Mayaro Virus: The State-of-the-Art for Antiviral Drug Development
Abstract
:1. Introduction
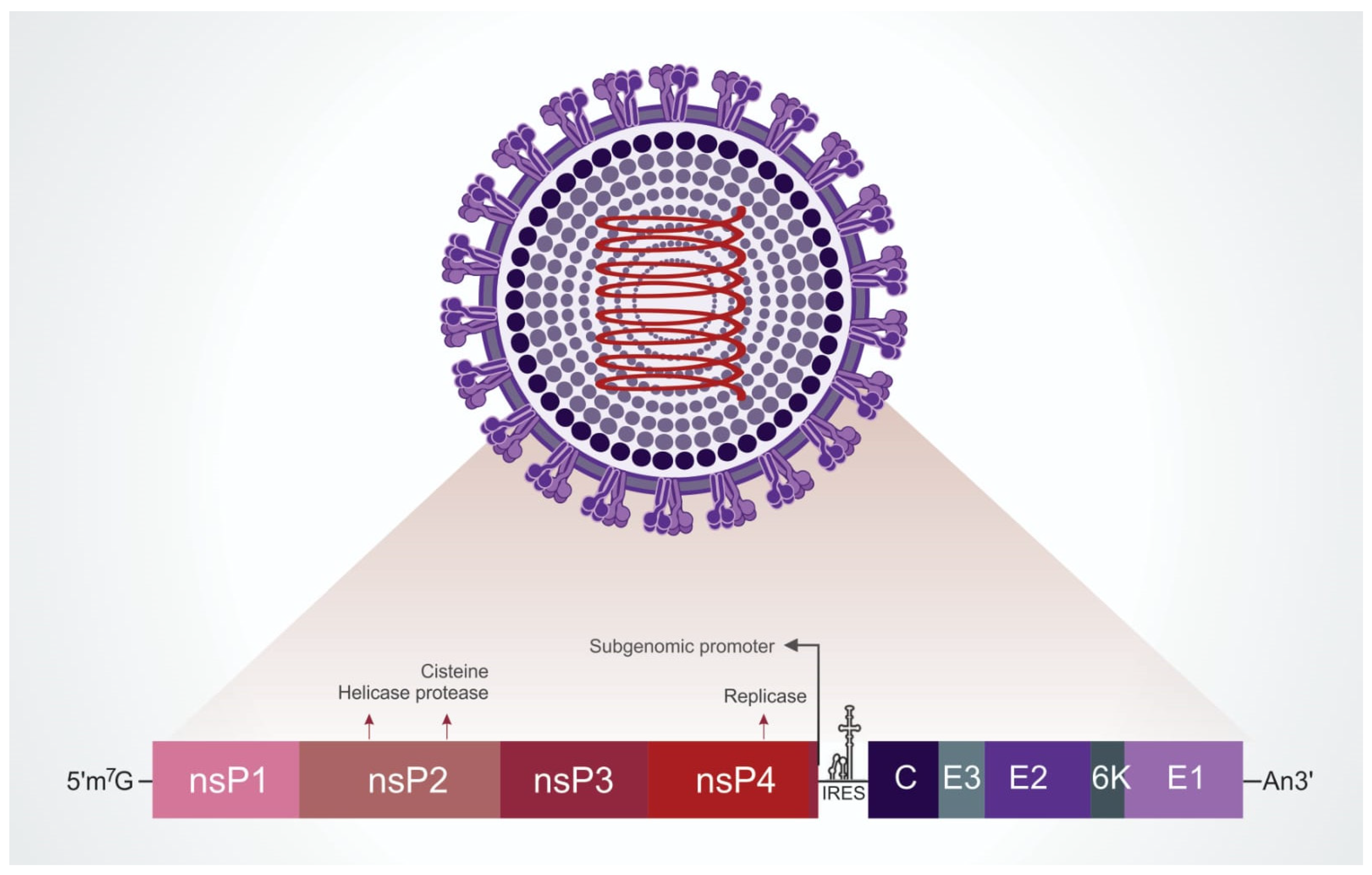
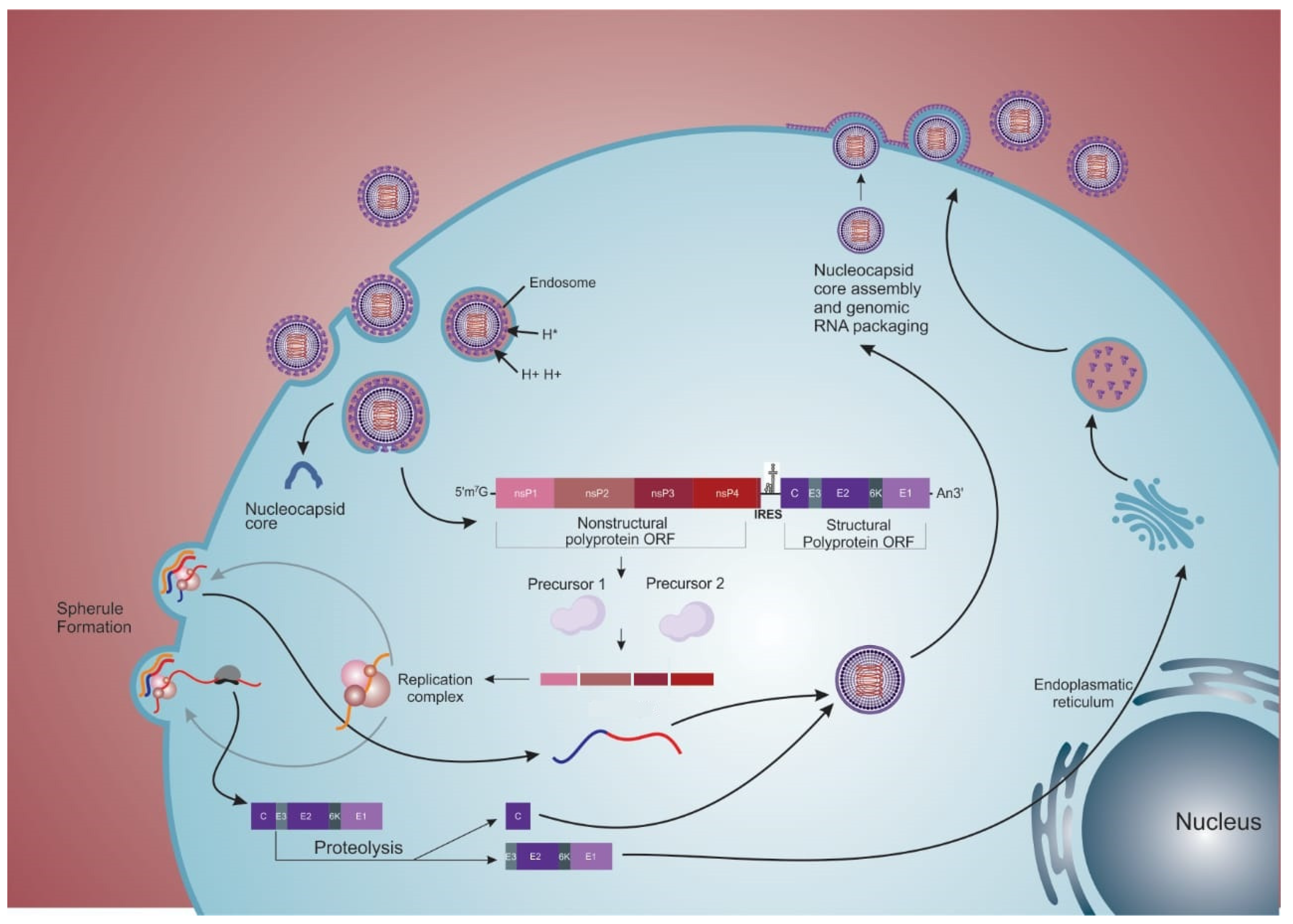
2. Viral Structure and Replication Cycle
2.1. Viral Particle
2.2. Internalization
2.3. Replication
2.4. Exit
2.5. “Cell-to-Cell” Transmission
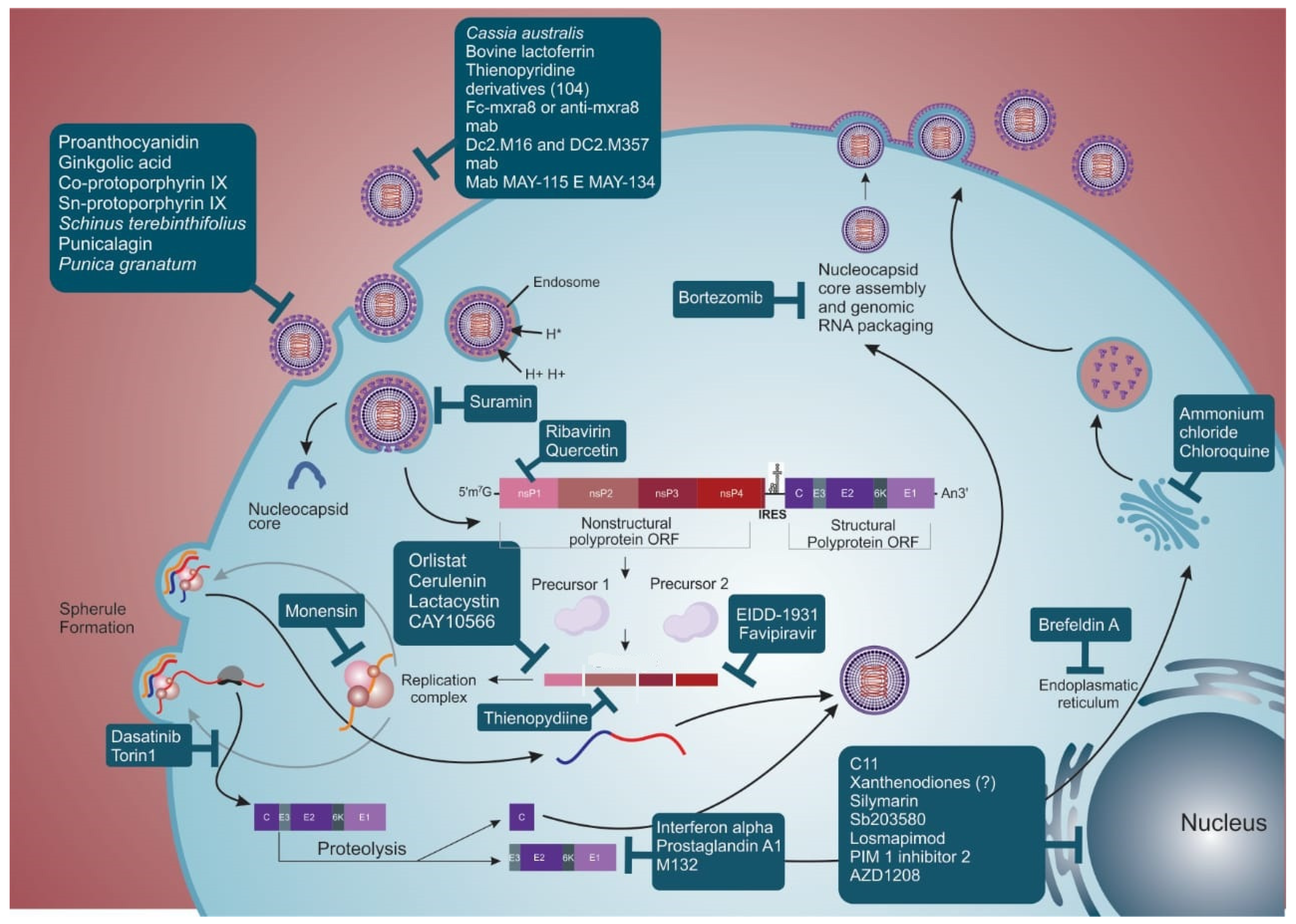
3. Antivirals
3.1. Virucidal Compounds
3.2. Compounds That Interfere with Adsorption and Internalization of the Virus Particle
3.3. Compounds That Interfere with Replication, Morphogenesis, and Viral Exit
3.4. Drugs That Modulate Host Response to Viral Infection
4. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Calisher, C.H.; Karabatsos, N. Arbovirus Serogroups: Definition and Distribution. In The Arboviruses: Epidemiology and Ecology; Monath, T.P., Ed.; CRC Press: New York, NY, USA, 1988; pp. 19–57. [Google Scholar]
- Knipe, D.M.; Howley, P.M. Fields Virology, 6th ed.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2013; ISBN 9780849340871. [Google Scholar]
- da Saúde, M. Boletim de Vigilância Laboratorial Dos Arbovírus. Bol. Epidemiológico 2021, 52, 7–18. [Google Scholar]
- Coffey, L.L.; Forrester, N.; Tsetsarkin, K.; Vasilakis, N.; Weaver, S.C. Factors Shaping the Adaptive Landscape for Arboviruses: Implications for the Emergence of Disease. Future Microbiol. 2013, 8, 155–176. [Google Scholar] [CrossRef] [PubMed]
- Travassos da Rosa, A.; Shope, R.; Pinheiro, F.; Travassos da Rosa, J.; Vasconcelos, P.; Herve, J.; Degallier, N. Arbovirus Research in the Brazilian Amazon. In Proceedings of the Fifth Symposium on Arbovírus Research in Australia, Brisbane, Australia, 28 August–1 September 1989; pp. 4–8. [Google Scholar]
- Zanluca, C.; De Melo, V.C.A.; Mosimann, A.L.P.; Dos Santos, G.I.V.; dos Santos, C.N.D.; Luz, K. First Report of Autochthonous Transmission of Zika Virus in Brazil. Mem. Inst. Oswaldo Cruz 2015, 110, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, I.G.C.D.; Marandino, R.; Mendonça, A.P.; Nogueira, R.M.R.; Vasconcelos, P.F.D.C.; Guerra, L.R.; Brandão, B.C.; Mendonça, A.P.P.; Aguiar, G.R.; Bacco, P.A.M.D. Chikungunya Virus Infection: Report of the First Case Diagnosed in Rio de Janeiro, Brazil. Rev. Soc. Bras. Med. Trop. 2012, 45, 128–129. [Google Scholar] [CrossRef]
- Pezzi, L.; Rodriguez-Morales, A.J.; Reusken, C.B.; Ribeiro, G.S.; LaBeaud, A.D.; Lourenço-de-Oliveira, R.; Brasil, P.; Lecuit, M.; Failloux, A.B.; Gallian, P.; et al. GloPID-R Report on Chikungunya, o’nyong-Nyong and Mayaro Virus, Part 3: Epidemiological Distribution of Mayaro Virus. Antivir. Res. 2019, 172, 104610. [Google Scholar] [CrossRef]
- Lopes, N.; Nozawa, C.; Linhares, R.E.C. Características Gerais e Epidemiologia Dos Arbovírus Emergentes No Brasil. Rev. Pan-Amazônica Saúde 2014, 5, 55–64. [Google Scholar] [CrossRef]
- Saatkamp, C.J.; Rodrigues, L.R.R.; Pereira, A.M.N.; Coelho, J.A.; Marques, R.G.B.; Souza, V.C.D.; Nascimento, V.A.D.; Saatkamp, J.G.D.S.; Naveca, F.G.; Figueiredo, R.M.P.D. Mayaro Virus Detection in the Western Region of Pará State, Brazil. Rev. Soc. Bras. Med. Trop. 2021, 54, 1–3. [Google Scholar] [CrossRef]
- Romeiro, M.F.; Fumagalli, M.J.; dos Anjos, A.B.; Figueiredo, L.T.M. Serological Evidence of Mayaro Virus Infection in Blood Donors from São Carlos, São Paulo, Brazil. Trans. R. Soc. Trop. Med. Hyg. 2020, 114, 693–696. [Google Scholar] [CrossRef]
- Brunini, S.; França, D.D.S.; Silva, J.B.; Silva, L.N.; Silva, F.P.A.; Spadoni, M.; Rezza, G. High Frequency of Mayaro Virus IgM among Febrile Patients, Central Brazil. Emerg. Infect. Dis. 2017, 23, 1025–1026. [Google Scholar] [CrossRef]
- Coimbra, T.L.M.; Santos, C.L.S.; Suzuki, A.; Petrella, S.M.C.; Bisordi, I.; Nagamori, A.H.; Marti, A.T.; Santos, R.N.; Fialho, D.M.; Lavigne, S.; et al. Mayaro Virus: Imported Cases of Human Infection in São Paulo State, Brazil. Rev. Inst. Med. Trop. Sao Paulo 2007, 49, 221–224. [Google Scholar] [CrossRef]
- de Paula Silveira-Lacerda, E.; Laschuk Herlinger, A.; Tanuri, A.; Rezza, G.; Anunciação, C.E.; Ribeiro, J.P.; Tannous, I.P.; Abrantes, G.R.; da Silva, E.G.; Arruda, K.F.; et al. Molecular Epidemiological Investigation of Mayaro Virus in Febrile Patients from Goiania City, 2017–2018. Infect. Genet. Evol. 2021, 95, 104981. [Google Scholar] [CrossRef] [PubMed]
