.K.; Durojaiye, O.A. Detection of rotavirus antigen in diarrhoeic and non-diarrhoeic piglets in Nigeria. Rev. Elev. Med. Vet. Pays Trop. 1990, 42, 494–496. [Google Scholar] [PubMed]
- Zhou, Y.; Wu, Y.; Zhu, J.; Tong, W.; Yu, H.; Jiang, Y.; Tong, G. Complete genome sequence of a virulent porcine epidemic diarrhea virus strain. J. Virol. 2012, 86, 13862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerdts, V.; Zakhartchouk, A. Vaccines for porcine epidemic diarrhea virus and other swine coronaviruses. Vet. Microbiol. 2017, 206, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Pascual-Iglesias, A.; Sanchez, C.M.; Penzes, Z.; Sola, I.; Enjuanes, L.; Zuñiga, S. Recombinant Chimeric Transmissible Gastroenteritis Virus (TGEV)—Porcine Epidemic Diarrhea Virus (PEDV) Virus Provides Protection against Virulent PEDV. Viruses 2019, 11, 682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, T.; Gao, C.; Hao, J.; Lu, X.; Xie, K.; Wang, X.; Li, J.; Zhou, H.; Cui, W.; Shan, Z.; et al. Strategy of Developing Oral Vaccine Candidates Against Co-infection of Porcine Diarrhea Viruses Based on a Lactobacillus Delivery System. Front. Microbiol. 2022, 13, 872550. [Google Scholar] [CrossRef]
- Meng, F.; Ren, Y.; Suo, S.; Sun, X.; Li, X.; Li, P.; Yang, W.; Li, G.; Li, L.; Schwegmann-Wessels, C.; et al. Evaluation on the efficacy and immunogenicity of recombinant DNA plasmids expressing spike genes from porcine transmissible gastroenteritis virus and porcine epidemic diarrhea virus. PLoS ONE 2013, 8, e57468. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Fang, L.; Xiao, S. Porcine epidemic diarrhea in China. Virus Res. 2016, 226, 7–13. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Sun, Y.; Jiang, C.; Cao, H.; Zeng, W.; Zhang, X.; Li, Z.; He, Q. Porcine circovirus type 2 infection activates NF-κB pathway and cellular inflammatory responses through circPDCD4/miR-21/PDCD4 axis in porcine kidney 15 cell. Virus Res. 2021, 298, 198385. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.N.; Wang, L.; Zheng, D.Z.; Chen, S.; Shi, W.; Qiao, X.Y.; Jiang, Y.P.; Tang, L.J.; Xu, Y.G.; Li, Y.J. Oral immunization with a Lactobacillus casei-based anti-porcine epidemic diarrhoea virus (PEDV) vaccine expressing microfold cell-targeting peptide Co1 fused with the COE antigen of PEDV. J. Appl. Microbiol. 2018, 124, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Su, M.; Li, C.; Zhang, B.; Qi, S.; Sun, D.; Yin, B. Isolation and characterization of a variant subgroup GII-a porcine epidemic diarrhea virus strain in China. Microb. Pathog. 2020, 140, 103922. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Amimo, J.O.; Saif, L.J. Porcine Rotaviruses: Epidemiology, Immune Responses and Control Strategies. Viruses 2017, 9, 48. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Q.; Zhang, L.; Zhou, P.; Yang, J.; Fang, Y.; Dong, Z.; Zhao, D.; Li, W.; Feng, J.; et al. A newly isolated Chinese virulent genotype GIIb porcine epidemic diarrhea virus strain: Biological characteristics, pathogenicity and immune protective effects as an inactivated vaccine candidate. Virus Res. 2019, 259, 18–27. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G.; Wang, J.; Man, K.; Yang, Q. Cell attenuated porcine epidemic diarrhea virus strain Zhejiang08 provides effective immune protection attributed to dendritic cell stimulation. Vaccine 2017, 35, 7033–7041. [Google Scholar] [CrossRef]
