Progress towards the Development of a Universal Influenza Vaccine
Abstract
:1. Introduction
2. The Need for a Universal Influenza Vaccine
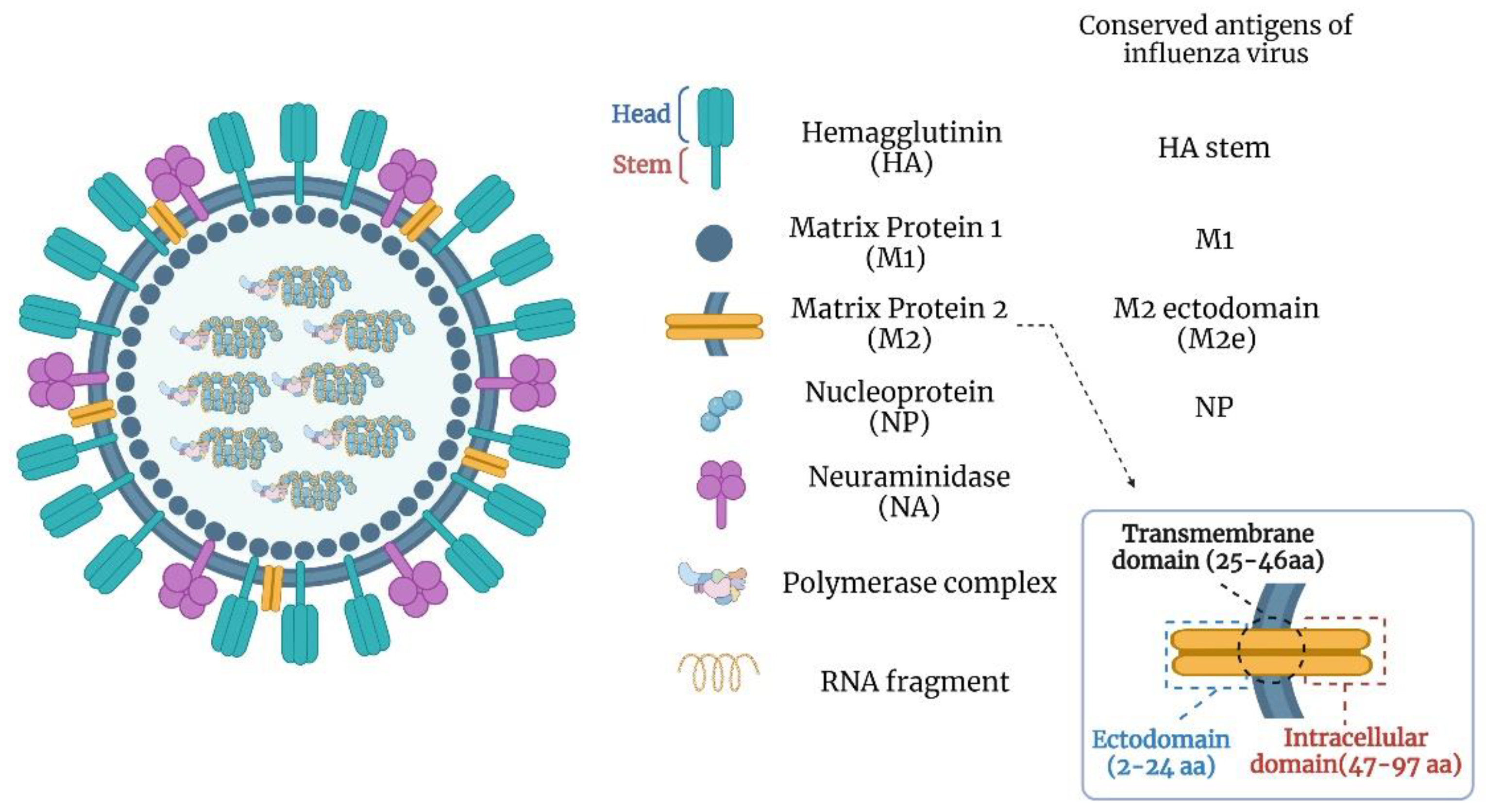
3. Universal Influenza Vaccine Targets
3.1. HA Stalk Domain
3.2. Matrix Protein 2 Ectodomain (M2e)
3.3. Nucleoprotein (NP)
3.4. Neuraminidase (NA)
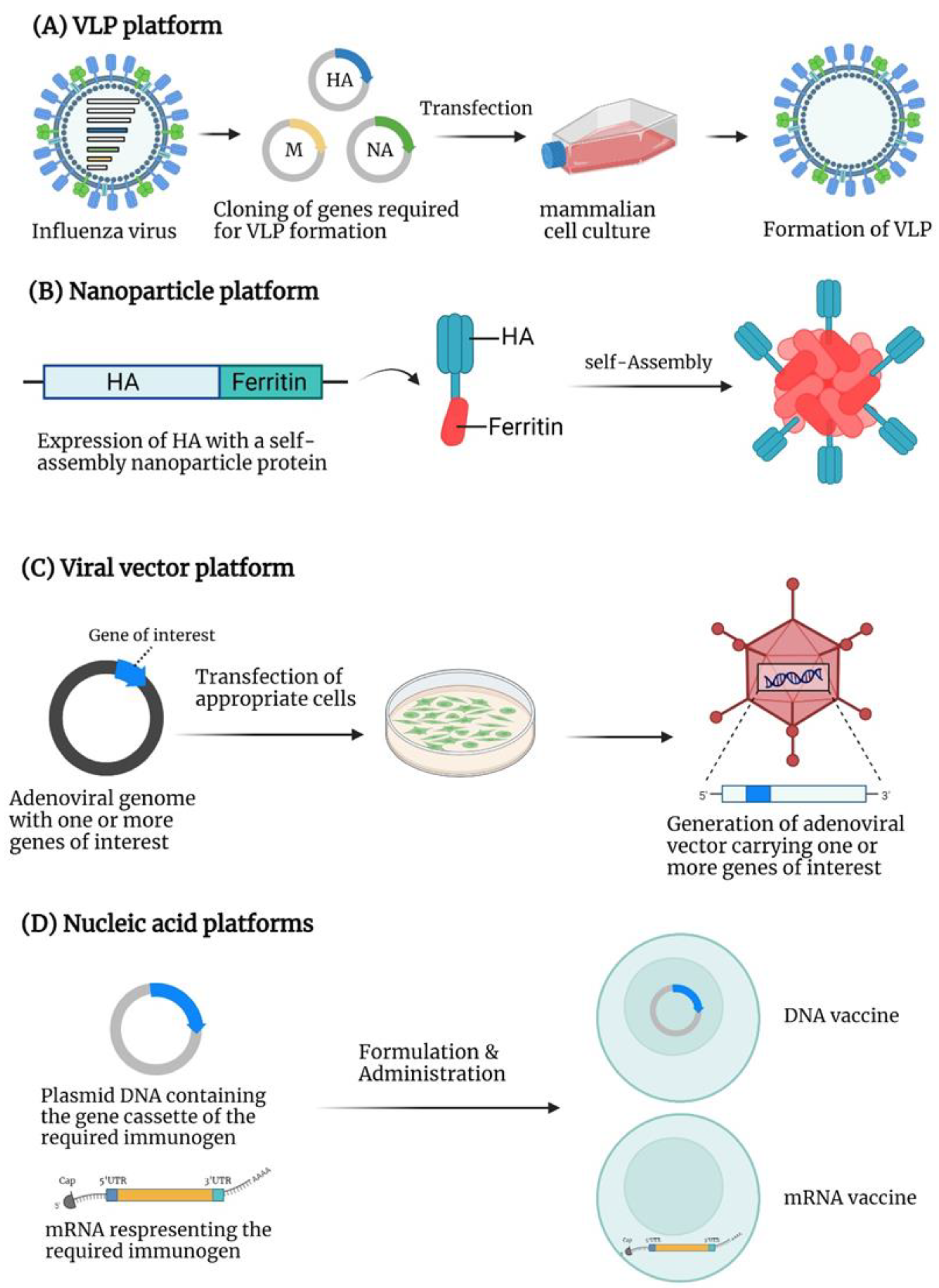
4. Virus-like Particle (VLP)-Based Vaccines
5. Nanoparticle-Based Vaccines
6. Viral Vector-Based Vaccines
7. Nucleic Acid-Based Vaccines
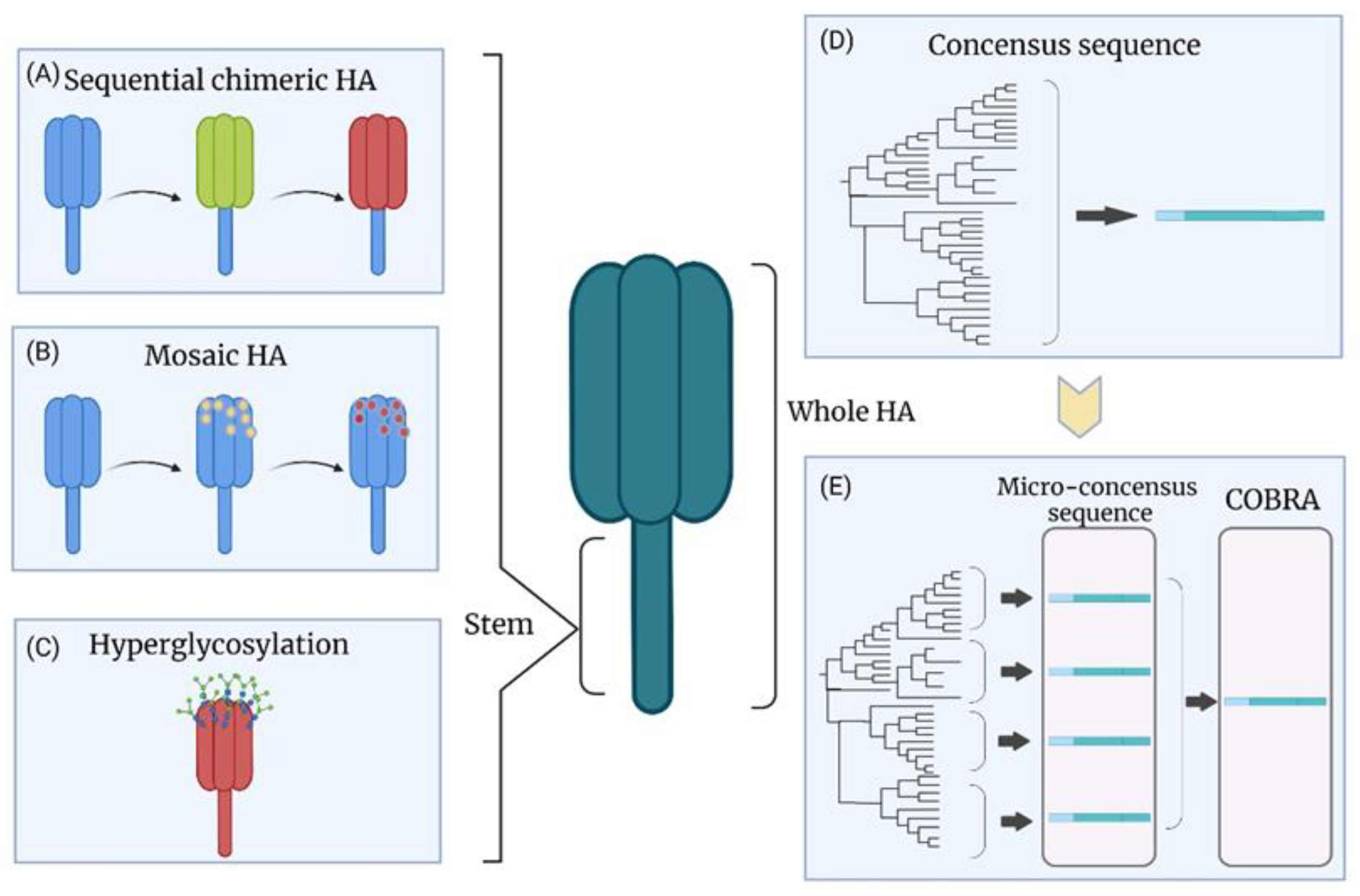
8. New Vaccine Strategies on Existing Platforms
8.1. Chimeric HA
8.2. Hyperglycosylation of HA
8.3. M2-Modified Live Attenuated Influenza Vaccine (LAIV)
8.4. Epitope-Based Influenza Vaccine
9. Conclusions
10. Future Direction
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Disclaimer
References
- WHO. Influenza (Seasonal) Fact sheet. Available online: https://www.who.int/news-room/fact-sheets/detail/influenza-(seasonal) (accessed on 6 November 2018).
- Czaja, C.A.; Miller, L.; Alden, N.; Wald, H.L.; Cummings, C.N.; Rolfes, M.A.; Anderson, E.J.; Bennett, N.M.; Billing, L.M.; Chai, S.J.; et al. Age-Related Differences in Hospitalization Rates, Clinical Presentation, and Outcomes Among Older Adults Hospitalized With Influenza-U.S. Influenza Hospitalization Surveillance Network (FluSurv-NET). Open Forum Infect. Dis. 2019, 6, ofz225. [Google Scholar] [CrossRef] [PubMed]
- Holstein, R.; Dawood, F.S.; O’Halloran, A.; Cummings, C.; Ujamaa, D.; Daily Kirley, P.; Yousey-Hindes, K.; Fawcett, E.; Monroe, M.L.; Kim, S.; et al. Characteristics and Outcomes of Hospitalized Pregnant Women With Influenza, 2010 to 2019: A Repeated Cross-Sectional Study. Ann. Intern. Med. 2022, 175, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Olson, S.M.; Newhams, M.M.; Halasa, N.B.; Feldstein, L.R.; Novak, T.; Weiss, S.L.; Coates, B.M.; Schuster, J.E.; Schwarz, A.J.; Maddux, A.B.; et al. Vaccine Effectiveness Against Life-Threatening Influenza Illness in US Children. Clin. Infect. Dis. 2022, ciab931. [Google Scholar] [CrossRef]
- Simmerman, J.M.; Lertiendumrong, J.; Dowell, S.F.; Uyeki, T.; Olsen, S.J.; Chittaganpitch, M.; Chunsutthiwat, S.; Tangcharoensathien, V. The cost of influenza in Thailand. Vaccine 2006, 24, 4417–4426. [Google Scholar] [CrossRef]
- Putri, W.; Muscatello, D.J.; Stockwell, M.S.; Newall, A.T. Economic burden of seasonal influenza in the United States. Vaccine 2018, 36, 3960–3966. [Google Scholar] [CrossRef]
- Gagnon, A.; Acosta, E.; Hallman, S.; Bourbeau, R.; Dillon, L.Y.; Ouellette, N.; Earn, D.J.D.; Herring, D.A.; Inwood, K.; Madrenas, J.; et al. Pandemic Paradox: Early Life H2N2 Pandemic Influenza Infection Enhanced Susceptibility to Death during the 2009 H1N1 Pandemic. mBio 2018, 9, e02091-17. [Google Scholar] [CrossRef] [Green Version]
- Martini, M.; Gazzaniga, V.; Bragazzi, N.L.; Barberis, I. The Spanish Influenza Pandemic: A lesson from history 100 years after 1918. J. Prev. Med. Hyg. 2019, 60, E64–E67. [Google Scholar] [CrossRef]
- Butt, K.M.; Smith, G.J.; Chen, H.; Zhang, L.J.; Leung, Y.H.; Xu, K.M.; Lim, W.; Webster, R.G.; Yuen, K.Y.; Peiris, J.S.; et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J. Clin. Microbiol. 2005, 43, 5760–5767. [Google Scholar] [CrossRef] [Green Version]
- Bi, Y.; Tan, S.; Yang, Y.; Wong, G.; Zhao, M.; Zhang, Q.; Wang, Q.; Zhao, X.; Li, L.; Yuan, J.; et al. Clinical and Immunological Characteristics of Human Infections With H5N6 Avian Influenza Virus. Clin. Infect. Dis. 2019, 68, 1100–1109. [Google Scholar] [CrossRef]
- Watanabe, T.; Kiso, M.; Fukuyama, S.; Nakajima, N.; Imai, M.; Yamada, S.; Murakami, S.; Yamayoshi, S.; Iwatsuki-Horimoto, K.; Sakoda, Y.; et al. Characterization of H7N9 influenza A viruses isolated from humans. Nature 2013, 501, 551–555. [Google Scholar] [CrossRef] [Green Version]
- Gou, X.; Wu, X.; Shi, Y.; Zhang, K.; Huang, J. A systematic review and meta-analysis of cross-reactivity of antibodies induced by H7 influenza vaccine. Hum. Vaccin. Immunother. 2020, 16, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Shinya, K.; Deng, G.; Jiang, Y.; Li, Z.; Guan, Y.; Tian, G.; Li, Y.; Shi, J.; et al. Identification of amino acids in HA and PB2 critical for the transmission of H5N1 avian influenza viruses in a mammalian host. PLoS Pathog. 2009, 5, e1000709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, L.; Shi, J.; Kong, X.; Ma, S.; Zhang, Y.; Yin, X.; He, X.; Liu, L.; Suzuki, Y.; Li, C.; et al. H3N2 avian influenza viruses detected in live poultry markets in China bind to human-type receptors and transmit in guinea pigs and ferrets. Emerg. Microbes. Infect. 2019, 8, 1280–1290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouvier, N.M.; Palese, P. The biology of influenza viruses. Vaccine 2008, 26 (Suppl. 4), D49–D53. [Google Scholar] [CrossRef] [Green Version]
- Ferhadian, D.; Contrant, M.; Printz-Schweigert, A.; Smyth, R.P.; Paillart, J.C.; Marquet, R. Structural and Functional Motifs in Influenza Virus RNAs. Front. Microbiol. 2018, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Hause, B.M.; Collin, E.A.; Liu, R.; Huang, B.; Sheng, Z.; Lu, W.; Wang, D.; Nelson, E.A.; Li, F. Characterization of a novel influenza virus in cattle and Swine: Proposal for a new genus in the Orthomyxoviridae family. mBio 2014, 5, e00031-14. [Google Scholar] [CrossRef] [Green Version]
- Henritzi, D.; Hoffmann, B.; Wacheck, S.; Pesch, S.; Herrler, G.; Beer, M.; Harder, T.C. A newly developed tetraplex real-time RT-PCR for simultaneous screening of influenza virus types A, B, C and D. Influenza Other Respir. Viruses 2019, 13, 71–82. [Google Scholar] [CrossRef]
- Green, N.; Alexander, H.; Olson, A.; Alexander, S.; Shinnick, T.M.; Sutcliffe, J.G.; Lerner, R.A. Immunogenic structure of the influenza virus hemagglutinin. Cell 1982, 28, 477–487. [Google Scholar] [CrossRef]
- Wiley, D.C.; Skehel, J.J. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu. Rev.. Biochem. 1987, 56, 365–394. [Google Scholar] [CrossRef] [PubMed]
- Stevens, J.; Blixt, O.; Tumpey, T.M.; Taubenberger, J.K.; Paulson, J.C.; Wilson, I.A. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science 2006, 312, 404–410. [Google Scholar] [CrossRef] [Green Version]
- Zost, S.J.; Wu, N.C.; Hensley, S.E.; Wilson, I.A. Immunodominance and Antigenic Variation of Influenza Virus Hemagglutinin: Implications for Design of Universal Vaccine Immunogens. J. Infect. Dis. 2019, 219, S38–S45. [Google Scholar] [CrossRef]
