Salmonella Enteritidis Bacteriophages Isolated from Kenyan Poultry Farms Demonstrate Time-Dependent Stability in Environments Mimicking the Chicken Gastrointestinal Tract
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
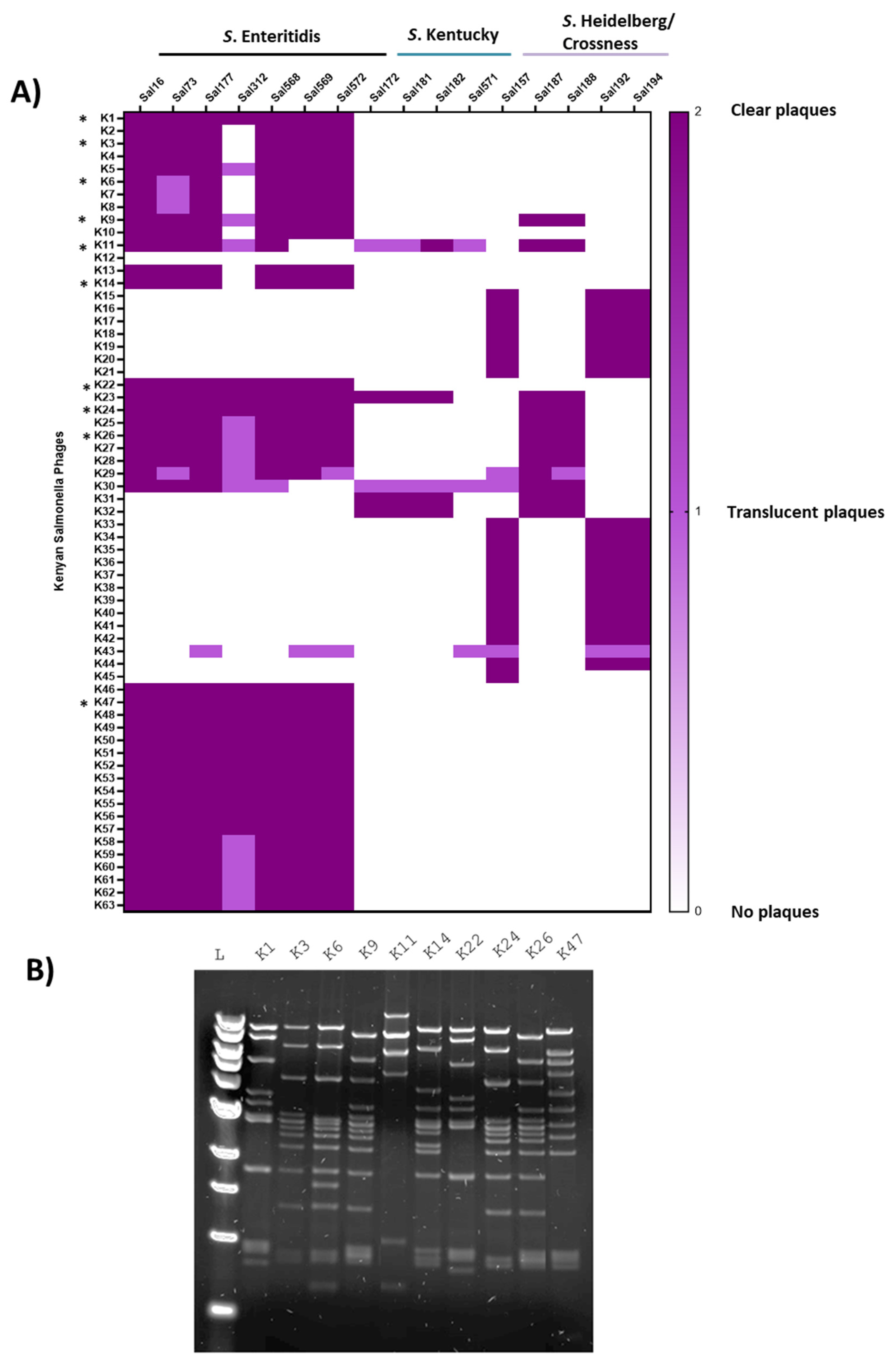
2.2. Phage Isolates
2.3. Phage Stability in pH-Adjusted Media
2.4. Phage Stability in Simulated Gastric and Intestinal Fluids
2.5. Phage Stability in Different Thermal Conditions
2.6. Control of Salmonella by Phages in pH-Adjusted Media
2.7. Control of Salmonella by Phages in SGF and SIF
2.8. Phage Replication in SGF
2.9. Phage Persistence in Different Water Sources
2.10. Data Analysis
3. Results
3.1. Isolating and Characterizing the Biological Materials
3.2. Assessing Phage Stability in Media at Low and High pH
3.3. Assessing Phage Stability in Simulated Gastric and Intestinal Fluids
3.4. Phage Stability at Different Temperatures
3.5. Control of Salmonella by Phages in pH-Adjusted Media
3.6. Control of Salmonella by Phages in SGF and SIF
3.7. Phage Replication in SGF
3.8. Phage Persistence in Water from Different Sources
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- OECD/FAO. Organisation for Economic Co-Operation and Development (OECD)-Food and Agricultural Organisation (FAO) Agricultural Outlook 2021–2020; OECD Publishing: Paris, France, 2021. [Google Scholar]
- Omiti, J.M.; Okuthe, S.O. An Overview of the Poultry Sector and Status of Highly Pathongenic Avian Influenza (HPAI) in Kenya-Background Paper; HPAI Africa/Indonesia Team Working Paper 4. IFPRI, ILRI and RVC: Washington, DC, USA, 2008. [Google Scholar]
- Hedman, H.D.; Vasco, K.A.; Zhang, L. A Review of Antimicrobial Resistance in Poultry Farming within Low-resource Settings. Animals 2020, 10, 1264. [Google Scholar] [CrossRef] [PubMed]
- Nadi, Z.R.; Salehi, T.Z.; Tamai, I.A.; Foroushani, A.R.; Sillanpaa, M.; Dallal, M.M.S. Evaluation of Antibiotic Resistance and Prevalence of Common Salmonella enterica Serovars Isolated from Foodborne Outbreaks. Microchem. J. 2020, 155, 104–660. [Google Scholar] [CrossRef]
- FAO. Non-Typhoidal Salmonella. Available online: https://www.who.int/news-room/fact-sheets/detail/salmonella-(non-typhoidal) (accessed on 23 July 2020).
- Havelaar, A.H.; Kirk, M.D.; Torgerson, P.R.; Gibb, H.J.; Hald, T.; Lake, R.J.; Praet, N.; Bellinger, D.C.; De Silva, N.R.; Gargouri, N.; et al. World Health Organization Global Estimates and Regional Comparisons of the Burden of Foodborne Disease in 2010. PLoS Med. 2015, 12, 1001–1023. [Google Scholar] [CrossRef]
- Marks, F.; von Kalckreuth, V.; Aaby, P.; Adu-Sarkodie, Y.; El Tayeb, M.A.; Ali, M.; Aseffa, A.; Baker, S.; Biggs, H.M.; Bjerregaard-Andersen, M.; et al. Incidence of Invasive Salmonella Disease in Sub-Saharan Africa: A Multicentre Population-Based Surveillance Study. Lancet Glob. Health 2017, 5, 310–323. [Google Scholar] [CrossRef]