- Esposito, D.L.A.; Fonseca, B.A.L.d. Will Mayaro Virus Be Responsible for the next Outbreak of an Arthropod-Borne Virus in Brazil? Braz. J. Infect. Dis. 2017, 21, 540–544. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, R.S.S.; Chiang, J.O.; Martins, L.C.; Viana, G.M.R. Monitoramento de Casos de Febre Do Mayaro e Febre Do Oropouche Até a Semana Epidemiológica 35, 2019. Bol. Epidemiológico 2019, 50, 14–16. [Google Scholar]
- Lorenz, C.; Ribeiro, A.F.; Chiaravalloti-neto, F. Mayaro Virus Distribution in South America. Acta Trop. 2019, 198, 105093. [Google Scholar] [CrossRef]
- Webb, E.M.; Azar, S.R.; Haller, S.L.; Langsjoen, R.M.; Cuthbert, C.E.; Ramjag, A.T.; Luo, H.; Plante, K.; Wang, T.; Simmons, G.; et al. Effects of Chikungunya Virus Immunity on Mayaro Virus Disease and Epidemic Potential. Sci. Rep. 2019, 9, 20399. [Google Scholar] [CrossRef]
- Fumagalli, M.J.; de Souza, W.M.; de Castro-Jorge, L.A.; de Carvalho, R.V.H.; Castro, Í.A.; de Almeida, L.G.N.; Consonni, S.R.; Zamboni, D.S.; Figueiredo, L.T.M. Chikungunya Virus Exposure Partially Cross-Protects against Mayaro Virus Infection in Mice. J. Virol. 2021, 95, e01122-21. [Google Scholar] [CrossRef]
- Malonis, R.J.; Earnest, J.T.; Kim, A.S.; Angeliadis, M.; Holtsberg, F.W.; Javad Aman, M.; Jangra, R.K.; Chandran, K.; Daily, J.P.; Diamond, M.S.; et al. Near-Germline Human Monoclonal Antibodies Neutralize and Protect against Multiple Arthritogenic Alphaviruses. Proc. Natl. Acad. Sci. USA 2021, 118, e2100104118. [Google Scholar] [CrossRef]
- de Figueiredo, M.L.G.; Figueiredo, L.T.M. Emerging Alphaviruses in the Americas: Chikungunya and Mayaro. Rev. Soc. Bras. Med. Trop. 2014, 47, 677–683. [Google Scholar] [CrossRef]
- Lima, W.G.; Pereira, R.S.; da Cruz Nizer, W.S.; Brito, J.C.M.; Godói, I.P.; Cardoso, V.N.; Fernandes, S.O.A.; Ferreira, J.M.S. Rate of Exposure to Mayaro Virus (MAYV) in Brazil between 1955 and 2018: A Systematic Review and Meta-Analysis. Arch. Virol. 2021, 166, 347–361. [Google Scholar] [CrossRef]
- da Costa, V.G.; de Rezende Féres, V.C.; Saivish, M.V.; de Lima Gimaque, J.B.; Moreli, M.L. Silent Emergence of Mayaro and Oropouche Viruses in Humans in Central Brazil. Int. J. Infect. Dis. 2017, 62, 84–85. [Google Scholar] [CrossRef]
- Filho, P.; de Paula, F. Epidemias Simultâneas de Mayaro e Febre Amarela Em Belterra, Pará. Bol. Epidemiológico 1978, 10, 146–152. [Google Scholar]
- Anderson, C.R.; Downs, W.G.; Wattley, G.H.; Ahin, N.W.; Reese, A.A. Mayaro Virus: A New Human Disease Agent: II. Isolation from Blood of Patients in Trinidad, B.W.I. Am. J. Trop. Med. Hyg. 1957, 6, 1012–1016. [Google Scholar] [CrossRef] [PubMed]
- Diagne, C.T.; Bengue, M.; Choumet, V.; Hamel, R.; Pompon, J.; Missé, D. Mayaro Virus Pathogenesis and Transmission Mechanisms. Pathogens 2020, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- McGill, P.E. Viral Infections: α-Viral Arthropathy. Bailliere’s Clin. Rheumatol. 1995, 9, 145–150. [Google Scholar] [CrossRef]
- Pinheiro, F.P.; Freitas, R.B.; Travassos da Rosa, J.F.; Gabbay, Y.B.; Mello, W.A.; LeDuc, J.W. An Outbreak of Mayaro Virus Disease in Belterra, Brazil. I. Clinical and Virological Findings. Am. J. Trop. Med. Hyg. 1981, 30, 674–681. [Google Scholar] [CrossRef]
- Theilacker, C.; Held, J.; Allering, L.; Emmerich, P.; Schmidt-Chanasit, J.; Kern, W.V.; Panning, M. Prolonged Polyarthralgia in a German Traveller with Mayaro Virus Infection without Inflammatory Correlates. BMC Infect. Dis. 2013, 13, 2011–2014. [Google Scholar] [CrossRef]
- do Rosário Casseb, A.; Casseb LM, N.; da Silva, S.P.; da Costa Vasconcelos, P.F. Arbovírus: Importante Zoonose Na Amazônia Brasileira. Vet. Zootec. 2013, 20, 9–21. [Google Scholar]
- Lednicky, J.; Beau De Rochars, V.M.; Elbadry, M.; Loeb, J.; Telisma, T.; Chavannes, S.; Anilis, G.; Cella, E.; Ciccozzi, M.; Okech, B.; et al. Mayaro Virus in Child with Acute Febrile Illness, Haiti, 2015. Emerg. Infect. Dis. 2016, 22, 2000–2002. [Google Scholar] [CrossRef]
- Aguilar-Luis, M.A.; del Valle-Mendoza, J.; Sandoval, I.; Silva-Caso, W.; Mazulis, F.; Carrillo-Ng, H.; Tarazona-Castro, Y.; Martins-Luna, J.; Aquino-Ortega, R.; Peña-Tuesta, I.; et al. A Silent Public Health Threat: Emergence of Mayaro Virus and Co-Infection with Dengue in Peru. BMC Res. Notes 2021, 14, 29. [Google Scholar] [CrossRef]