- Bi, J.; Zeng, S.; Xiao, S.; Chen, H.; Fang, L. Complete genome sequence of porcine epidemic diarrhea virus strain AJ1102 isolated from a suckling piglet with acute diarrhea in China. J. Virol. 2012, 86, 10910–10911. [Google Scholar] [CrossRef] [Green Version]
- Gao, X.; Liu, T.; Liu, Y.; Xiao, J.; Wang, H. Transmission of African swine fever in China Through Legal Trade of Live Pigs. Transbound. Emerg. Dis. 2021, 68, 355–360. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Qi, W.; Yang, Y.; Liu, Z.; An, T.; Wu, X.; Chen, J. Prevention and Control Strategies of African Swine Fever and Progress on Pig Farm Repopulation in China. Viruses 2021, 13, 2552. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.; Li, J.; Fan, X.; Liu, F.; Li, L.; Wang, Q.; Ren, W.; Bao, J.; Liu, C.; Wang, H.; et al. Molecular Characterization of African Swine Fever Virus, China, 2018. Emerg. Infect. Dis. 2018, 24, 2131–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, Z.; Wang, L.; Cauchemez, S.; Xu, X.; Wang, X.; Cowling, B.J.; Meyers, L.A. Risk for Transportation of Coronavirus Disease from Wuhan to Other Cities in China. Emerg. Infect. Dis. 2020, 26, 1049–1052. [Google Scholar] [CrossRef] [Green Version]
- Flahault, A. Has China faced only a herald wave of SARS-CoV-2? Lancet 2020, 395, 947. [Google Scholar] [CrossRef]
- Li, Z.-L.; Zhu, L.; Ma, J.-Y.; Zhou, Q.-F.; Song, Y.-H.; Sun, B.-L.; Chen, R.-A.; Xie, Q.-M.; Bee, Y.-Z. Molecular characterization and phylogenetic analysis of porcine epidemic diarrhea virus (PEDV) field strains in south China. Virus Genes 2012, 45, 181–185. [Google Scholar] [CrossRef]
- Luo, L.; Chen, J.; Li, X.; Qiao, D.; Wang, Z.; Wu, X.; Du, Q.; Tong, D.; Huang, Y. Establishment of method for dual simultaneous detection of PEDV and TGEV by combination of magnetic micro-particles and nanoparticles. J. Infect. Chemother. Off. J. Jpn. Soc. Chemother. 2020, 26, 523–526. [Google Scholar] [CrossRef]
- Kong, F.; Xu, Y.; Ran, W.; Yin, B.; Feng, L.; Sun, D. Cold Exposure-Induced Up-Regulation of Hsp70 Positively Regulates PEDV mRNA Synthesis and Protein Expression In Vitro. Pathogens 2020, 9, 246. [Google Scholar] [CrossRef] [Green Version]
- Hsueh, F.-C.; Lin, C.-N.; Chiou, H.-Y.; Chia, M.-Y.; Chiou, M.-T.; Haga, T.; Kao, C.-F.; Chang, Y.-C.; Chang, C.-Y.; Jeng, C.-R.; et al. Updated phylogenetic analysis of the spike gene and identification of a novel recombinant porcine epidemic diarrhoea virus strain in Taiwan. Transbound. Emerg. Dis. 2020, 67, 417–430. [Google Scholar] [CrossRef]
- He, W.-T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography Reveals Association between Swine Trade and the Spread of Porcine Epidemic Diarrhea Virus in China and across the World. Mol. Biol. Evol. 2022, 39, msab364. [Google Scholar] [CrossRef]