- Roesel, K.; Grace, D. Food Safety and Informal Markets: Animal Products in Sub-Saharan Africa Participatory Rural Appraisals Focus Group Discussions Key Informant Interviews Key Informant Interviews Mapping: Slaughter Process Mapping: Dairy Supply Chain Proportional Piling; Routledge: London, UK, 2014. [Google Scholar]
- Kingsley, R.A.; Msefula, C.L.; Thomson, N.R.; Kariuki, S.; Holt, K.E.; Gordon, M.A.; Harris, D.; Clarke, L.; Whitehead, S.; Sangal, V.; et al. Epidemic Multiple Drug Resistant Salmonella typhimurium Causing Invasive Disease in Sub-Saharan Africa Have a Distinct Genotype. Genome Res. 2009, 19, 2279–2287. [Google Scholar] [CrossRef]
- Muthumbi, E.; Morpeth, S.C.; Ooko, M.; Mwanzu, A.; Mwarumba, S.; Mturi, N.; Etyang, A.O.; Berkley, J.A.; Williams, T.N.; Kariuki, S.; et al. Invasive Salmonellosis in Kilifi, Kenya. Clin. Infect. Dis. 2015, 61, 290–301. [Google Scholar] [CrossRef]
- Ajayi, A.; Smith, S.I.; Kalpy, J.C.; Bode-Sojobi, I.O.; René, Y.K.; Adeleye, A.I. Molecular Diversity and Antibiotic Resistance Gene Profile of Salmonella enterica Serovars Isolated from Humans and Food Animals in Lagos, Nigeria. Acta Microbiol. Immunol. Hung. 2019, 66, 509–527. [Google Scholar] [CrossRef] [PubMed]
- Browne, A.J.; Kashef Hamadani, B.H.; Kumaran, E.A.P.; Rao, P.; Longbottom, J.; Harriss, E.; Moore, C.E.; Dunachie, S.; Basnyat, B.; Baker, S.; et al. Drug-Resistant Enteric Fever Worldwide, 1990 to 2018: A Systematic Review and Meta-Analysis. BMC Med. 2020, 18, 629–655. [Google Scholar] [CrossRef] [PubMed]
- Gadotti, C.; Forghani, F.; Diez-Gonzalez, F. Evaluation of Single and Combined Antimicrobial Treatments to Inhibit Salmonella in Queso Fresco. Food Microbiol. 2020, 85, 1032–1046. [Google Scholar] [CrossRef]
- Mehdi, Y.; Létourneau-Montminy, M.P.; Gaucher, M.L.; Chorfi, Y.; Suresh, G.; Rouissi, T.; Brar, S.K.; Côté, C.; Ramirez, A.A.; Godbout, S. Use of Antibiotics in Broiler Production: Global Impacts and Alternatives. Anim. Nutr. 2018, 4, 170–178. [Google Scholar] [CrossRef]
- Costa, M.C.; Bessegatto, J.A.; Alfieri, A.A.; Weese, J.S.; Filho, J.A.B.; Oba, A. Different Antibiotic Growth Promoters Induce Specific Changes in the Cecal Microbiota Membership of Broiler Chicken. PLoS ONE 2017, 12, 2193–2201. [Google Scholar] [CrossRef] [PubMed]
- Nyachieo, A.; Alafi, S.; Mutai, I.J.; Ngolobe, B.; Nabunje, R.; Nakavuma, J.L. Isolation and Characterization of Novel Lytic Phages to Combat Multidrug-Resistant E. Coli and Salmonella Spp. J. Microbiol. Infect. Dis. 2021, 11, 183–190. [Google Scholar] [CrossRef]