- Slegers, C.A.D.; Keuter, M.; Günther, S.; Schmidt-Chanasit, J.; van der Ven, A.J.; de Mast, Q. Persisting Arthralgia Due to Mayaro Virus Infection in a Traveler from Brazil: Is There a Risk for Attendants to the 2014 FIFA World Cup? J. Clin. Virol. 2014, 60, 317–319. [Google Scholar] [CrossRef]
- Llagonne-Barets, M.; Icard, V.; Leparc-Goffart, I.; Prat, C.; Perpoint, T.; André, P.; Ramière, C. A Case of Mayaro Virus Infection Imported from French Guiana. J. Clin. Virol. 2016, 77, 66–68. [Google Scholar] [CrossRef]
- Vieira, C.J.d.S.P.; da Silva, D.J.F.; Barreto, E.S.; Siqueira, C.E.H.; Colombo, T.E.; Ozanic, K.; Schmidt, D.J.; Drumond, B.P.; Mondini, A.; Nogueira, M.L.; et al. Detection of Mayaro Virus Infections during a Dengue Outbreak in Mato Grosso, Brazil. Acta Trop. 2015, 147, 12–16. [Google Scholar] [CrossRef]
- Pereira, T.N.; Carvalho, F.D.; De Mendonça, S.F.; Rocha, M.N.; Moreira, L.A. Vector Competence of Aedes Aegypti, Aedes Albopictus, and Culex Quinquefasciatus Mosquitoes for Mayaro Virus. PLoS Negl. Trop. Dis. 2020, 14, e0007518. [Google Scholar] [CrossRef] [PubMed]
- PAHO; WHO. Epidemiological Alert: Mayaro Fever; PAHO/WHO: Washington, DC, USA, 2019; pp. 1–5. [Google Scholar]
- Mello, M.V.P.; Domingos, T.F.S.; Ferreira, D.F.; Ribeiro, M.M.J.; Ribeiro, T.P.; Rodrigues, C.R.; Souza, A.M.T. Antiviral Drug Discovery and Development for Mayaro Fever—What Do We Have so Far? Mini-Rev. Med. Chem. 2020, 20, 921–928. [Google Scholar] [CrossRef] [PubMed]
- Lima-Camara, T.N. Arboviroses Emergentes e Novos Desafios Para a Saúde Pública No Brasil. Rev. Saude Publica 2016, 50, 1–7. [Google Scholar] [CrossRef]
- Mezencio, J.M.; de Souza, W.; Fonseca, M.E.; Rebello, M.A. Replication of Mayaro Virus in Aedes Albopictus Cells: An Electron Microscopic Study. Arch. Virol. 1989, 104, 299–308. [Google Scholar] [CrossRef]
- Ribeiro-Filho, H.V.; Coimbra, L.D.; Cassago, A.; Rocha, R.P.F.; Guerra, J.V.d.S.; de Felicio, R.; Carnieli, C.M.; Leme, L.; Padilha, A.C.; Paes Leme, A.F.; et al. Cryo-EM Structure of the Mature and Infective Mayaro Virus at 4.4 Å Resolution Reveals Features of Arthritogenic Alphaviruses. Nat. Commun. 2021, 12, 3038. [Google Scholar] [CrossRef]
- Ramsey, J.; Mukhopadhyay, S. Disentangling the Frames, the State of Research on the Alphavirus 6K and TF Proteins. Viruses 2017, 9, 228. [Google Scholar] [CrossRef]
- Espósito, D.L.A.; da Fonseca, B.A.L. Complete Genome Sequence of Mayaro Virus (Togaviridae, Alphavirus) Strain BeAr 20290 from Brazil. Genome Announc. 2015, 3, 141660. [Google Scholar] [CrossRef]
- Jose, J.; Snyder, J.E.; Kuhn, R.J. A Structural and Functional Perspective of Alphavirus Replication and Assembly. Future Microbiol. 2009, 4, 837–856. [Google Scholar] [CrossRef]
- Mezencio, J.M.; Rebello, M.A. Mayaro Virus Proteins. Mem. Inst. Oswaldo Cruz 1992, 88, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G. The Alphaviruses: Gene Expression, Replication, and Evolution. Microbiol. Rev. 1994, 58, 491–562. [Google Scholar] [CrossRef] [PubMed]
- Lavergne, A.; Thoisy, D.; Lacoste, V.; Tolou, H.; Dussart, P.; Morvan, J.; Talarmin, A.; Kazanji, M. Mayaro Virus: Complete Nucleotide Sequence and Phylogenetic Relationships with Other Alphaviruses. Virus Res. 2006, 117, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Bakar, F.A.; Ng, L.F.P. Nonstructural Proteins of Alphavirus–Potential Targets for Drug Development. Viruses 2018, 10, 71. [Google Scholar] [CrossRef]
- Carvalho, C.A.M.; Silva, J.L.; Oliveira, A.C.; Gomes, A.M.O. On the Entry of an Emerging Arbovirus into Host Cells: Mayaro Virus Takes the Highway to the Cytoplasm through Fusion with Early Endosomes and Caveolae-Derived Vesicles. PeerJ 2017, 5, e3245. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Kim, A.S.; Fox, J.M.; Nair, S.; Basore, K.; Klimstra, W.B.; Rimkunas, R.; Fong, R.H.; Lin, H.; Poddar, S.; et al. Mxra8 Is a Receptor for Multiple Arthritogenic Alphaviruses. Nature 2018, 557, 570–574. [Google Scholar] [CrossRef] [PubMed]
- Kielian, M.; Chanel-vos, C.; Liao, M. Alphavirus Entry and Membrane Fusion. Viruses 2010, 2, 796–825. [Google Scholar] [CrossRef]
- Kononchik, J.P., Jr.; Hernandez, R.; Brown, D.T. An Alternative Pathway for Alphavirus Entry. Virol. J. 2011, 8, 304. [Google Scholar] [CrossRef]