- Harding, A.T.; Heaton, N.S. Efforts to Improve the Seasonal Influenza Vaccine. Vaccines 2018, 6, 19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tisa, V.; Barberis, I.; Faccio, V.; Paganino, C.; Trucchi, C.; Martini, M.; Ansaldi, F. Quadrivalent influenza vaccine: A new opportunity to reduce the influenza burden. J. Prev. Med. Hyg. 2016, 57, E28–E33. [Google Scholar] [PubMed]
- Grohskopf, L.A.; Alyanak, E.; Ferdinands, J.M.; Broder, K.R.; Blanton, L.H.; Talbot, H.K.; Fry, A.M. Prevention and Control of Seasonal Influenza with Vaccines: Recommendations of the Advisory Committee on Immunization Practices, United States, 2021–2022 Influenza Season. MMWR Recomm. Rep.. 2021, 70, 1–28. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.A.; Ennis, F.A.; Gaerlan, P.F.; Denson, L.J.; Denning, C.R.; Schiffman, D. A controlled double-blind comparison of reactogenicity, immunogenicity, and protective efficacy of whole-virus and split-product influenza vaccines in children. J. Infect. Dis. 1977, 136, 623–632. [Google Scholar] [CrossRef]
- O’Gorman, W.E.; Huang, H.; Wei, Y.L.; Davis, K.L.; Leipold, M.D.; Bendall, S.C.; Kidd, B.A.; Dekker, C.L.; Maecker, H.T.; Chien, Y.H.; et al. The Split Virus Influenza Vaccine rapidly activates immune cells through Fcγ receptors. Vaccine 2014, 32, 5989–5997. [Google Scholar] [CrossRef] [Green Version]
- Dunkle, L.M.; Izikson, R. Recombinant hemagglutinin influenza vaccine provides broader spectrum protection. Expert Rev. Vaccines 2016, 15, 957–966. [Google Scholar] [CrossRef]
- Carter, N.J.; Curran, M.P. Live attenuated influenza vaccine (FluMist®; Fluenz™): A review of its use in the prevention of seasonal influenza in children and adults. Drugs 2011, 71, 1591–1622. [Google Scholar] [CrossRef]
- Gerdil, C. The annual production cycle for influenza vaccine. Vaccine 2003, 21, 1776–1779. [Google Scholar] [CrossRef]
- De Jong, J.C.; Beyer, W.E.; Palache, A.M.; Rimmelzwaan, G.F.; Osterhaus, A.D. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J. Med. Virol. 2000, 61, 94–99. [Google Scholar] [CrossRef]
- Skowronski, D.M.; Masaro, C.; Kwindt, T.L.; Mak, A.; Petric, M.; Li, Y.; Sebastian, R.; Chong, M.; Tam, T.; De Serres, G. Estimating vaccine effectiveness against laboratory-confirmed influenza using a sentinel physician network: Results from the 2005-2006 season of dual A and B vaccine mismatch in Canada. Vaccine 2007, 25, 2842–2851. [Google Scholar] [CrossRef] [PubMed]
- Flannery, B.; Kondor, R.J.G.; Chung, J.R.; Gaglani, M.; Reis, M.; Zimmerman, R.K.; Nowalk, M.P.; Jackson, M.L.; Jackson, L.A.; Monto, A.S.; et al. Spread of Antigenically Drifted Influenza A(H3N2) Viruses and Vaccine Effectiveness in the United States During the 2018-2019 Season. J. Infect. Dis. 2020, 221, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Zhou, H.; Ye, D.; Kemble, G.; Jin, H. Improvement of influenza A/Fujian/411/02 (H3N2) virus growth in embryonated chicken eggs by balancing the hemagglutinin and neuraminidase activities, using reverse genetics. J. Virol. 2005, 79, 6763–6771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Widjaja, L.; Ilyushina, N.; Webster, R.G.; Webby, R.J. Molecular changes associated with adaptation of human influenza A virus in embryonated chicken eggs. Virology 2006, 350, 137–145. [Google Scholar] [CrossRef] [Green Version]
- Skowronski, D.M.; Janjua, N.Z.; De Serres, G.; Sabaiduc, S.; Eshaghi, A.; Dickinson, J.A.; Fonseca, K.; Winter, A.L.; Gubbay, J.B.; Krajden, M.; et al. Low 2012-13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS ONE 2014, 9, e92153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Cumulative Number of Confirmed Human Cases for Avian Influenza A(H5N1) Reported to WHO, 2003–2021. 15 April 2021. Available online: https://www.who.int/publications/m/item/cumulative-number-of-confirmed-human-cases-for-avian-influenza-a(h5n1)-reported-to-who-2003-2021-15-april-2021 (accessed on 15 April 2021).
- FAO. H7N9 situation update-Emergency Prevention System for Animal Health (EMPRES-AH). Available online: http://www.fao.org/ag/againfo/programmes/en/empres/h7n9/situation_update.html (accessed on 5 May 2022).
- USDA. Impacts of the 2014–2015 Highly Pathogenic Avian Influenza Outbreak on the U.S. Poultry Sector. Available online: https://www.ers.usda.gov/publications/pub-details/?pubid=86281. (accessed on 21 May 2022).
- Sun, H.; Xiao, Y.; Liu, J.; Wang, D.; Li, F.; Wang, C.; Li, C.; Zhu, J.; Song, J.; Sun, H.; et al. Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc. Natl Acad Sci USA 2020, 117, 17204–17210. [Google Scholar] [CrossRef]
- Imai, M.; Watanabe, T.; Hatta, M.; Das, S.C.; Ozawa, M.; Shinya, K.; Zhong, G.; Hanson, A.; Katsura, H.; Watanabe, S.; et al. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 2012, 486, 420–428. [Google Scholar] [CrossRef] [Green Version]
- Herfst, S.; Schrauwen, E.J.; Linster, M.; Chutinimitkul, S.; de Wit, E.; Munster, V.J.; Sorrell, E.M.; Bestebroer, T.M.; Burke, D.F.; Smith, D.J.; et al. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 2012, 336, 1534–1541. [Google Scholar] [CrossRef] [Green Version]
- Subbarao, K.; Joseph, T. Scientific barriers to developing vaccines against avian influenza viruses. Nat. Rev. Immunol 2007, 7, 267–278. [Google Scholar] [CrossRef]
- Singh, N.; Pandey, A.; Mittal, S.K. Avian influenza pandemic preparedness: Developing prepandemic and pandemic vaccines against a moving target. Expert Rev. Mol. Med. 2010, 12, e14. [Google Scholar] [CrossRef] [Green Version]
- Atmar, R.L.; Keitel, W.A.; Quarles, J.M.; Cate, T.R.; Patel, S.M.; Nino, D.; Wells, J.; Arden, N.; Guo, K.; Hill, H.; et al. Evaluation of age-related differences in the immunogenicity of a G9 H9N2 influenza vaccine. Vaccine 2011, 29, 8066–8072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, R.J.; Madhun, A.S.; Hauge, S.; Sjursen, H.; Major, D.; Kuhne, M.; Höschler, K.; Saville, M.; Vogel, F.R.; Barclay, W.; et al. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 2009, 27, 1889–1897. [Google Scholar] [CrossRef] [PubMed]
- Blanchfield, K.; Kamal, R.P.; Tzeng, W.P.; Music, N.; Wilson, J.R.; Stevens, J.; Lipatov, A.S.; Katz, J.M.; York, I.A. Recombinant influenza H7 hemagglutinins induce lower neutralizing antibody titers in mice than do seasonal hemagglutinins. Influenza Other Respir. Viruses 2014, 8, 628–635. [Google Scholar] [CrossRef] [PubMed]