- Jung, L.S.; Ding, T.; Ahn, J. Evaluation of Lytic Bacteriophages for Control of Multidrug-Resistant Salmonella typhimurium. Ann. Clin. Microbiol. Antimicrob. 2017, 16, 66–76. [Google Scholar] [CrossRef]
- Aranaga, C.; Pantoja, L.D.; Andr, E.; Falco, A. Phage Therapy in the Era of Multidrug Resistance in Bacteria: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 4577. [Google Scholar] [CrossRef]
- Yongsheng, M.; Pacan, J.C.; Wang, Q.; Xu, Y.; Huang, X.; Korenevsky, A.; Sabour, P.M. Microencapsulation of Bacteriophage Felix O1 into Chitosan-Alginate Microspheres for Oral Delivery. Appl. Environ. Microbiol. 2008, 74, 4799–4805. [Google Scholar] [CrossRef]
- Mody, K.T.; Mahony, D.; Cavallaro, A.S.; Zhang, J.; Zhang, B.; Mahony, T.J.; Yu, C.; Mitter, N. Silica Vesicle Nanovaccine Formulations Stimulate Long-Term Immune Responses to the Bovine Viral Diarrhoea Virus E2 Protein. PLoS ONE 2015, 10, 325–332. [Google Scholar] [CrossRef]
- Vinner, G.K.; Rezaie-Yazdi, Z.; Leppanen, M.; Stapley, A.G.F.; Leaper, M.C.; Malik, D.J. Microencapsulation of Salmonella-Specific Bacteriophage Felix O1 Using Spray-Drying in a Ph-Responsive Formulation and Direct Compression Tableting of Powders into a Solid Oral Dosage Form. Pharmaceuticals 2019, 12, 43. [Google Scholar] [CrossRef]
- Ma, Y.H.; Islam, G.S.; Wu, Y.; Sabour, P.M.; Chambers, J.R.; Wang, Q.; Wu, S.X.Y.; Griffiths, M.W. Temporal Distribution of Encapsulated Bacteriophages during Passage through the Chick Gastrointestinal Tract. Poult. Sci. 2016, 95, 2911–2920. [Google Scholar] [CrossRef]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Aríñez-Soriano, J.; Cortés, P.; Maspoch, D.; Llagostera, M. Microencapsulation with Alginate/CaCO3: A Strategy for Improved Phage Therapy. Sci. Rep. 2017, 7, 271–283. [Google Scholar] [CrossRef]
- Clavijo, V.; Baquero, D.; Hernandez, S.; Farfan, J.C.; Arias, J.; Arévalo, A.; Donado-Godoy, P.; Vives-Flores, M. Phage Cocktail SalmoFREE® Reduces Salmonella on a Commercial Broiler Farm. Poult. Sci. 2019, 98, 5054–5063. [Google Scholar] [CrossRef]
- Li, Z.; Ma, W.; Li, W.; Ding, Y.; Zhang, Y.; Yang, Q.; Wang, J.; Wang, X. A Broad-Spectrum Phage Controls Multidrug-Resistant Salmonella in Liquid Eggs. Food Res. Int. 2020, 132, 109011. [Google Scholar] [CrossRef] [PubMed]
- Moye, Z.D.; Woolston, J.; Sulakvelidze, A. Bacteriophage Applications for Food Production and Processing. Viruses 2018, 10, 205. [Google Scholar] [CrossRef] [PubMed]
- Viertel, T.M.; Ritter, K.; Horz, H.-P. Viruses versus Bacteria-Novel Approaches to Phage Therapy as a Tool against Multidrug-Resistant Pathogens. Antimicrob. Chemother. 2014, 69, 2326–2336. [Google Scholar] [CrossRef]
- Abdelsattar, A.S.; Abdelrahman, F.; Dawoud, A.; Connerton, I.F.; El-Shibiny, A. Encapsulation of E. Coli Phage ZCEC5 in Chitosan–Alginate Beads as a Delivery System in Phage Therapy. AMB Express 2019, 9, 87–98. [Google Scholar] [CrossRef]