- Singh, I.; Helenius, A. Role of Ribosomes in Semliki Forest Virus Nucleocapsid Uncoating. J. Virol. 1992, 66, 7049–7058. [Google Scholar] [CrossRef]
- Wengler, G.; Wengler, G. Identification of a Transfer of Viral Core Protein to Cellular Ribosomes during the Early Stages of Alphavirus Infection. Virology 1984, 134, 435–442. [Google Scholar] [CrossRef]
- Wengler, G.; Würkner, D.; Wengler, G. Identification of a Sequence Element in the Alphavirus Core Protein Which Mediates Interaction of Cores with Ribosomes and the Disassembly of Cores. Virology 1992, 191, 880–888. [Google Scholar] [CrossRef]
- Glanville, N.; Ranki, M.; Morser, J. Initiation of Translocation Directed by 42S and 26S RNAs from Semliki Forest Virus in Vitro. Proc. Natl. Acad. Sci. USA 1976, 73, 3059–3063. [Google Scholar] [CrossRef] [PubMed]
- Pietilä, M.K.; Hellström, K.; Ahola, T. Alphavirus Polymerase and RNA Replication. Virus Res. 2017, 234, 44–57. [Google Scholar] [CrossRef]
- Grimley, P.M.; Berezesky, I.K.; Friedman, R.M. Cytoplasmic Structures Associated with an Arbovirus Infection: Loci of Viral Ribonucleic Acid Synthesis. J. Virol. 1968, 2, 1326–1338. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.M.; Levin, J.G.; Grimley, P.M.; Berezesky, I.K. Membrane-Associated Replication Complex in Arbovirus Infection. J. Virol. 1972, 10, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Rice, C.M. The Signal for Translational Readthrough of a UGA Codon in Sindbis Virus RNA Involves a Single Cytidine Residue Immediately Downstream of the Termination Codon. J. Virol. 1993, 67, 5062–5067. [Google Scholar] [CrossRef]
- Kallio, K.; Hellström, K.; Jokitalo, E.; Ahola, T. RNA Replication and Membrane Modification Require the Same Functions of Alphavirus Nonstructural Proteins. J. Virol. 2016, 90, 1687–1692. [Google Scholar] [CrossRef]
- Shirako, Y.; Strauss, J.H. Cleavage between NsP1 and NsP2 Initiates the Processing Pathway of Sindbis Virus Nonstructural Polyprotein P123. Virology 1990, 177, 54–64. [Google Scholar] [CrossRef]
- Lemm, J.A.; Rice, C.M. Assembly of Functional Sindbis Virus RNA Replication Complexes: Requirement for Coexpression of P123 and P34. J. Virol. 1993, 67, 1905–1915. [Google Scholar] [CrossRef]
- Lemm, J.A.; Rümenapf, T.; Strauss, E.G.; Strauss, J.H.; Mrice, C. Polypeptide Requirements for Assembly of Functional Sindbis Virus Replication Complexes: A Model for the Temporal Regulation of Minus- and plus-Strand RNA Synthesis. EMBO J. 1994, 13, 2925–2934. [Google Scholar] [CrossRef]
- Sawicki, D.L.; Sawicki, S.G. Short-Lived Minus-Strand Polymerase for Semliki Forest Virus. J. Virol. 1980, 34, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Sawicki, D.L.; Sawicki, S.G. Alphavirus Positive and Negative Strand RNA Synthesis and the Role of Polyproteins in Formation of Viral Replication Complexes. Arch. Virol. Suppl. 1994, 9, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Ishida, R.; Cole, J.; Lopez-Orozco, J.; Fayad, N.; Felix-Lopez, A.; Elaish, M.; Luo, S.Y.; Julien, O.; Kumar, A.; Hobman, T.C. Mayaro Virus Non-Structural Protein 2 Circumvents the Induction of Interferon in Part by Depleting Host Transcription Initiation Factor IIE Subunit 2. Cells 2021, 10, 3510. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA Synthesis and Non-Structural Protein Functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef] [PubMed]
- Adkins, S.; Stawicki, S.S.; Faurote, G.; Siegel, R.W.; Kao, C.C. Mechanistic Analysis of RNA Synthesis by RNA-Dependent RNA Polymerase from Two Promoters Reveals Similarities to DNA-Dependent RNA Polymerase. RNA 1998, 4, 455–470. [Google Scholar] [PubMed]
- Fros, J.J.; Pijlman, G.P. Alphavirus Infection: Host Cell Shut-Off and Inhibition of Antiviral Responses. Viruses 2016, 8, 166. [Google Scholar] [CrossRef]
- Mezencio, J.M.; de Souza, W.; Fonseca, M.E.; Rebello, M.A. Ultrastructural Study of Mayaro Virus Replication in BHK-21 Cells. Arch. Virol. 1990, 114, 229–235. [Google Scholar] [CrossRef]
- Nieva, J.L.; Madan, V.; Carrasco, L. Viroporins: Structure and Biological Functions. Nat. Rev. Microbiol. 2012, 10, 563–574. [Google Scholar] [CrossRef]
- Snyder, J.E.; Kulcsar, K.A.; Schultz, K.L.W.; Riley, C.P.; Neary, J.T.; Marr, S.; Jose, J.; Griffin, D.E.; Kuhn, R.J. Functional Characterization of the Alphavirus TF Protein. J. Virol. 2013, 87, 8511–8523. [Google Scholar] [CrossRef]