- Kamal, R.P.; Blanchfield, K.; Belser, J.A.; Music, N.; Tzeng, W.P.; Holiday, C.; Burroughs, A.; Sun, X.; Maines, T.R.; Levine, M.Z.; et al. Inactivated H7 Influenza Virus Vaccines Protect Mice despite Inducing Only Low Levels of Neutralizing Antibodies. J. Virol. 2017, 91, e01202-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eggink, D.; Goff, P.H.; Palese, P. Guiding the immune response against influenza virus hemagglutinin toward the conserved stalk domain by hyperglycosylation of the globular head domain. J. Virol. 2014, 88, 699–704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, L.Y.; Yang, J.; Liu, S. Investigational hemagglutinin-targeted influenza virus inhibitors. Expert Opin. Investig. Drugs 2017, 26, 63–73. [Google Scholar] [CrossRef]
- Russell, C.J.; Hu, M.; Okda, F.A. Influenza Hemagglutinin Protein Stability, Activation, and Pandemic Risk. Trends Microbiol. 2018, 26, 841–853. [Google Scholar] [CrossRef]
- Ekiert, D.C.; Friesen, R.H.; Bhabha, G.; Kwaks, T.; Jongeneelen, M.; Yu, W.; Ophorst, C.; Cox, F.; Korse, H.J.; Brandenburg, B.; et al. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 2011, 333, 843–850. [Google Scholar] [CrossRef] [Green Version]
- Throsby, M.; van den Brink, E.; Jongeneelen, M.; Poon, L.L.; Alard, P.; Cornelissen, L.; Bakker, A.; Cox, F.; van Deventer, E.; Guan, Y.; et al. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 2008, 3, e3942. [Google Scholar] [CrossRef] [Green Version]
- Nachbagauer, R.; Feser, J.; Naficy, A.; Bernstein, D.I.; Guptill, J.; Walter, E.B.; Berlanda-Scorza, F.; Stadlbauer, D.; Wilson, P.C.; Aydillo, T.; et al. A chimeric hemagglutinin-based universal influenza virus vaccine approach induces broad and long-lasting immunity in a randomized, placebo-controlled phase I trial. Nat. Med. 2021, 27, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Meldgaard, T.S.; Bertholet, S. Novel Platforms for the Development of a Universal Influenza Vaccine. Front. Immunol. 2018, 9, 600. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.M.; Tebianian, M. Influenza A viruses: Why focusing on M2e-based universal vaccines. Virus Genes 2011, 42, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kolpe, A.; Schepens, B.; Fiers, W.; Saelens, X. M2-based influenza vaccines: Recent advances and clinical potential. Expert Rev. Vaccines 2017, 16, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, M.; Mozdzanowska, K.; Zharikova, D.; Hoff, H.; Wunner, W.; Couch, R.B.; Gerhard, W. Influenza A virus infection engenders a poor antibody response against the ectodomain of matrix protein 2. Virol. J. 2006, 3, 102. [Google Scholar] [CrossRef] [Green Version]
- Fiers, W.; De Filette, M.; El Bakkouri, K.; Schepens, B.; Roose, K.; Schotsaert, M.; Birkett, A.; Saelens, X. M2e-based universal influenza A vaccine. Vaccine 2009, 27, 6280–6283. [Google Scholar] [CrossRef]
- Schotsaert, M.; De Filette, M.; Fiers, W.; Saelens, X. Universal M2 ectodomain-based influenza A vaccines: Preclinical and clinical developments. Expert Rev. Vaccines 2009, 8, 499–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zebedee, S.L.; Lamb, R.A. Influenza A virus M2 protein: Monoclonal antibody restriction of virus growth and detection of M2 in virions. J. Virol 1988, 62, 2762–2772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.N.; Kim, M.C.; Lee, Y.T.; Kim, Y.J.; Kang, S.M. Mechanisms of Cross-protection by Influenza Virus M2-based Vaccines. Immune. Netw. 2015, 15, 213–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jegerlehner, A.; Schmitz, N.; Storni, T.; Bachmann, M.F. Influenza A vaccine based on the extracellular domain of M2: Weak protection mediated via antibody-dependent NK cell activity. J. Immunol. 2004, 172, 5598–5605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cianci, C.; Gerritz, S.W.; Deminie, C.; Krystal, M. Influenza nucleoprotein: Promising target for antiviral chemotherapy. Antivir. Chem. Chemother. 2012, 23, 77–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisfeld, A.J.; Neumann, G.; Kawaoka, Y. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 2015, 13, 28–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portela, A.; Digard, P. The influenza virus nucleoprotein: A multifunctional RNA-binding protein pivotal to virus replication. J. Gen. Virol. 2002, 83, 723–734. [Google Scholar] [CrossRef] [PubMed]
- Townsend, A.R.; Gotch, F.M.; Davey, J. Cytotoxic T cells recognize fragments of the influenza nucleoprotein. Cell 1985, 42, 457–467. [Google Scholar] [CrossRef]
- Yewdell, J.W.; Bennink, J.R.; Smith, G.L.; Moss, B. Influenza A virus nucleoprotein is a major target antigen for cross-reactive anti-influenza A virus cytotoxic T lymphocytes. Proc. Natl. Acad. Sci. USA 1985, 82, 1785–1789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voeten, J.T.; Bestebroer, T.M.; Nieuwkoop, N.J.; Fouchier, R.A.; Osterhaus, A.D.; Rimmelzwaan, G.F. Antigenic drift in the influenza A virus (H3N2) nucleoprotein and escape from recognition by cytotoxic T lymphocytes. J. Virol. 2000, 74, 6800–6807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, L.E.; Kelso, A. Prospects for an influenza vaccine that induces cross-protective cytotoxic T lymphocytes. Immunol. Cell Biol. 2009, 87, 300–308. [Google Scholar] [CrossRef]
- Chen, Y.Q.; Wohlbold, T.J.; Zheng, N.Y.; Huang, M.; Huang, Y.; Neu, K.E.; Lee, J.; Wan, H.; Rojas, K.T.; Kirkpatrick, E.; et al. Influenza Infection in Humans Induces Broadly Cross-Reactive and Protective Neuraminidase-Reactive Antibodies. Cell 2018, 173, 417–429.e410. [Google Scholar] [CrossRef] [Green Version]
- Eichelberger, M.C.; Wan, H. Influenza Neuraminidase as a Vaccine Antigen. In Influenza Pathogenesis and Control—Volume II; Oldstone, M.B.A., Compans, R.W., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 275–299. [Google Scholar]
- Vogel, O.A.; Manicassamy, B. Broadly Protective Strategies Against Influenza Viruses: Universal Vaccines and Therapeutics. Front. Microbiol. 2020, 11, 135. [Google Scholar] [CrossRef] [Green Version]
- Maier, H.E.; Nachbagauer, R.; Kuan, G.; Ng, S.; Lopez, R.; Sanchez, N.; Stadlbauer, D.; Gresh, L.; Schiller, A.; Rajabhathor, A.; et al. Pre-existing Antineuraminidase Antibodies Are Associated With Shortened Duration of Influenza A(H1N1)pdm Virus Shedding and Illness in Naturally Infected Adults. Clin. Infect. Dis. 2020, 70, 2290–2297. [Google Scholar] [CrossRef] [PubMed]