- Klein, S. The Use of Biorelevant Dissolution Media to Forecast the In Vivo Performance of a Drug. AAPS J. 2010, 12, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Śliwka, P.; Mituła, P.; Mituła, A.; Skaradziński, G.; Choińska-Pulit, A.; Niezgoda, N.; Weber-Dąbrowska, B.; Żaczek, M.; Skaradzińska, A. Encapsulation of Bacteriophage T4 in Mannitol-Alginate Dry Macrospheres and Survival in Simulated Gastrointestinal Conditions. LWT 2019, 99, 238–243. [Google Scholar] [CrossRef]
- Dressman, J.B.; Reppas, C. In Vitro-in Vivo Correlations for Lipophilic, Poorly Water-Soluble Drugs. Eur. J. Pharm. Sci. 2000, 11, 73–80. [Google Scholar] [CrossRef]
- Shi, L.E.; Zheng, W.; Zhang, Y.; Tang, Z.X. Milk-Alginate Microspheres: Protection and Delivery of Enterococcus faecalis HZNU P2. LWT-Food Sci. Technol. 2016, 65, 840–844. [Google Scholar] [CrossRef]
- Jiang, T.; Singh, B.; Maharjan, S.; Li, H.S.; Kang, S.K.; Bok, J.D.; Cho, C.S.; Choi, Y.J. Oral Delivery of Probiotic Expressing M Cell Homing Peptide Conjugated BmpB Vaccine Encapsulated into Alginate/Chitosan/Alginate Microcapsules. Eur. J. Pharm. Biopharm. 2014, 88, 768–777. [Google Scholar] [CrossRef]
- Morris, G.K.; Steele, C.D.; Wells, J.G. Evaluation of Plastic Multi-Well Plates for Serological Screening of Salmonella Cultures with Spicer-Edwards Pooled Antisera. Appl. Microbiol. 1972, 24, 846–848. [Google Scholar] [CrossRef]
- Shanmugasamy, M.; Velayutham, T.; Rajeswar, J. Inv a Gene Specific Pcr for Detection of Salmonella from Broilers. Vet. World 2011, 4, 562–564. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Gao, Y.; Li, C.; Ma, B.; Wang, H. Antimicrobial Resistance and CRISPR Typing Among Salmonella Isolates From Poultry Farms in China. Front. Microbiol. 2021, 12, 7300–7316. [Google Scholar] [CrossRef] [PubMed]
- Maszewska, A.; Wójcik, E.; Ciurzyńska, A.; Wojtasik, A.; Piatkowska, I.; Dastych, J.; Rózalski, A. Differentiation of Polyvalent Bacteriophages Specific to Uropathogenic Proteus Mirabilis Strains Based on the Host Range Pattern and RFLP. Acta Biochim. Pol. 2016, 63, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Duc, H.M.; Son, H.M.; Yi, H.P.S.; Sato, J.; Ngan, P.H.; Masuda, Y.; Honjoh, K.I.; Miyamoto, T. Isolation, Characterization and Application of a Polyvalent Phage Capable of Controlling Salmonella and Escherichia coli O157:H7 in Different Food Matrices. Food Res. Int. 2020, 131, 108977. [Google Scholar] [CrossRef]
- Lu, M.; Liu, H.; Lu, H.; Liu, R.; Liu, X. Characterization and Genome Analysis of a Novel Salmonella Phage VB_SenS_SE1. Curr. Microbiol. 2020, 77, 1308–1315. [Google Scholar] [CrossRef]