- Firth, A.E.; Chung, B.Y.W.; Fleeton, M.N.; Atkins, J.F. Discovery of Frameshifting in Alphavirus 6K Resolves a 20-Year Enigma. Virol. J. 2008, 5, 108. [Google Scholar] [CrossRef]
- Rogers, K.J.; Jones-Burrage, S.; Maury, W.; Mukhopadhyay, S. TF Protein of Sindbis Virus Antagonizes Host Type I Interferon Responses in a Palmitoylation-Dependent Manner. Virology 2020, 542, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Llamas-González, Y.Y.; Campos, D.; Pascale, J.M.; Arbiza, J.; González-Santamaría, J. A Functional Ubiquitin-Proteasome System Is Required for Efficient Replication of New World Mayaro and Una Alphaviruses. Viruses 2019, 11, 370. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.T.D.O.; Ribeiro, M.R.; Vedovello, D.; Nogueira, M.L. Mayaro Virus: A Neglected Arbovirus of the Americas. Future Virol. 2015, 10, 1109–1122. [Google Scholar] [CrossRef]
- Brown, R.S.; Wan, J.J.; Kielian, M. The Alphavirus Exit Pathway: What We Know and What We Wish We Knew. Viruses 2018, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Brown, D.T. Phosphorylation and Dephosphorylation Events Play Critical Roles in Sindbis Virus Maturation. Virology 1993, 196, 703–711. [Google Scholar] [CrossRef] [PubMed]
- Strauss, J.H.; Strauss, E.G.; Kuhn, R.J. Budding of Alphaviruses. Trends Microbiol. 1995, 3, 346–350. [Google Scholar] [CrossRef]
- Zhao, H.; Garoff, H. Role of Cell Surface Spikes in Alphavirus Budding. J. Virol. 1992, 66, 7089–7095. [Google Scholar] [CrossRef]
- West, J.; Hernandez, R.; Ferreira, D.; Brown, D.T. Mutations in the Endodomain of Sindbis Virus Glycoprotein E2 Define Sequences Critical for Virus Assembly. J. Virol. 2006, 80, 4458–4468. [Google Scholar] [CrossRef]
- Jose, J.; Taylor, A.B.; Kuhn, R.J. Spatial and Temporal Analysis of Alphavirus Replication and Assembly in Mammalian and Mosquito Cells. MBio 2017, 8, e02294-16. [Google Scholar] [CrossRef]
- Hahon, N.; Zimmerman, W.D. Chikungunya Virus Infection of Cell Monolayers by Cell-to-Cell and Extracellular Transmission. Appl. Microbiol. 1970, 19, 389–391. [Google Scholar] [CrossRef]
- Radoshitzky, S.R.; Pegoraro, G.; Chī, X.; Dǒng, L.; Chiang, C.Y.; Jozwick, L.; Clester, J.C.; Cooper, C.L.; Courier, D.; Langan, D.P.; et al. SiRNA Screen Identifies Trafficking Host Factors That Modulate Alphavirus Infection. PLoS Pathog. 2016, 12, e1005466. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Taylor, A. Arthritogenic Alphavirus Capsid Protein. Life 2021, 11, 230. [Google Scholar] [CrossRef] [PubMed]
- Mahauad-Fernandez, W.D.; Jones, P.H.; Okeoma, C.M. Critical Role for Bone Marrow Stromal Antigen 2 in Acute Chikungunya Virus Infection. J. Gen. Virol. 2014, 95, 2450–2461. [Google Scholar] [CrossRef]
- Ooi, Y.S.; Dubé, M.; Kielian, M. BST2/Tetherin Inhibition of Alphavirus Exit. Viruses 2015, 7, 2147–2167. [Google Scholar] [CrossRef]
- Jones, P.H.; Maric, M.; Madison, M.N.; Maury, W.; Roller, R.J.; Okeoma, C.M. BST-2/Tetherin-Mediated Restriction of Chikungunya (CHIKV) VLP Budding Is Counteracted by CHIKV Non-Structural Protein 1 (NsP1). Virology 2013, 438, 37–49. [Google Scholar] [CrossRef]
- Martinez, M.G.; Snapp, E.-L.; Perumal, G.S.; Macaluso, F.P.; Kielian, M. Imaging the Alphavirus Exit Pathway. J. Virol. 2014, 88, 6922–6933. [Google Scholar] [CrossRef]
- Jose, J.; Przybyla, L.; Edwards, T.J.; Perera, R.; Burgner, J.W.; Kuhn, R.J. Interactions of the Cytoplasmic Domain of Sindbis Virus E2 with Nucleocapsid Cores Promote Alphavirus Budding. J. Virol. 2012, 86, 2585–2599. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.G.; Kielian, M. Intercellular Extensions Are Induced by the Alphavirus Structural Proteins and Mediate Virus Transmission. PLoS Pathog. 2016, 12, e1006061. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.Y.; Kam, Y.W.; Fric, J.; Malleret, B.; Koh, E.G.L.; Prakash, C.; Huang, W.; Lee, W.W.L.; Lin, C.; Lin, R.T.P.; et al. Chikungunya Virus Neutralization Antigens and Direct Cell-to-Cell Transmission Are Revealed by Human Antibody-Escape Mutants. PLoS Pathog. 2011, 7, e1002390. [Google Scholar] [CrossRef]
- Labadie, K.; Larcher, T.; Joubert, C.; Mannioui, A.; Delache, B.; Brochard, P.; Guigand, L.; Dubreil, L.; Lebon, P.; Verrier, B.; et al. Chikungunya Disease in Nonhuman Primates Involves Long-Term Viral Persistence in Macrophages. J. Clin. Investig. 2010, 120, 894–906. [Google Scholar] [CrossRef]