- Monto, A.S.; Petrie, J.G.; Cross, R.T.; Johnson, E.; Liu, M.; Zhong, W.; Levine, M.; Katz, J.M.; Ohmit, S.E. Antibody to influenza virus neuraminidase: An independent correlate of protection. J. Infect. Dis. 2015, 212, 1191–1199. [Google Scholar] [CrossRef]
- Wohlbold, T.J.; Krammer, F. In the shadow of hemagglutinin: A growing interest in influenza viral neuraminidase and its role as a vaccine antigen. Viruses 2014, 6, 2465–2494. [Google Scholar] [CrossRef] [PubMed]
- Gravel, C.; Li, C.; Wang, J.; Hashem, A.M.; Jaentschke, B.; Xu, K.W.; Lorbetskie, B.; Gingras, G.; Aubin, Y.; Van Domselaar, G.; et al. Qualitative and quantitative analyses of virtually all subtypes of influenza A and B viral neuraminidases using antibodies targeting the universally conserved sequences. Vaccine 2010, 28, 5774–5784. [Google Scholar] [CrossRef] [PubMed]
- Doyle, T.M.; Hashem, A.M.; Li, C.; Van Domselaar, G.; Larocque, L.; Wang, J.; Smith, D.; Cyr, T.; Farnsworth, A.; He, R.; et al. Universal anti-neuraminidase antibody inhibiting all influenza A subtypes. Antiviral Res. 2013, 100, 567–574. [Google Scholar] [CrossRef]
- Nooraei, S.; Bahrulolum, H.; Hoseini, Z.S.; Katalani, C.; Hajizade, A.; Easton, A.J.; Ahmadian, G. Virus-like particles: Preparation, immunogenicity and their roles as nanovaccines and drug nanocarriers. J. Nanobiotechnology 2021, 19, 59. [Google Scholar] [CrossRef] [PubMed]
- Sautto, G.A.; Kirchenbaum, G.A.; Abreu, R.B.; Ecker, J.W.; Pierce, S.R.; Kleanthous, H.; Ross, T.M. A Computationally Optimized Broadly Reactive Antigen Subtype-Specific Influenza Vaccine Strategy Elicits Unique Potent Broadly Neutralizing Antibodies against Hemagglutinin. J. Immunol. 2020, 204, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Bullard, B.L.; Weaver, E.A. Strategies Targeting Hemagglutinin as a Universal Influenza Vaccine. Vaccines 2021, 9, 257. [Google Scholar] [CrossRef]
- Huang, Y.; França, M.S.; Allen, J.D.; Shi, H.; Ross, T.M. Next Generation of Computationally Optimized Broadly Reactive HA Vaccines Elicited Cross-Reactive Immune Responses and Provided Protection against H1N1 Virus Infection. Vaccines 2021, 9, 793. [Google Scholar] [CrossRef]
- Giles, B.M.; Ross, T.M. A computationally optimized broadly reactive antigen (COBRA) based H5N1 VLP vaccine elicits broadly reactive antibodies in mice and ferrets. Vaccine 2011, 29, 3043–3054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giles, B.M.; Crevar, C.J.; Carter, D.M.; Bissel, S.J.; Schultz-Cherry, S.; Wiley, C.A.; Ross, T.M. A computationally optimized hemagglutinin virus-like particle vaccine elicits broadly reactive antibodies that protect nonhuman primates from H5N1 infection. J. Infect. Dis. 2012, 205, 1562–1570. [Google Scholar] [CrossRef] [PubMed]
- Carter, D.M.; Darby, C.A.; Lefoley, B.C.; Crevar, C.J.; Alefantis, T.; Oomen, R.; Anderson, S.F.; Strugnell, T.; Cortés-Garcia, G.; Vogel, T.U.; et al. Design and Characterization of a Computationally Optimized Broadly Reactive Hemagglutinin Vaccine for H1N1 Influenza Viruses. J. Virol. 2016, 90, 4720–4734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, D.M.; Darby, C.A.; Johnson, S.K.; Carlock, M.A.; Kirchenbaum, G.A.; Allen, J.D.; Vogel, T.U.; Delagrave, S.; DiNapoli, J.; Kleanthous, H.; et al. Elicitation of Protective Antibodies against a Broad Panel of H1N1 Viruses in Ferrets Preimmune to Historical H1N1 Influenza Viruses. J. Virol. 2017, 91, e01283-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, J.D.; Ross, T.M. Next generation methodology for updating HA vaccines against emerging human seasonal influenza A(H3N2) viruses. Sci. Rep. 2021, 11, 4554. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Mohan, T.; Zhu, W.; Wang, C.; Deng, L.; Wang, B.Z. Sequential Immunizations with heterosubtypic virus-like particles elicit cross protection against divergent influenza A viruses in mice. Sci. Rep. 2018, 8, 4577. [Google Scholar] [CrossRef] [Green Version]
- Schwartzman, L.M.; Cathcart, A.L.; Pujanauski, L.M.; Qi, L.; Kash, J.C.; Taubenberger, J.K. An Intranasal Virus-Like Particle Vaccine Broadly Protects Mice from Multiple Subtypes of Influenza A Virus. mBio 2015, 6, e01044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, M.C.; Song, J.M.; Eunju, O.; Kwon, Y.M.; Lee, Y.J.; Compans, R.W.; Kang, S.M. Virus-like particles containing multiple M2 extracellular domains confer improved cross-protection against various subtypes of influenza virus. Mol. Ther. 2013, 21, 485–492. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Lee, J.W.; Choi, H.J.; Lee, Y.N.; Hwang, H.S.; Lee, J.; Kim, C.; Lee, J.S.; Montemagno, C.; Prausnitz, M.R.; et al. Microneedle patch delivery to the skin of virus-like particles containing heterologous M2e extracellular domains of influenza virus induces broad heterosubtypic cross-protection. J. Control. Release 2015, 210, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Kirsteina, A.; Akopjana, I.; Bogans, J.; Lieknina, I.; Jansons, J.; Skrastina, D.; Kazaka, T.; Tars, K.; Isakova-Sivak, I.; Mezhenskaya, D.; et al. Construction and Immunogenicity of a Novel Multivalent Vaccine Prototype Based on Conserved Influenza Virus Antigens. Vaccines 2020, 8, 197. [Google Scholar] [CrossRef]
- Sokolova, V.; Westendorf, A.M.; Buer, J.; Überla, K.; Epple, M. The potential of nanoparticles for the immunization against viral infections. J. Mater. Chem. B. 2015, 3, 4767–4779. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Wang, B.Z. A Perspective on Nanoparticle Universal Influenza Vaccines. ACS Infect. Dis. 2018, 4, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.A.; Vanhecke, D.; Michen, B.; Blank, F.; Gehr, P.; Petri-Fink, A.; Rothen-Rutishauser, B. Different endocytotic uptake mechanisms for nanoparticles in epithelial cells and macrophages. Beilstein J. Nanotechnol. 2014, 5, 1625–1636. [Google Scholar] [CrossRef] [Green Version]
- Manolova, V.; Flace, A.; Bauer, M.; Schwarz, K.; Saudan, P.; Bachmann, M.F. Nanoparticles target distinct dendritic cell populations according to their size. Eur. J. Immunol. 2008, 38, 1404–1413. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Seth, A.; Wibowo, N.; Zhao, C.-X.; Mitter, N.; Yu, C.; Middelberg, A.P.J. Nanoparticle vaccines. Vaccine 2014, 32, 327–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sia, Z.R.; Miller, M.S.; Lovell, J.F. Engineered Nanoparticle Applications for Recombinant Influenza Vaccines. Mol. Pharm. 2021, 18, 576–592. [Google Scholar] [CrossRef] [PubMed]