- Verthé, K.; Possemiers, S.; Boon, N.; Vaneechoutte, M.; Verstraete, W. Stability and Activity of an Enterobacter aerogenes-Specific Bacteriophage under Simulated Gastro-Intestinal Conditions. Appl. Microbiol. Biotechnol. 2004, 65, 465–472. [Google Scholar] [CrossRef]
- Koo, J.; DePaola, A.; Marshall, D.L. Effect of Simulated Gastric Fluid and Bile on Survival of Vibrio Vulnificus and Vibrio Vulnificus Phage. J. Food Prot. 2000, 63, 1665–1669. [Google Scholar] [CrossRef]
- Ramirez, K.; Cazarez-Montoya, C.; Lopez-Moreno, H.S.; Castro-del Campo, N. Bacteriophage Cocktail for Biocontrol of Escherichia coli O157:H7: Stability and Potential Allergenicity Study. PLoS ONE 2018, 13, e0195023. [Google Scholar] [CrossRef]
- Liu, A.; Liu, Y.; Peng, L.; Cai, X.; Shen, L.; Duan, M.; Ning, Y.; Liu, S.; Li, C.; Liu, Y.; et al. Characterization of the Narrow-Spectrum Bacteriophage LSE7621 towards Salmonella Enteritidis and Its Biocontrol Potential on Lettuce and Tofu. LWT 2020, 118, 1087–1091. [Google Scholar] [CrossRef]
- Tang, F.; Zhang, P.; Zhang, Q.; Xue, F.; Ren, J.; Sun, J.; Qu, Z.; Zhuge, X.; Li, D.; Wang, J.; et al. Isolation and Characterization of a Broad-Spectrum Phage of Multiple Drug Resistant Salmonella and Its Therapeutic Utility in Mice. Microb. Pathog. 2019, 126, 193–198. [Google Scholar] [CrossRef]
- Yan, T.; Liang, L.; Yin, P.; Zhou, Y.; Sharoba, A.M.; Lu, Q.; Dong, X.; Liu, K.; Connerton, I.F.; Li, J. Application of a Novel Phage LPSEYT for Biological Control of Salmonella in Foods. Microorganisms 2020, 8, 400. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Virk, S.M.; Shi, J.; Zhou, Y.; Willias, S.P.; Morsy, M.K.; Abdelnabby, H.E.; Liu, J.; Wang, X.; Li, J. Isolation, Characterization, and Application of Bacteriophage LPSE1 against Salmonella enterica in Ready to Eat (RTE) Foods. Front. Microbiol. 2018, 9, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Larock, D.L.; Chaudhary, A.; Miller, S.I. Salmonellae Interactions with Host Processes. Nat. Rev. Microbiol. 2015, 13, 191–205. [Google Scholar] [CrossRef]
- Gundy, P.M.; Gerba, C.P.; Pepper, I.L. Survival of Coronaviruses in Water and Wastewater. Food Environ. Virol. 2009, 1, 10–14. [Google Scholar] [CrossRef]
- Pinon, A.; Vialette, M. Survival of Viruses in Water. Intervirology 2019, 61, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Grabow, W.O.K. Bacteriophages: Update on Application as Models for Viruses in Water. Water SA 2001, 27, 251–268. [Google Scholar] [CrossRef]
- Ravindran, V. Feed Enzymes: The Science, Practice, and Metabolic Realities. J. Appl. Poult. Res. 2013, 22, 628–636. [Google Scholar] [CrossRef]
- KMD. Kenya Weather Forecast and Climate; Kenya Meteorogical Department: Nairobi, Kenya, 2022; Volume 254, pp. 4–8.