- Hoarau, J.-J.; Jaffar Bandjee, M.-C.; Krejbich Trotot, P.; Das, T.; Li-Pat-Yuen, G.; Dassa, B.; Denizot, M.; Guichard, E.; Ribera, A.; Henni, T.; et al. Persistent Chronic Inflammation and Infection by Chikungunya Arthritogenic Alphavirus in Spite of a Robust Host Immune Response. J. Immunol. 2010, 184, 5914–5927. [Google Scholar] [CrossRef] [PubMed]
- Fung, M.; Thornton, A.; Mybeck, K.; wu, J.H.H.; Hornbuckle, K.; Muniz, E. Evaluation of the Characteristics of Safety Withdrawal of Prescription Drugs from Worldwide Pharmaceutical Markets-1960 to 1999. Ther. Innov. Regul. Sci. 2001, 35, 293–317. [Google Scholar] [CrossRef]
- Martin, H.L.; Adams, M.; Higgins, J.; Bond, J.; Morrison, E.E.; Bell, S.M.; Warriner, S.; Nelson, A.; Tomlinson, D.C. High-Content, High-Throughput Screening for the Identification of Cytotoxic Compounds Based on Cell Morphology and Cell Proliferation Markers. PLoS ONE 2014, 9, e88338. [Google Scholar] [CrossRef] [PubMed]
- Cruz, D.J.M.; Koishi, A.C.; Taniguchi, J.B.; Li, X.; Milan Bonotto, R.; No, J.H.; Kim, K.H.; Baek, S.; Kim, H.Y.; Windisch, M.P.; et al. High Content Screening of a Kinase-Focused Library Reveals Compounds Broadly-Active against Dengue Viruses. PLoS Negl. Trop. Dis. 2013, 7, e2073. [Google Scholar] [CrossRef] [PubMed]
- Aguiar, A.C.C.; Murce, E.; Cortopassi, W.A.; Pimentel, A.S.; Almeida, M.M.F.S.; Barros, D.C.S.; Guedes, J.S.; Meneghetti, M.R.; Krettli, A.U. Chloroquine Analogs as Antimalarial Candidates with Potent in Vitro and in Vivo Activity. Int. J. Parasitol. Drugs Drug Resist. 2018, 8, 459–464. [Google Scholar] [CrossRef]
- Quartuccio, L.; Zabotti, A.; Del Zotto, S.; Zanier, L.; De Vita, S.; Valent, F. Risk of Serious Infection among Patients Receiving Biologics for Chronic Inflammatory Diseases: Usefulness of Administrative Data. J. Adv. Res. 2019, 15, 87–93. [Google Scholar] [CrossRef]
- Ferreira, P.G.; Ferraz, A.C.; Figueiredo, J.E.; Lima, C.F.; Rodrigues, V.G.; Taranto, A.G.; Ferreira, J.M.S.; Brandão, G.C.; Vieira-Filho, S.A.; Duarte, L.P.; et al. Detection of the Antiviral Activity of Epicatechin Isolated from Salacia Crassifolia (Celastraceae) against Mayaro Virus Based on Protein C Homology Modelling and Virtual Screening. Arch. Virol. 2018, 163, 1567–1576. [Google Scholar] [CrossRef]
- Campos, D.; Navarro, S.; Llamas-González, Y.Y.; Sugasti, M.; González-Santamaría, J. Broad Antiviral Activity of Ginkgolic Acid against Chikungunya, Mayaro, Una, and Zika Viruses. Viruses 2020, 12, 449. [Google Scholar] [CrossRef]
- Neris, R.L.S.; Figueiredo, C.M.; Higa, L.M.; Araujo, D.F.; Carvalho, C.A.M.; Verçoza, B.R.F.; Silva, M.O.L.; Carneiro, F.A.; Tanuri, A.; Gomes, A.M.O.; et al. Co-Protoporphyrin IX and Sn-Protoporphyrin IX Inactivate Zika, Chikungunya and Other Arboviruses by Targeting the Viral Envelope. Sci. Rep. 2018, 8, 9805. [Google Scholar] [CrossRef]
- Salles, T.S.; Meneses, M.D.F.; Yamamoto, K.A.; Sá-Guimarães, T.E.; Caldas, L.A.; Silva, J.H.S.; da Silva Ferreira, P.; Amaral, A.C.F.; Ventura, J.A.; Azevedo, R.C.; et al. Chemical Composition and Anti-Mayaro Virus Activity of Schinus Terebinthifolius Fruits. VirusDisease 2021, 32, 526–534. [Google Scholar] [CrossRef]
- Salles, T.S.; Meneses, M.D.F.; Caldas, L.A.; Sá-Guimarães, T.E.; de Oliveira, D.M.; Ventura, J.A.; Azevedo, R.C.; Kuster, R.M.; Soares, M.R.; Ferreira, D.F. Virucidal and Antiviral Activities of Pomegranate (Punica Granatum) Extract against the Mosquito-Borne Mayaro Virus. Parasites Vectors 2021, 14, 443. [Google Scholar] [CrossRef] [PubMed]
- Du, R.; Cooper, L.; Chen, Z.; Lee, H.; Rong, L.; Cui, Q. Discovery of Chebulagic Acid and Punicalagin as Novel Allosteric Inhibitors of SARS-CoV-2 3CLpro. Antivir. Res. 2021, 190, 105075. [Google Scholar] [CrossRef] [PubMed]
- Spindola, K.C.W.; Simas, N.K.; Salles, T.S.; De Meneses, M.D.F.; Sato, A.; Ferreira, D.; Romão, W.; Kuster, R.M. Anti-Mayaro Virus Activity of Cassia Australis Extracts (Fabaceae, Leguminosae). Parasites Vectors 2014, 7, 537. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, C.A.M.; Sousa, I.P.; Silva, J.L.; Oliveira, A.C.; Gonçalves, R.B.; Gomes, A.M.O. Inhibition of Mayaro Virus Infection by Bovine Lactoferrin. Virology 2014, 452–453, 297–302. [Google Scholar] [CrossRef]