- Kanekiyo, M.; Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.; Rao, S.S.; Kong, W.P.; Wang, L.; Nabel, G.J. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Joyce, M.G.; Gillespie, R.A.; Gallagher, J.R.; Andrews, S.F.; Yassine, H.M.; Wheatley, A.K.; Fisher, B.E.; Ambrozak, D.R.; Creanga, A.; et al. Mosaic nanoparticle display of diverse influenza virus hemagglutinins elicits broad B cell responses. Nat. Immunol. 2019, 20, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Boyoglu-Barnum, S.; Ellis, D.; Gillespie, R.A.; Hutchinson, G.B.; Park, Y.J.; Moin, S.M.; Acton, O.J.; Ravichandran, R.; Murphy, M.; Pettie, D.; et al. Quadrivalent influenza nanoparticle vaccines induce broad protection. Nature 2021, 592, 623–628. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.; Liu, Y.; Flyer, D.; Massare, M.J.; Zhou, B.; Patel, N.; Ellingsworth, L.; Lewis, M.; Cummings, J.F.; Glenn, G. Novel hemagglutinin nanoparticle influenza vaccine with Matrix-M™ adjuvant induces hemagglutination inhibition, neutralizing, and protective responses in ferrets against homologous and drifted A(H3N2) subtypes. Vaccine 2017, 35, 5366–5372. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, L.; Gonzalez, G.X.; Luthra, L.; Dong, C.; Ma, Y.; Zou, J.; Kang, S.M.; Wang, B.Z. Double-Layered M2e-NA Protein Nanoparticle Immunization Induces Broad Cross-Protection against Different Influenza Viruses in Mice. Adv. Healthc. Mater. 2020, 9, e1901176. [Google Scholar] [CrossRef] [Green Version]
- Jabbal-Gill, I.; Watts, P.; Smith, A. Chitosan-based delivery systems for mucosal vaccines. Expert Opin. Drug Deliv. 2012, 9, 1051–1067. [Google Scholar] [CrossRef]
- Illum, L.; Jabbal-Gill, I.; Hinchcliffe, M.; Fisher, A.N.; Davis, S.S. Chitosan as a novel nasal delivery system for vaccines. Adv. Drug Deliv. Rev. 2001, 51, 81–96. [Google Scholar] [CrossRef]
- Van der Lubben, I.M.; Verhoef, J.C.; Borchard, G.; Junginger, H.E. Chitosan and its derivatives in mucosal drug and vaccine delivery. Eur. J. Pharm. Sci. 2001, 14, 201–207. [Google Scholar] [CrossRef]
- Gogev, S.; de Fays, K.; Versali, M.F.; Gautier, S.; Thiry, E. Glycol chitosan improves the efficacy of intranasally administrated replication defective human adenovirus type 5 expressing glycoprotein D of bovine herpesvirus 1. Vaccine 2004, 22, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, M.Y.E.; Kim, T.H.; Uddin, M.B.; Kim, J.H.; Hewawaduge, C.Y.; Ferdowshi, Z.; Sung, M.H.; Kim, C.J.; Lee, J.S. Mucosal vaccination of conserved sM2, HA2 and cholera toxin subunit A1 (CTA1) fusion protein with poly gamma-glutamate/chitosan nanoparticles (PC NPs) induces protection against divergent influenza subtypes. Vet. Microbiol. 2017, 201, 240–251. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, S.; Renu, S.; Ghimire, S.; Shaan Lakshmanappa, Y.; Hogshead, B.T.; Feliciano-Ruiz, N.; Lu, F.; HogenEsch, H.; Krakowka, S.; Lee, C.W.; et al. Mucosal Immunity and Protective Efficacy of Intranasal Inactivated Influenza Vaccine Is Improved by Chitosan Nanoparticle Delivery in Pigs. Front. Immunol. 2018, 9, 934. [Google Scholar] [CrossRef] [PubMed]
- Hiremath, J.; Kang, K.I.; Xia, M.; Elaish, M.; Binjawadagi, B.; Ouyang, K.; Dhakal, S.; Arcos, J.; Torrelles, J.B.; Jiang, X.; et al. Entrapment of H1N1 Influenza Virus Derived Conserved Peptides in PLGA Nanoparticles Enhances T Cell Response and Vaccine Efficacy in Pigs. PLoS ONE 2016, 11, e0151922. [Google Scholar] [CrossRef] [Green Version]
- Price, G.E.; Soboleski, M.R.; Lo, C.Y.; Misplon, J.A.; Quirion, M.R.; Houser, K.V.; Pearce, M.B.; Pappas, C.; Tumpey, T.M.; Epstein, S.L. Single-dose mucosal immunization with a candidate universal influenza vaccine provides rapid protection from virulent H5N1, H3N2 and H1N1 viruses. PLoS ONE 2010, 5, e13162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soboleski, M.R.; Gabbard, J.D.; Price, G.E.; Misplon, J.A.; Lo, C.Y.; Perez, D.R.; Ye, J.; Tompkins, S.M.; Epstein, S.L. Cold-adapted influenza and recombinant adenovirus vaccines induce cross-protective immunity against pH1N1 challenge in mice. PLoS ONE 2011, 6, e21937. [Google Scholar] [CrossRef]
- Lo, C.Y.; Misplon, J.A.; Li, X.; Price, G.E.; Ye, Z.; Epstein, S.L. Universal influenza vaccine based on conserved antigens provides long-term durability of immune responses and durable broad protection against diverse challenge virus strains in mice. Vaccine 2021, 39, 4628–4640. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kang, J.O.; Kim, J.Y.; Jung, H.E.; Lee, H.K.; Chang, J. Single mucosal vaccination targeting nucleoprotein provides broad protection against two lineages of influenza B virus. Antiviral Res. 2019, 163, 19–28. [Google Scholar] [CrossRef]
- Hassan, A.O.; Amen, O.; Sayedahmed, E.E.; Vemula, S.V.; Amoah, S.; York, I.; Gangappa, S.; Sambhara, S.; Mittal, S.K. Adenovirus vector-based multi-epitope vaccine provides partial protection against H5, H7, and H9 avian influenza viruses. PLoS ONE 2017, 12, e0186244. [Google Scholar] [CrossRef] [Green Version]
- DiLillo, D.J.; Palese, P.; Wilson, P.C.; Ravetch, J.V. Broadly neutralizing anti-influenza antibodies require Fc receptor engagement for in vivo protection. J. Clin. Investig. 2016, 126, 605–610. [Google Scholar] [CrossRef]
- Bliss, C.M.; Freyn, A.W.; Caniels, T.G.; Leyva-Grado, V.H.; Nachbagauer, R.; Sun, W.; Tan, G.S.; Gillespie, V.L.; McMahon, M.; Krammer, F.; et al. A single-shot adenoviral vaccine provides hemagglutinin stalk-mediated protection against heterosubtypic influenza challenge in mice. Mol. Ther. 2022, 30, 2024–2047. [Google Scholar] [CrossRef] [PubMed]
- Assarsson, E.; Bui, H.H.; Sidney, J.; Zhang, Q.; Glenn, J.; Oseroff, C.; Mbawuike, I.N.; Alexander, J.; Newman, M.J.; Grey, H.; et al. Immunomic analysis of the repertoire of T-cell specificities for influenza A virus in humans. J. Virol. 2008, 82, 12241–12251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, E.; Wu, C.; Chan, K.F.; Eckle, S.; Bharadwaj, M.; Zou, Q.M.; Kedzierska, K.; Chen, W. Nucleoprotein of influenza A virus is a major target of immunodominant CD8+ T-cell respon.nses. Immunol. Cell Biol. 2013, 91, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Coughlan, L.; Mullarkey, C.; Gilbert, S. Adenoviral vectors as novel vaccines for influenza. J. Pharm. Pharmacol. 2015, 67, 382–399. [Google Scholar] [CrossRef] [PubMed]