- Kentucky University Poultry Production Manual. Poultry House Evaluation Service; University of Kentucky: Lexington, KY, USA, 2019. [Google Scholar]
- Malik, D.J. Bacteriophage Encapsulation Using Spray Drying for Phage Therapy. Curr. Issues Mol. Biol. 2021, 40, 303–316. [Google Scholar] [CrossRef]
- Kittler, S.; Mengden, R.; Korf, I.H.E.; Bierbrodt, A.; Wittmann, J.; Plötz, M.; Jung, A.; Lehnherr, T.; Rohde, C.; Lehnherr, H.; et al. Impact of Bacteriophage-Supplemented Drinking Water on the E. coli Population in the Chicken Gut. Pathogens 2020, 9, 293. [Google Scholar] [CrossRef]
- Vinner, G.K.; Malik, D.J. High Precision Microfluidic Microencapsulation of Bacteriophages for Enteric Delivery. Res. Microbiol. 2018, 169, 522–530. [Google Scholar] [CrossRef]
- Jończyk, E.; Kłak, M.; Międzybrodzki, R.; Górski, A. The Influence of External Factors on Bacteriophages-Review. Folia Microbiol. 2011, 56, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Nobrega, F.L.; Costa, A.R.; Santos, J.F.; Siliakus, M.F.; Van Lent, J.W.M.; Kengen, S.W.M.; Azeredo, J.; Kluskens, L.D. Genetically Manipulated Phages with Improved PH Resistance for Oral Administration in Veterinary Medicine. Sci. Rep. 2016, 6, 3923–3925. [Google Scholar] [CrossRef] [PubMed]
- Stanford, K.; McAllister, T.A.; Niu, Y.D.; Stephens, T.P.; Mazzocco, A.; Waddell, T.E.; Johnson, R.P. Oral Delivery Systems for Encapsulated Bacteriophages Targeted Escherichia coli O157: H7 in Feedlot Cattle. J. Food Prot. 2010, 73, 1304–1312. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Pacan, J.C.; Wang, Q.; Sabour, P.M.; Huang, X.; Xu, Y. Enhanced Alginate Microspheres as Means of Oral Delivery of Bacteriophage for Reducing Staphylococcus aureus Intestinal Carriage. Food Hydrocoll. 2012, 26, 434–440. [Google Scholar] [CrossRef]
- Tang, Z.; Huang, X.; Sabour, P.M.; Chambers, J.R.; Wang, Q. Preparation and Characterization of Dry Powder Bacteriophage K for Intestinal Delivery through Oral Administration. LWT-Food Sci. Technol. 2015, 60, 263–270. [Google Scholar] [CrossRef]
- Salo, R.J.; Cliver, D.O. Effect of Acid PH, Salts, and Temperature on the Infectivity and Physical Integrity of Enteroviruses. Arch. Virol. 1976, 52, 269–282. [Google Scholar] [CrossRef]
- Dąbrowska, K. Phage Therapy: What Factors Shape Phage Pharmacokinetics and Bioavailability? Systematic and Critical Review. Med. Res. Rev. 2019, 39, 2000–2006. [Google Scholar] [CrossRef]
- Tokman, J.I.; Kent, D.J.; Wiedmann, M.; Denes, T. Temperature Significantly Affects the Plaquing and Adsorption Efficiencies of Listeria Phages. Front. Microbiol. 2016, 7, 631–641. [Google Scholar] [CrossRef]
- Seeley, N.D.; Primrose, S.B. The Effect of Temperature on the Ecology of Aquatic Bacteriophages. J. Gen. Virol. 1980, 46, 87–95. [Google Scholar] [CrossRef]
- Cvirkaite-Krupovič, V.; Krupovič, M.; Daugelavičius, R.; Bamford, D.H. Calcium Ion-Dependent Entry of the Membrane-Containing Bacteriophage PM2 into Its Pseudoalteromonas Host. Virology 2010, 405, 120–128. [Google Scholar] [CrossRef]