- Zuo, Q.; Zhao, R.; Liu, J.; Zhao, Q.; Zhu, L.; Zhang, B.; Bi, J.; Yang, G.; Liu, J.; Yin, G. Epidemiology and phylogeny of spike gene of porcine epidemic diarrhea virus from Yunnan, China. Virus Res. 2018, 249, 45–51. [Google Scholar] [CrossRef]
- Yu, J.; Chai, X.; Cheng, Y.; Xing, G.; Liao, A.; Du, L.; Wang, Y.; Lei, J.; Gu, J.; Zhou, J. Molecular characteristics of the spike gene of porcine epidemic diarrhoea virus strains in Eastern China in 2016. Virus Res. 2018, 247, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Li, Y.; He, J.; Hu, Y.; Cai, X.; Liu, W.; Liu, T.; Wang, J.; Li, Z.; Yuan, X.; et al. Epidemic and genetic characterization of porcine epidemic diarrhea virus strains circulating in the regions around Hunan, China, during 2017-2018. Arch. Virol. 2020, 165, 877–889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, Y.; Yuan, W.; Peng, Q.; Zhang, F.; Ye, Y.; Huang, D.; Ding, Z.; Lin, L.; He, H.; et al. Evaluation of Cross-Protection between G1a- and G2a-Genotype Porcine Epidemic Diarrhea Viruses in Suckling Piglets. Animals 2020, 10, 1674. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.-T.; Qiao, H.; Hou, C.-Y.; Zheng, H.-H.; Li, X.-S.; Zheng, L.-L.; Chen, H.-Y. Characteristics of the spike and ORF3 genes of porcine epidemic diarrhea virus in Henan and Shanxi provinces of China. Arch. Virol. 2020, 165, 2323–2333. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Han, F.; Yan, X.; Liu, L.; Shu, X.; Hu, H. Prevalence and phylogenetic analysis of spike gene of porcine epidemic diarrhea virus in Henan province, China in 2015–2019. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2021, 88, 104709. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Shang, Y.; Wu, J.; Dong, Z.; Cao, X.; Liu, Y.; Lan, X. Rapid differentiation of PEDV wild-type strains and classical attenuated vaccine strains by fluorescent probe-based reverse transcription recombinase polymerase amplification assay. BMC Vet. Res. 2020, 16, 208. [Google Scholar] [CrossRef]
- Tian, Y.; Yu, Z.; Cheng, K.; Liu, Y.; Huang, J.; Xin, Y.; Li, Y.; Fan, S.; Wang, T.; Huang, G.; et al. Molecular characterization and phylogenetic analysis of new variants of the porcine epidemic diarrhea virus in Gansu, China in 2012. Viruses 2013, 5, 1991–2004. [Google Scholar] [CrossRef] [Green Version]
- Wen, F.; Yang, J.; Li, A.; Gong, Z.; Yang, L.; Cheng, Q.; Wang, C.; Zhao, M.; Yuan, S.; Chen, Y.; et al. Genetic characterization and phylogenetic analysis of porcine epidemic diarrhea virus in Guangdong, China, between 2018 and 2019. PLoS ONE 2021, 16, e0253622. [Google Scholar] [CrossRef]
- Li, D.; Li, Y.; Liu, Y.; Chen, Y.; Jiao, W.; Feng, H.; Wei, Q.; Wang, J.; Zhang, Y.; Zhang, G. Isolation and Identification of a Recombinant Porcine Epidemic Diarrhea Virus With a Novel Insertion in S1 Domain. Front. Microbiol. 2021, 12, 667084. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Zhang, Q.; Zhou, P.; Fang, Y.; Zhao, D.; Feng, J.; Li, W.; Zhang, Y.; Wang, Y. Evaluation and comparison of immunogenicity and cross-protective efficacy of two inactivated cell culture-derived GIIa- and GIIb-genotype porcine epidemic diarrhea virus vaccines in suckling piglets. Vet. Microbiol. 2019, 230, 278–282. [Google Scholar] [CrossRef]