- Van Kampen, K.R.; Shi, Z.; Gao, P.; Zhang, J.; Foster, K.W.; Chen, D.T.; Marks, D.; Elmets, C.A.; Tang, D.C. Safety and immunogenicity of adenovirus-vectored nasal and epicutaneous influenza vaccines in humans. Vaccine 2005, 23, 1029–1036. [Google Scholar] [CrossRef]
- Kim, S.H.; Kim, J.Y.; Choi, Y.; Nguyen, H.H.; Song, M.K.; Chang, J. Mucosal vaccination with recombinant adenovirus encoding nucleoprotein provides potent protection against influenza virus infection. PLoS ONE 2013, 8, e75460. [Google Scholar] [CrossRef] [Green Version]
- Bricker, T.L.; Darling, T.L.; Hassan, A.O.; Harastani, H.H.; Soung, A.; Jiang, X.; Dai, Y.N.; Zhao, H.; Adams, L.J.; Holtzman, M.J.; et al. A single intranasal or intramuscular immunization with chimpanzee adenovirus-vectored SARS-CoV-2 vaccine protects against pneumonia in hamsters. Cell Rep. 2021, 36, 109400. [Google Scholar] [CrossRef] [PubMed]
- Barnes, E.; Folgori, A.; Capone, S.; Swadling, L.; Aston, S.; Kurioka, A.; Meyer, J.; Huddart, R.; Smith, K.; Townsend, R.; et al. Novel adenovirus-based vaccines induce broad and sustained T cell responses to HCV in man. Sci. Transl. Med. 2012, 4, 115ra111. [Google Scholar] [CrossRef] [Green Version]
- Ledgerwood, J.E.; DeZure, A.D.; Stanley, D.A.; Coates, E.E.; Novik, L.; Enama, M.E.; Berkowitz, N.M.; Hu, Z.; Joshi, G.; Ploquin, A.; et al. Chimpanzee Adenovirus Vector Ebola Vaccine. N. Engl. J. Med. 2014, 376, 928–938. [Google Scholar] [CrossRef]
- Van Doremalen, N.; Lambe, T.; Spencer, A.; Belij-Rammerstorfer, S.; Purushotham, J.N.; Port, J.R.; Avanzato, V.A.; Bushmaker, T.; Flaxman, A.; Ulaszewska, M.; et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature 2020, 586, 578–582. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Wu, T.L.; Lasaro, M.O.; Latimer, B.P.; Parzych, E.M.; Bian, A.; Li, Y.; Li, H.; Erikson, J.; Xiang, Z.; et al. A universal influenza A vaccine based on adenovirus expressing matrix-2 ectodomain and nucleoprotein protects mice from lethal challenge. Mol. Ther. 2010, 18, 2182–2189. [Google Scholar] [CrossRef] [PubMed]
- Asthagiri Arunkumar, G.; McMahon, M.; Pavot, V.; Aramouni, M.; Ioannou, A.; Lambe, T.; Gilbert, S.; Krammer, F. Vaccination with viral vectors expressing NP, M1 and chimeric hemagglutinin induces broad protection against influenza virus challenge in mice. Vaccine 2019, 37, 5567–5577. [Google Scholar] [CrossRef]
- McMahon, M.; Asthagiri Arunkumar, G.; Liu, W.C.; Stadlbauer, D.; Albrecht, R.A.; Pavot, V.; Aramouni, M.; Lambe, T.; Gilbert, S.C.; Krammer, F. Vaccination With Viral Vectors Expressing Chimeric Hemagglutinin, NP and M1 Antigens Protects Ferrets Against Influenza Virus Challenge. Front. Immunol. 2019, 10, 2005. [Google Scholar] [CrossRef] [PubMed]
- Hessel, A.; Savidis-Dacho, H.; Coulibaly, S.; Portsmouth, D.; Kreil, T.R.; Crowe, B.A.; Schwendinger, M.G.; Pilz, A.; Barrett, P.N.; Falkner, F.G.; et al. MVA vectors expressing conserved influenza proteins protect mice against lethal challenge with H5N1, H9N2 and H7N1 viruses. PLoS ONE 2014, 9, e88340. [Google Scholar] [CrossRef] [PubMed]
- Kamlangdee, A.; Kingstad-Bakke, B.; Anderson, T.K.; Goldberg, T.L.; Osorio, J.E. Broad protection against avian influenza virus by using a modified vaccinia Ankara virus expressing a mosaic hemagglutinin gene. J. Virol. 2014, 88, 13300–13309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Hong, Y.; Chen, W.; Wang, C. Polymers for DNA Vaccine Delivery. ACS Biomater. Sci. Eng. 2017, 3, 108–125. [Google Scholar] [CrossRef] [PubMed]
- Suschak, J.J.; Williams, J.A.; Schmaljohn, C.S. Advancements in DNA vaccine vectors, non-mechanical delivery methods, and molecular adjuvants to increase immunogenicity. Hum. Vaccin. Immunother. 2017, 13, 2837–2848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peachman, K.K.; Rao, M.; Alving, C.R. Immunization with DNA through the skin. Methods 2003, 31, 232–242. [Google Scholar] [CrossRef]
- Farris, E.; Brown, D.M.; Ramer-Tait, A.E.; Pannier, A.K. Micro- and nanoparticulates for DNA vaccine delivery. Exp. Biol. Med. 2016, 241, 919–929. [Google Scholar] [CrossRef] [Green Version]
- Lambricht, L.; Lopes, A.; Kos, S.; Sersa, G.; Préat, V.; Vandermeulen, G. Clinical potential of electroporation for gene therapy and DNA vaccine delivery. Expert Opin. Drug Deliv. 2016, 13, 295–310. [Google Scholar] [CrossRef] [PubMed]
- Hasson, S.S.A.A.; Al-Busaidi, J.K.Z.; Sallam, T.A. The past, current and future trends in DNA vaccine immunisations. Asian Pac. J. Trop. Biomed. 2015, 5, 344–353. [Google Scholar] [CrossRef] [Green Version]
- Lim, M.; Badruddoza, A.Z.M.; Firdous, J.; Azad, M.; Mannan, A.; Al-Hilal, T.A.; Cho, C.S.; Islam, M.A. Engineered Nanodelivery Systems to Improve DNA Vaccine Technologies. Pharmaceutics 2020, 12, 30. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.A. DNA vaccines: An historical perspective and view to the future. Immunol. Rev. 2011, 239, 62–84. [Google Scholar] [CrossRef]
- Hobernik, D.; Bros, M. DNA Vaccines-How Far From Clinical Use? Int. J. Mol. Sci. 2018, 19, 3605. [Google Scholar] [CrossRef] [Green Version]
- Ping, X.; Hu, W.; Xiong, R.; Zhang, X.; Teng, Z.; Ding, M.; Li, L.; Chang, C.; Xu, K. Generation of a broadly reactive influenza H1 antigen using a consensus HA sequence. Vaccine 2018, 36, 4837–4845. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.W.; Cheng, T.J.; Huang, Y.; Jan, J.T.; Ma, S.H.; Yu, A.L.; Wong, C.H.; Ho, D.D. A consensus-hemagglutinin-based DNA vaccine that protects mice against divergent H5N1 influenza viruses. Proc. Natl. Acad. Sci. USA 2008, 105, 13538–13543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Morrow, M.P.; Chu, J.S.; Racine, T.; Reed, C.C.; Khan, A.S.; Broderick, K.E.; Kim, J.J.; Kobinger, G.P.; Sardesai, N.Y.; et al. Broad cross-protective anti-hemagglutination responses elicited by influenza microconsensus DNA vaccine. Vaccine 2018, 36, 3079–3089. [Google Scholar] [CrossRef] [PubMed]
- Elliott, S.T.C.; Keaton, A.A.; Chu, J.D.; Reed, C.C.; Garman, B.; Patel, A.; Yan, J.; Broderick, K.E.; Weiner, D.B. A Synthetic Micro-Consensus DNA Vaccine Generates Comprehensive Influenza A H3N2 Immunity and Protects Mice Against Lethal Challenge by Multiple H3N2 Viruses. Hum. Gene Ther. 2018, 29, 1044–1055. [Google Scholar] [CrossRef]