- Chhibber, S.; Kaur, T.; Kaur, S. Essential Role of Calcium in the Infection Process of Broad-Spectrum Methicillin-Resistant Staphylococcus aureus Bacteriophage. J. Basic Microbiol. 2014, 54, 775–780. [Google Scholar] [CrossRef]
- Raza, T.; Andleeb, S.; Ullah, S.R.; Jamal, M.; Mehmood, K.; Ali, M. Isolation and Characterization of a Phage to Control Vancomycin Resistant Enterococcus faecium. Open Life Sci. 2018, 13, 553–560. [Google Scholar] [CrossRef]
- Armon, R.; Araujo, R.; Kott, Y.; Lucena, F.; Jofre, J. Bacteriophages of Enteric Bacteria in Drinking Water, Comparison of Their Distribution in Two Countries. J. Appl. Microbiol. 1997, 83, 627–633. [Google Scholar] [CrossRef]
- Seeley, N.D.; Primrose, S.B. The Isolation of Bacteriophages from the Environment. J. Appl. Bacteriol. 1982, 53, 1365–1382. [Google Scholar] [CrossRef]
- Lorenzo-Rebenaque, L.; Malik, D.J.; Catalá-Gregori, P.; Marin, C.; Sevilla-Navarro, S. Gastrointestinal Dynamics of Non-Encapsulated and Microencapsulated Salmonella Bacteriophages in Broiler Production. Animals 2022, 12, 144. [Google Scholar] [CrossRef]
- White, H.E.; Sherman, M.B.; Brasilès, S.; Jacquet, E.; Seavers, P.; Tavares, P.; Orlova, E.V. Capsid Structure and Its Stability at the Late Stages of Bacteriophage SPP1 Assembly. J. Virol. 2012, 86, 6768–6777. [Google Scholar] [CrossRef]
- Parker, N.; Schneegurt, M.; Tu, A.-H.T.; Lister, P.; Forster, B.M. The Effects of PH and Temperature on Microbial Growth. Microbiology 2016, 1, 317–323. [Google Scholar]
- Ma, L.; Green, S.I.; Trautner, B.W.; Ramig, R.F.; Maresso, A.W. Metals Enhance the Killing of Bacteria by Bacteriophage in Human Blood. Sci. Rep. 2018, 8, 2326–2331. [Google Scholar] [CrossRef]








| Sal 16 | Sal 73 | Sal 157 | Sal 172 | Sal 177 | Sal 181 | Sal 182 | Sal 187 | Sal 188 | Sal 192 | Sal 194 | Sal 312 | Sal 568 | Sal 569 | Sal 571 | Sal 572 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| invA | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Group O (A-S) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Poly H (Phase 1 & 2) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| Group D (9) | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| CRISPR 1 | E | E | C | K | E | K | K | H | H | C | C | E | E | E | K | E |
| CRISPR 2 | E | E | H | K | E | K | K | H | H | H | H | E | E | E | K | E |
| Phages | Original Salmonella Strain | Identity of Poultry Farm (PF)/Slaughter House (SH) | Region | Remarks |
|---|---|---|---|---|
| ILRI_K1 | Sal16 | PF_16 | Kiambu (Peri-Urban) | Salmonella isolated |
| ILRI_K3 | Sal16 | SH_6 | Nairobi (Urban) | Salmonella absent |
| ILRI_K6 | Sal16 | SH_7 | Nairobi (Urban) | Salmonella absent |
| ILRI_K9 | Sal16 | PF-58 | Nairobi (Urban) | Salmonella absent |
| ILRI_K11 | Sal73 | SH_1 | Kiambu (Peri-Urban) | Salmonella isolated |
| ILRI_K14 | Sal73 | PF_33 | Kiambu (Peri-Urban) | Salmonella isolated |
| ILRI_K22 | Sal177 | PF_16 | Kiambu (Peri-Urban) | Salmonella isolated |