- Langendries, L.; Abdelnabi, R.; Neyts, J.; Delang, L. Repurposing Drugs for Mayaro Virus: Identification of Eidd-1931, Favipiravir and Suramin as Mayaro Virus Inhibitors. Microorganisms 2021, 9, 734. [Google Scholar] [CrossRef]
- Albulescu, I.C.; White-Scholten, L.; Tas, A.; Hoornweg, T.E.; Ferla, S.; Kovacikova, K.; Smit, J.M.; Brancale, A.; Snijder, E.J.; van Hemert, M.J. Suramin Inhibits Chikungunya Virus Replication by Interacting with Virions and Blocking the Early Steps of Infection. Viruses 2020, 12, 314. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Ryman, K.D.; Johnston, R.E. Adaptation of Sindbis Virus to BHK Cells Selects for Use of Heparan Sulfate as an Attachment Receptor. J. Virol. 1998, 72, 7357–7366. [Google Scholar] [CrossRef] [PubMed]
- Klimstra, W.B.; Nangle, E.M.; Smith, M.S.; Yurochko, A.D.; Ryman, K.D. DC-SIGN and L-SIGN Can Act as Attachment Receptors for Alphaviruses and Distinguish between Mosquito Cell- and Mammalian Cell-Derived Viruses. J. Virol. 2004, 78, 7862. [Google Scholar] [CrossRef]
- La Linn, M.; Eble, J.A.; Lübken, C.; Slade, R.W.; Heino, J.; Davies, J.; Suhrbier, A. An Arthritogenic Alphavirus Uses the A1β1 Integrin Collagen Receptor. Virology 2005, 336, 229–239. [Google Scholar] [CrossRef]
- Earnest, J.T.; Basore, K.; Roy, V.; Bailey, A.L.; Wang, D.; Alter, G.; Fremont, D.H.; Diamond, M.S. Neutralizing Antibodies against Mayaro Virus Require Fc Effector Functions for Protective Activity. J. Exp. Med. 2019, 2282–2301. [Google Scholar] [CrossRef]
- Fox, J.M.; Long, F.; Edeling, M.A.; Lin, H.; Van Duijl-Richter, M.K.S.; Fong, R.H.; Kahle, K.M.; Smit, J.M.; Jin, J.; Simmons, G.; et al. Broadly Neutralizing Alphavirus Antibodies Bind an Epitope on E2 and Inhibit Entry and Egress. Cell 2015, 163, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Kim, A.S.; Austin, S.K.; Gardner, C.L.; Zuiani, A.; Reed, D.S.; Trobaugh, D.W.; Sun, C.; Basore, K.; Williamson, L.E.; Crowe, J.E.; et al. Protective Antibodies against Eastern Equine Encephalitis Virus Bind to Epitopes in Domains A and B of the E2 Glycoprotein. Nat. Microbiol. 2019, 4, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Grabarz, F.; Paulo, A.; Lopes, Y.; Barazzone, G.C.; Santos, J.C.; Botosso, V.F.; Attie, S.; Jorge, C.; Lucia, A.; Oller, T.; et al. Strategies for the Production of Soluble Interferon-Alpha Consensus and Potential Application in Arboviruses and SARS-CoV-2. Life 2021, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Maheshwari, R.K.; Sidhu, G.S.; Singh, A.K.; Sivaram, S.S.; Kinchington, P.R.; Hay, J.; Friedman, R.M. Defective Transport of Herpes Simplex Virus Glycoprotein in Interferon-Treated Cells: Role of Intracellular PH. J. Interferon Res. 1994, 14, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, D.F.; Rebello, M.C.S. Interferon Selectively Inhibits the Synthesis of Mayaro Virus Glycoproteins. Rev. Microbiol. 1998, 29, 219–221. [Google Scholar] [CrossRef]
- Rebello, M.C.; Fonseca, M.E.; Marinho, J.O.; Rebello, M.A. Studies on the Replication of Mayaro Virus Grown in Interferon Treated Cells. Mem. Inst. Oswaldo Cruz 1994, 89, 619–623. [Google Scholar] [CrossRef]
- Maheshwari, R.K.; Sidhu, G.S.; Bhartiya, D.; Friedman, R.M. Primary Amines Enhance the Antiviral Activity of Interferon against a Membrane Virus: Role of Intracellular PH. J. Gen. Virol. 1991, 72, 2143–2152. [Google Scholar] [CrossRef]
- Li, J.; Kemper, T.; Broering, R.; Chen, J.; Yuan, Z.; Wang, X.; Lu, M. Interferon Alpha Induces Cellular Autophagy and Modulates Hepatitis B Virus Replication. Front. Cell. Infect. Microbiol. 2022, 12, 27. [Google Scholar] [CrossRef]
- Murgue, B.; Domart, Y.; Coudrier, D.; Rollin, P.E.; Darchis, J.P. Efficacy of Interferon Alpha-2b and Ribavirin against West Nile Virus In Vitro. Emerg. Infect. Dis. 2002, 8, 107–108. [Google Scholar]
- Livonesi, M.C.; De Moro Sousa, R.L.; Badra, S.J.; Figueiredo, L.T.M. In Vitro and in Vivo Studies of Ribavirin Action on Brazilian Orthobunyavirus. Am. J. Trop. Med. Hyg. 2006, 75, 1011–1016. [Google Scholar] [CrossRef]