- Verbeke, R.; Lentacker, I.; De Smedt, S.C.; Dewitte, H. The dawn of mRNA vaccines: The COVID-19 case. J. Control Release 2021, 333, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Probst, J.; Weide, B.; Scheel, B.; Pichler, B.J.; Hoerr, I.; Rammensee, H.G.; Pascolo, S. Spontaneous cellular uptake of exogenous messenger RNA in vivo is nucleic acid-specific, saturable and ion dependent. Gene Ther. 2007, 14, 1175–1180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steinle, H.; Behring, A.; Schlensak, C.; Wendel, H.P.; Avci-Adali, M. Concise Review: Application of In Vitro Transcribed Messenger RNA for Cellular Engineering and Reprogramming: Progress and Challenges. Stem Cells 2017, 35, 68–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef]
- Igyártó, B.Z.; Jacobsen, S.; Ndeupen, S. Future considerations for the mRNA-lipid nanoparticle vaccine platform. Curr. Opin. Virol. 2021, 48, 65–72. [Google Scholar] [CrossRef]
- Freyn, A.W.; Ramos da Silva, J.; Rosado, V.C.; Bliss, C.M.; Pine, M.; Mui, B.L.; Tam, Y.K.; Madden, T.D.; de Souza Ferreira, L.C.; Weissman, D.; et al. A Multi-Targeting, Nucleoside-Modified mRNA Influenza Virus Vaccine Provides Broad Protection in Mice. Mol. Ther. 2020, 28, 1569–1584. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Parkhouse, K.; Kirkpatrick, E.; McMahon, M.; Zost, S.J.; Mui, B.L.; Tam, Y.K.; Karikó, K.; Barbosa, C.J.; Madden, T.D.; et al. Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat. Commun. 2018, 9, 3361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magini, D.; Giovani, C.; Mangiavacchi, S.; Maccari, S.; Cecchi, R.; Ulmer, J.B.; De Gregorio, E.; Geall, A.J.; Brazzoli, M.; Bertholet, S. Self-Amplifying mRNA Vaccines Expressing Multiple Conserved Influenza Antigens Confer Protection against Homologous and Heterosubtypic Viral Challenge. PLoS ONE 2016, 11, e0161193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hai, R.; Krammer, F.; Tan, G.S.; Pica, N.; Eggink, D.; Maamary, J.; Margine, I.; Albrecht, R.A.; Palese, P. Influenza viruses expressing chimeric hemagglutinins: Globular head and stalk domains derived from different subtypes. J. Virol. 2012, 86, 5774–5781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nachbagauer, R.; Liu, W.C.; Choi, A.; Wohlbold, T.J.; Atlas, T.; Rajendran, M.; Solórzano, A.; Berlanda-Scorza, F.; García-Sastre, A.; Palese, P.; et al. A universal influenza virus vaccine candidate confers protection against pandemic H1N1 infection in preclinical ferret studies. NPJ Vaccines 2017, 2, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, A.; Bouzya, B.; Cortés Franco, K.D.; Stadlbauer, D.; Rajabhathor, A.; Rouxel, R.N.; Mainil, R.; Van der Wielen, M.; Palese, P.; García-Sastre, A.; et al. Chimeric Hemagglutinin-Based Influenza Virus Vaccines Induce Protective Stalk-Specific Humoral Immunity and Cellular Responses in Mice. Immunohorizons 2019, 3, 133–148. [Google Scholar] [CrossRef] [Green Version]
- Krammer, F.; Pica, N.; Hai, R.; Margine, I.; Palese, P. Chimeric hemagglutinin influenza virus vaccine constructs elicit broadly protective stalk-specific antibodies. J. Virol. 2013, 87, 6542–6550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margine, I.; Krammer, F.; Hai, R.; Heaton, N.S.; Tan, G.S.; Andrews, S.A.; Runstadler, J.A.; Wilson, P.C.; Albrecht, R.A.; García-Sastre, A.; et al. Hemagglutinin stalk-based universal vaccine constructs protect against group 2 influenza A viruses. J. Virol. 2013, 87, 10435–10446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermler, M.E.; Kirkpatrick, E.; Sun, W.; Hai, R.; Amanat, F.; Chromikova, V.; Palese, P.; Krammer, F. Chimeric Hemagglutinin Constructs Induce Broad Protection against Influenza B Virus Challenge in the Mouse Model. J. Virol. 2017, 91, e00286-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, W.; Kirkpatrick, E.; Ermler, M.; Nachbagauer, R.; Broecker, F.; Krammer, F.; Palese, P. Development of Influenza B Universal Vaccine Candidates Using the “Mosaic” Hemagglutinin Approach. J. Virol. 2019, 93, e00333-19. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.J.; Boyington, J.C.; Dai, K.; Houser, K.V.; Pearce, M.B.; Kong, W.P.; Yang, Z.Y.; Tumpey, T.M.; Nabel, G.J. Cross-neutralization of 1918 and 2009 influenza viruses: Role of glycans in viral evolution and vaccine design. Sci. Transl. Med. 2010, 2, 24ra21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, R.A.; Stertz, S.; Manicassamy, B.; Zimmermann, P.; Sun, X.; Albrecht, R.A.; Uusi-Kerttula, H.; Zagordi, O.; Belshe, R.B.; Frey, S.E.; et al. Glycosylations in the globular head of the hemagglutinin protein modulate the virulence and antigenic properties of the H1N1 influenza viruses. Sci. Transl. Med. 2013, 5, 187ra170. [Google Scholar] [CrossRef] [Green Version]
- Jang, Y.H.; Seong, B.L. Options and obstacles for designing a universal influenza vaccine. Viruses 2014, 6, 3159–3180. [Google Scholar] [CrossRef]
- Sarawar, S.; Hatta, Y.; Watanabe, S.; Dias, P.; Neumann, G.; Kawaoka, Y.; Bilsel, P. M2SR, a novel live single replication influenza virus vaccine, provides effective heterosubtypic protection in mice. Vaccine 2016, 34, 5090–5098. [Google Scholar] [CrossRef] [Green Version]
- Hatta, Y.; Boltz, D.; Sarawar, S.; Kawaoka, Y.; Neumann, G.; Bilsel, P. M2SR, a novel live influenza vaccine, protects mice and ferrets against highly pathogenic avian influenza. Vaccine 2017, 35, 4177–4183. [Google Scholar] [CrossRef]
- Wang, L.; Liu, S.Y.; Chen, H.W.; Xu, J.; Chapon, M.; Zhang, T.; Zhou, F.; Wang, Y.E.; Quanquin, N.; Wang, G.; et al. Generation of a Live Attenuated Influenza Vaccine that Elicits Broad Protection in Mice and Ferrets. Cell Host Microbe. 2017, 21, 334–343. [Google Scholar] [CrossRef] [Green Version]
- Park, B.R.; Kim, K.H.; Kotomina, T.; Kim, M.C.; Kwon, Y.M.; Jeeva, S.; Jung, Y.J.; Bhatnagar, N.; Isakova-Sivak, I.; Mezhenskaya, D.; et al. Broad cross protection by recombinant live attenuated influenza H3N2 seasonal virus expressing conserved M2 extracellular domain in a chimeric hemagglutinin. Sci. Rep. 2021, 11, 4151. [Google Scholar] [CrossRef] [PubMed]
- Kotomina, T.; Isakova-Sivak, I.; Kim, K.H.; Park, B.R.; Jung, Y.J.; Lee, Y.; Mezhenskaya, D.; Matyushenko, V.; Kang, S.M.; Rudenko, L. Generation and Characterization of Universal Live-Attenuated Influenza Vaccine Candidates Containing Multiple M2e Epitopes. Vaccines 2020, 8, 648. [Google Scholar] [CrossRef] [PubMed]
- Gottlieb, T.; Ben-Yedidia, T. Epitope-based approaches to a universal influenza vaccine. J. Autoimmun. 2014, 54, 15–20. [Google Scholar] [CrossRef]