| ILRI_K24 | Sal177 | SH_6 | Nairobi (Urban) | Salmonella absent |
| ILRI_K26 | Sal177 | SH_6 | Nairobi (Urban) | Salmonella absent |
| ILRI_K47 | Sal312 | SH_6 | Nairobi (Urban) | Salmonella absent |
| Most Similar Kenyan Phage Genome | Most Similar Phage Genome from the NCBI Public Database | ||||
|---|---|---|---|---|---|
| Phage Name | Nucleotide Similarity % (Aligned Nucleotide %) | Phage Name | NCBI Accession Number | Nucleotide Similarity % (Aligned Nucleotide %) | |
| ILRI_K1 * | ILRI_K22 | 99.99 (100%) | Salmonella phage wast | MT074451.1 | 93.72% (90%) |
| ILRI_K3 | ILRI_K24 | 99.79% (100%) | Salmonella phage wast | MT074451.1 | 92.18% (90%) |
| ILRI_K6 | ILRI_K24 | 98.23% (97%) | Salmonella phage wast | MT074451.1 | 92.30% (86%) |
| ILRI_K9 | ILRI_K26 | 99.99 (100%) | Salmonella phage wast | MT074451.1 | 92.70% (90%) |
| ILRI_K11 | None | -- | Salmonella phage SP6 | AY288927.2 | 89.67% (90%) |
| ILRI_K14 | ILRI_K1 & _K22 | 97.27% (96%) | Salmonella phage wast | MT074451.1 | 93.59% (92%) |
| ILRI_K22 | ILRI_K1 | 99.99 (100%) | Salmonella phage wast | MT074451.1 | 93.72% (90%) |
| ILRI_K24 | ILRI_K3 | 99.79% (100%) | Salmonella phage wast | MT074451.1 | 92.41% (90%) |
| ILRI_K26 ** | ILRI_K9 | 99.99 (93%) | Salmonella phage wast | MT074451.1 | 92.55% (90%) |
| ILRI_K47 | ILRI_K9 | 96.74% (93%) | Salmonella phage wast | MT074451.1 | 92.44% (91%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhone, A.L.; Makumi, A.; Odaba, J.; Guantai, L.; Gunathilake, K.M.D.; Loignon, S.; Ngugi, C.W.; Akhwale, J.K.; Moineau, S.; Svitek, N. Salmonella Enteritidis Bacteriophages Isolated from Kenyan Poultry Farms Demonstrate Time-Dependent Stability in Environments Mimicking the Chicken Gastrointestinal Tract. Viruses 2022, 14, 1788. https://doi.org/10.3390/v14081788
Mhone AL, Makumi A, Odaba J, Guantai L, Gunathilake KMD, Loignon S, Ngugi CW, Akhwale JK, Moineau S, Svitek N. Salmonella Enteritidis Bacteriophages Isolated from Kenyan Poultry Farms Demonstrate Time-Dependent Stability in Environments Mimicking the Chicken Gastrointestinal Tract. Viruses. 2022; 14(8):1788. https://doi.org/10.3390/v14081788
Chicago/Turabian StyleMhone, Amos Lucky, Angela Makumi, Josiah Odaba, Linda Guantai, K. M. Damitha Gunathilake, Stéphanie Loignon, Caroline Wangari Ngugi, Juliah Khayeli Akhwale, Sylvain Moineau, and Nicholas Svitek. 2022. "Salmonella Enteritidis Bacteriophages Isolated from Kenyan Poultry Farms Demonstrate Time-Dependent Stability in Environments Mimicking the Chicken Gastrointestinal Tract" Viruses 14, no. 8: 1788. https://doi.org/10.3390/v14081788
APA StyleMhone, A. L., Makumi, A., Odaba, J., Guantai, L., Gunathilake, K. M. D., Loignon, S., Ngugi, C. W., Akhwale, J. K., Moineau, S., & Svitek, N. (2022). Salmonella Enteritidis Bacteriophages Isolated from Kenyan Poultry Farms Demonstrate Time-Dependent Stability in Environments Mimicking the Chicken Gastrointestinal Tract. Viruses, 14(8), 1788. https://doi.org/10.3390/v14081788