- Xue, R.; Tian, Y.; Zhang, Y.; Zhang, M.; Li, Z.; Chen, S.; Liu, Q. Diversity of group A rotavirus of porcine rotavirus in Shandong province China. Acta Virol. 2018, 62, 229–234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maneekarn, N.; Khamrin, P. Rotavirus associated gastroenteritis in Thailand. Virusdisease 2014, 25, 201–207. [Google Scholar] [CrossRef] [Green Version]
- Mitui, M.T.; Chandrasena, T.N.; Chan, P.K.; Rajindrajith, S.; Nelson, E.A.S.; Leung, T.F.; Nishizono, A.; Ahmed, K. Inaccurate identification of rotavirus genotype G9 as genotype G3 strains due to primer mismatch. Virol. J. 2012, 9, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, N.; Tang, C.; Kan, R.; Feng, F.; Yue, H. Genome analysis of a G9P[23] group A rotavirus isolated from a dog with diarrhea in China. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 2019, 70, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Yan, N.; Yue, H.; Wang, Y.; Zhang, B.; Tang, C. Genomic analysis reveals G3P[13] porcine rotavirus A interspecific transmission to human from pigs in a swine farm with diarrhoea outbreak. J. Gen. Virol. 2021, 102, 001532. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.-W.; Yang, Y.-L.; Shi, C.-C.; Chen, Z.; Li, C.; Wang, Y.-M.; Li, L.-B.; Chen, J.-H. Molecular epidemiological characteristics of the virus in 96 children with acute diarrhea in Changdu of Tibet, China. Zhongguo Dang Dai Er Ke Za Zhi 2022, 24, 266–272. [Google Scholar] [CrossRef]
- Tian, Y.; Gao, Z.; Li, W.; Liu, B.; Chen, Y.; Jia, L.; Yan, H.; Wang, Q. Group A rotavirus prevalence and genotypes among adult outpatients with diarrhea in Beijing, China, 2011–2018. J. Med. Virol. 2021, 93, 6191–6199. [Google Scholar] [CrossRef]
- Zhao, L.; Shi, X.; Meng, D.; Guo, J.; Li, Y.; Liang, L.; Guo, X.; Tao, R.; Zhang, X.; Gao, R.; et al. Prevalence and genotype distribution of group A rotavirus circulating in Shanxi Province, China during 2015–2019. BMC Infect. Dis. 2021, 21, 94. [Google Scholar] [CrossRef]
- Jing, Z.; Zhang, X.; Shi, H.; Chen, J.; Shi, D.; Dong, H.; Feng, L. A G3P[13] porcine group A rotavirus emerging in China is a reassortant and a natural recombinant in the VP4 gene. Transbound. Emerg. Dis. 2018, 65, e317–e328. [Google Scholar] [CrossRef]
- Ahmed, S.F.; Mansour, A.M.; Klena, J.D.; Husain, T.S.; Hassan, K.A.; Mohamed, F.; Steele, D. Rotavirus genotypes associated with acute diarrhea in Egyptian infants. Pediatr. Infect. Dis. J. 2014, 33 (Suppl. S1), S62–S68. [Google Scholar] [CrossRef]
- Sadiq, A.; Bostan, N.; Bokhari, H.; Yinda, K.C.; Matthijnssens, J. Whole Genome Analysis of Selected Human Group A Rotavirus Strains Revealed Evolution of DS-1-Like Single- and Double-Gene Reassortant Rotavirus Strains in Pakistan During 2015–2016. Front. Microbiol. 2019, 10, 2641. [Google Scholar] [CrossRef] [PubMed]
- Phua, K.B.; Emmanuel, S.C.; Goh, P.; Quak, S.H.; Lee, B.W.; Han, H.H.; Ward, R.L.; Bernstein, D.I.; De Vos, B.; Bock, H.L. A rotavirus vaccine for infants: The Asian experience. Ann. Acad. Med. Singap. 2006, 35, 38–44. [Google Scholar] [PubMed]