- Adar, Y.; Singer, Y.; Levi, R.; Tzehoval, E.; Perk, S.; Banet-Noach, C.; Nagar, S.; Arnon, R.; Ben-Yedidia, T. A universal epitope-based influenza vaccine and its efficacy against H5N1. Vaccine 2009, 27, 2099–2107. [Google Scholar] [CrossRef] [PubMed]
- Atsmon, J.; Kate-Ilovitz, E.; Shaikevich, D.; Singer, Y.; Volokhov, I.; Haim, K.Y.; Ben-Yedidia, T. Safety and Immunogenicity of Multimeric-001—a Novel Universal Influenza Vaccine. J. Clin. Immunol. 2012, 32, 595–603. [Google Scholar] [CrossRef]
- Wilkinson, T.M.; Li, C.K.; Chui, C.S.; Huang, A.K.; Perkins, M.; Liebner, J.C.; Lambkin-Williams, R.; Gilbert, A.; Oxford, J.; Nicholas, B.; et al. Preexisting influenza-specific CD4+ T cells correlate with disease protection against influenza challenge in humans. Nat. Med. 2012, 18, 274–280. [Google Scholar] [CrossRef]
- Stoloff, G.A.; Caparros-Wanderley, W. Synthetic multi-epitope peptides identified in silico induce protective immunity against multiple influenza serotypes. Eur. J. Immunol. 2007, 37, 2441–2449. [Google Scholar] [CrossRef] [PubMed]
- Pleguezuelos, O.; Robinson, S.; Fernández, A.; Stoloff, G.A.; Mann, A.; Gilbert, A.; Balaratnam, G.; Wilkinson, T.; Lambkin-Williams, R.; Oxford, J.; et al. A Synthetic Influenza Virus Vaccine Induces a Cellular Immune Response That Correlates with Reduction in Symptomatology and Virus Shedding in a Randomized Phase Ib Live-Virus Challenge in Humans. Clin. Vaccine Immunol. 2015, 22, 828–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pleguezuelos, O.; James, E.; Fernandez, A.; Lopes, V.; Rosas, L.A.; Cervantes-Medina, A.; Cleath, J.; Edwards, K.; Neitzey, D.; Gu, W.; et al. Efficacy of FLU-v, a broad-spectrum influenza vaccine, in a randomized phase IIb human influenza challenge study. NPJ Vaccines 2020, 5, 22. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.T.; Tan, G.S.; Hai, R.; Pica, N.; Ngai, L.; Ekiert, D.C.; Wilson, I.A.; García-Sastre, A.; Moran, T.M.; Palese, P. Vaccination with a synthetic peptide from the influenza virus hemagglutinin provides protection against distinct viral subtypes. Proc. Natl. Acad. Sci. USA 2010, 107, 18979–18984. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Qiao, Y.; Xu, Y.; Li, P.; Nie, J.; Zhao, Q.; Chai, W.; Shi, Y.; Kong, W.; Shan, Y. Identification of Linear Peptide Immunogens with Verified Broad-spectrum Immunogenicity from the Conserved Regions within the Hemagglutinin Stem Domain of H1N1 Influenza Virus. Immunol. Invest. 2022, 51, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, L.A.; Kotlyarov, R.Y.; Kovaleva, A.A.; Potapchuk, M.V.; Korotkov, A.V.; Sergeeva, M.V.; Kasianenko, M.A.; Kuprianov, V.V.; Ravin, N.V.; Tsybalova, L.M.; et al. Protection against multiple influenza A virus strains induced by candidate recombinant vaccine based on heterologous M2e peptides linked to flagellin. PLoS ONE 2015, 10, e0119520. [Google Scholar] [CrossRef] [PubMed]
- Tsybalova, L.M.; Stepanova, L.A.; Shuklina, M.A.; Mardanova, E.S.; Kotlyarov, R.Y.; Potapchuk, M.V.; Petrov, S.A.; Blokhina, E.A.; Ravin, N.V. Combination of M2e peptide with stalk HA epitopes of influenza A virus enhances protective properties of recombinant vaccine. PLoS ONE 2018, 13, e0201429. [Google Scholar] [CrossRef] [Green Version]
- Zykova, A.A.; Blokhina, E.A.; Stepanova, L.A.; Shuklina, M.A.; Tsybalova, L.M.; Kuprianov, V.V.; Ravin, N.V. Nanoparticles based on artificial self-assembling peptide and displaying M2e peptide and stalk HA epitopes of influenza A virus induce potent humoral and T-cell responses and protect against the viral infection. Nanomedicine 2022, 39, 102463. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Joshi, M.D.; Singhania, S.; Ramsey, K.H.; Murthy, A.K. Peptide Vaccine: Progress and Challenges. Vaccines 2014, 2, 515–536. [Google Scholar] [CrossRef] [Green Version]
- Nelde, A.; Rammensee, H.G.; Walz, J.S. The Peptide Vaccine of the Future. Mol. Cell Proteomics 2021, 20, 100022. [Google Scholar] [CrossRef]
- Bhatnager, R.; Bhasin, M.; Arora, J.; Dang, A.S. Epitope based peptide vaccine against SARS-COV2: An immune-informatics approach. J. Biomol. Struct. Dyn. 2021, 39, 5690–5705. [Google Scholar] [CrossRef]
| Platform | Vaccine Type | Target Antigen | Stage | Trial ID |
|---|---|---|---|---|
| LAIV/Inactivated virus | Single-replication virus | Whole virus (M2-deleted) | Phase I | NCT04960397 NCT02822105 NCT03999554 |
| Inactivated split virus | HA stem (chimeric) | Phase I | NCT03275389 | |
| Inactivated whole virus | Whole virus | Phase I | NCT05027932 | |
| LAIV + Inactivated split virus | HA stem (chimeric) | Phase I | NCT03300050 | |
| Subunit vaccine | Recombinant protein | M1, NP, HA | Phase I, II, III | NCT01419925 NCT00877448 NCT02293317 NCT03450915 NCT01146119 NCT02691130 |
| Recombinant protein | M2e | Phase I, II | NCT00921947 NCT00921973 NCT00921206 NCT00603811 | |
| Synthetic peptides | NP, M, PB1, PB2 | Phase I | NCT01265914 | |
| Synthetic peptides | M1, M2, NP | Phase II | NCT03180801 NCT02962908 NCT01226758 NCT01181336 | |
| VLP/Nanoparticle | Ferritin-based nanoparticles | HA stem | Phase I | NCT05155319 |
| Ferritin-based nanoparticles | HA stem | Phase I | NCT04579250 | |
| Computational design nanoparticles | HA | Phase I | NCT04896086 | |
| Oligomerization domain-based nanoparticles | NP | Phase II | NCT04192500 | |
| Hepatitis B VLP | M2e | Phase I | NCT00819013 | |
| Hepatitis B VLP | M2e | Phase I | NCT03789539 | |
| Viral vector | MVA | NP, M1 | Phase II | NCT03880474 NCT03883113 NCT00993083 |
| ChAd + MVA | NP, M1 | Phase I | NCT01818362 NCT01623518 | |
| Nucleic acid | DNA | HA, NA, M2e, NP | Phase I | NCT01184976 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.-C.; Sayedahmed, E.E.; Sambhara, S.; Mittal, S.K. Progress towards the Development of a Universal Influenza Vaccine. Viruses 2022, 14, 1684. https://doi.org/10.3390/v14081684
Wang W-C, Sayedahmed EE, Sambhara S, Mittal SK. Progress towards the Development of a Universal Influenza Vaccine. Viruses. 2022; 14(8):1684. https://doi.org/10.3390/v14081684
Chicago/Turabian StyleWang, Wen-Chien, Ekramy E. Sayedahmed, Suryaprakash Sambhara, and Suresh K. Mittal. 2022. "Progress towards the Development of a Universal Influenza Vaccine" Viruses 14, no. 8: 1684. https://doi.org/10.3390/v14081684
APA StyleWang, W.-C., Sayedahmed, E. E., Sambhara, S., & Mittal, S. K. (2022). Progress towards the Development of a Universal Influenza Vaccine. Viruses, 14(8), 1684. https://doi.org/10.3390/v14081684