| Year | Number of Samples | |||||||
|---|---|---|---|---|---|---|---|---|
| Henan | Hubei | Jiangsu | Shandong | Guangdong | Hunan | Jiangxi | Sichuan | |
| 2018 | 241 | 242 | 201 | 220 | 231 | 257 | 211 | 298 |
| 2019 | 225 | 217 | 177 | 211 | 203 | 221 | 197 | 271 |
| 2020 | 317 | 266 | 232 | 259 | 254 | 286 | 237 | 322 |
| 2021 | 188 | 147 | 141 | 167 | 153 | 171 | 148 | 196 |
| Primer Name | Sequence | Product Length | Target Gene |
|---|---|---|---|
| PDCoV-P1 | CCAGCAACCACTCGTGTTA | 620 bp | M gene |
| PDCoV-P2 | GTCCTTAGTTGGTTTGGTGGGT | ||
| PEDV-P2 | GTCTAACGGTTCTATTCCCG | 460 bp | M gene |
| PEDV-P3 | ATAGCCCTCTACAAGCAATG | ||
| TGEV-P5 | TTACAAACTCGCTATCGCATGG | 528 bp | N gene |
| TGEV-P6 | TCTTGTCACATCACCTTTACCTGC | ||
| PORVA-P7 | CCCCGGTATTGAATATACCACAGT | 333 bp | VP7 gene |
| PORVA-P8 | TTTCTGTTGGCCACCCTTTAGT |
| Primer Name | Sequence | Product Length | Purpose |
|---|---|---|---|
| S1-F | ATGAAGTCTTTAACGTACTTCTGG | 2208 bp | PEDV gene sequencing |
| S1-R | TAGAAGAAACCAGGCAACTCC | ||
| S2-F | ATGCATTCTAATGATGGCTCTAAT | 1953 bp | |
| S2-R | CTGCACGTGGACCTTTTCAAAAAC | ||
| ORF3-F | ATGTTTCTTGGACTTTTTCAGTACA | 675 bp | |
| ORF3-R | ACTAATTGTAGCATACTCGTCTAG |
| Year | Positive Rate (%) and (Number of Positive Samples/Number of Samples) | |||
|---|---|---|---|---|
| PEDV | TGEV | PoRVA | PDCoV | |
| 2018 | 61.02 (1160/1901) | 1.05 (20/1901) | 4.00 (76/1901) | 1.10 (21/1901) |
| 2019 | 57.03 (982/1722) | 0.93 (16/1722) | 9.06 (156/1722) | 2.56 (44/1722) |
| 2020 | 50.81 (1104/2173) | 0.87 (19/2173) | 9.94 (216/2173) | 1.89 (41/2173) |
| 2021 | 56.44 (740/1311) | 0.76 (10/1311) | 10.45 (137/1311) | 3.36 (44/1311) |
| Year | Positive Rate (%) and (Number of Positive Samples/Number of Samples) | |||
|---|---|---|---|---|
| Spring | Summer | Autumn | Winter | |
| 2018 | 63.14 (322/510) | 55.56 (210/378) | 59.34 (244/411) | 63.79 (384/602) |
| 2019 | 57.54 (271/471) | 54.76 (190/347) | 54.86 (209/381) | 59.66 (312/523) |
| 2020 | 52.87 (276/522) | 49.28 (239/485) | 49.60 (249/502) | 51.20 (340/664) |
| 2021 | 71.79 (313/436) | 36.88 (97/263) | 48.43 (139/287) | 58.76 (191/325) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, C.; Lu, H.; Geng, C.; Yang, K.; Liu, W.; Liu, Z.; Yuan, F.; Gao, T.; Wang, S.; Wen, P.; et al. Epidemic and Evolutionary Characteristics of Swine Enteric Viruses in South-Central China from 2018 to 2021. Viruses 2022, 14, 1420. https://doi.org/10.3390/v14071420
Li C, Lu H, Geng C, Yang K, Liu W, Liu Z, Yuan F, Gao T, Wang S, Wen P, et al. Epidemic and Evolutionary Characteristics of Swine Enteric Viruses in South-Central China from 2018 to 2021. Viruses. 2022; 14(7):1420. https://doi.org/10.3390/v14071420
Chicago/Turabian StyleLi, Chang, Hongyu Lu, Chao Geng, Keli Yang, Wei Liu, Zewen Liu, Fangyan Yuan, Ting Gao, Shuangshuang Wang, Ping Wen, and et al. 2022. "Epidemic and Evolutionary Characteristics of Swine Enteric Viruses in South-Central China from 2018 to 2021" Viruses 14, no. 7: 1420. https://doi.org/10.3390/v14071420