- Graci, J.D.; Cameron, C.E. Mechanisms of Action of Ribavirin against Distinct Viruses. Rev. Med. Virol. 2006, 16, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Cummings, K.J.; Lee, S.M.; West, E.S.; Cid-Ruzafa, J.; Fein, S.G.; Aoki, Y.; Sulkowski, M.S.; Goodman, S.N. Interferon and Ribavirin vs Interferon Alone in the Re-Treatment of Chronic Hepatitis C Previosly Nonresponsive to Interferon a Meta-Analysis of Randomized Trials. J. Am. Med. Assoc. 2001, 285, 193–199. [Google Scholar] [CrossRef]
- Ravichandran, R.; Manian, M. Ribavirin Therapy for Chikungunya Arthritis. J. Infect. Dev. Ctries. 2008, 2, 140–142. [Google Scholar] [CrossRef] [PubMed]
- Andrei, G.; De Clercq, E. Molecular Approaches for the Treatment of Hemorrhagic Fever Virus Infections. Antivir. Res. 1993, 22, 45–75. [Google Scholar] [CrossRef]
- Briolant, S.; Garin, D.; Scaramozzino, N.; Jouan, A.; Crance, J.M. In Vitro Inhibition of Chikungunya and Semliki Forest Viruses Replication by Antiviral Compounds: Synergistic Effect of Interferon-α and Ribavirin Combination. Antivir. Res. 2004, 61, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Rothan, H.A.; Bahrani, H.; Mohamed, Z.; Teoh, T.C.; Shankar, E.M.; Rahman, N.A.; Yusof, R. A Combination of Doxycycline and Ribavirin Alleviated Chikungunya Infection. PLoS ONE 2015, 10, e0126360. [Google Scholar] [CrossRef]
- Dos Santos, A.E.; Kuster, R.M.; Yamamoto, K.A.; Salles, T.S.; Campos, R.; De Meneses, M.D.F.; Soares, M.R.; Ferreira, D. Quercetin and Quercetin 3-O-Glycosides from Bauhinia Longifolia (Bong.) Steud. Show Anti-Mayaro Virus Activity. Parasites Vectors 2014, 7, 130. [Google Scholar] [CrossRef]
- Zandi, K.; Teoh, B.T.; Sam, S.S.; Wong, P.F.; Mustafa, M.; Abubakar, S. Antiviral Activity of Four Types of Bioflavonoid against Dengue Virus Type-2. Virol. J. 2011, 8, 560. [Google Scholar] [CrossRef]
- Amorim, R.; de Meneses, M.D.F.; Borges, J.C.; da Silva Pinheiro, L.C.; Caldas, L.A.; Cirne-Santos, C.C.; de Mello, M.V.P.; de Souza, A.M.T.; Castro, H.C.; de Palmer Paixão, I.C.N.; et al. Thieno[2,3-b]Pyridine Derivatives: A New Class of Antiviral Drugs against Mayaro Virus. Arch. Virol. 2017, 162, 1577–1587. [Google Scholar] [CrossRef]
- Ferreira, D.F.; Santo, M.P.E.; Rebello, M.A.; Rebello, M.C.S. Weak Bases Affect Late Stages of Mayaro Virus Replication Cycle in Vertebrate Cells. J. Med. Microbiol. 2000, 49, 313–318. [Google Scholar] [CrossRef]
- Ehteshami, M.; Tao, S.; Zandi, K.; Hsiao, H.-M.; Jiang, Y.; Hammond, E.; Amblard, F.; Russell, O.O.; Merits, A.; Schinazi, R.F. Characterization of B-D-N4-Hydroxycytidine as a Novel Inhibitor of Chikungunya Virus. Antimicrob. Agents Chemother. 2017, 61, e02395-16. [Google Scholar] [CrossRef] [PubMed]
- Delang, L.; Guerrero, N.S.; Tas, A.; Quérat, G.; Pastorino, B.; Froeyen, M.; Dallmeier, K.; Jochmans, D.; Herdewijn, P.; Bello, F.; et al. Mutations in the Chikungunya Virus Non-Structural Proteins Cause Resistance to Favipiravir (T-705), a Broad-Spectrum Antiviral. J. Antimicrob. Chemother. 2014, 69, 2770–2784. [Google Scholar] [CrossRef] [PubMed]
- Urakova, N.; Kuznetsova, V.; Crossman, D.K.; Sokratian, A.; Guthrie, D.B.; Kolykhalov, A.A.; Lockwood, M.A.; Natchus, M.G.; Crowley, M.R.; Painter, G.R.; et al. β-d-N4-Hydroxycytidine Is a Potent Anti-Alphavirus Compound That Induces a High Level of Mutations in the Viral Genome. J. Virol. 2018, 92, e01965-17. [Google Scholar] [CrossRef] [PubMed]
- Bengue, M.; Pintong, A.R.; Liegeois, F.; Nougairède, A.; Hamel, R.; Pompon, J.; de Lamballerie, X.; Roques, P.; Choumet, V.; Missé, D. Favipiravir Inhibits Mayaro Virus Infection in Mice. Viruses 2021, 13, 2213. [Google Scholar] [CrossRef]
- Abdelnabi, R.; Jochmans, D.; Verbeken, E.; Neyts, J.; Delang, L. Antiviral Treatment Efficiently Inhibits Chikungunya Virus Infection in the Joints of Mice during the Acute but Not during the Chronic Phase of the Infection. Antivir. Res. 2018, 149, 113–117. [Google Scholar] [CrossRef]
- Julander, J.G.; Smee, D.F.; Morrey, J.D.; Furuta, Y. Effect of T-705 Treatment on Western Equine Encephalitis in a Mouse Model. Antivir. Res. 2009, 82, 169–171. [Google Scholar] [CrossRef]
- Ertem, O.; Guner, O.; Incir, C.; Kalkan, S.; Gelal, A. The Outcomes of Favipiravir Exposure in Pregnancy: A Case Series. Arch. Gynecol. Obstet. 2022. [Google Scholar] [CrossRef]
- Ingley, E. Src Family Kinases: Regulation of Their Activities, Levels and Identification of New Pathways. Biochim. Biophys. Acta-Proteins Proteom. 2008, 1784, 56–65. [Google Scholar] [CrossRef]
- Bouhaddou, M.; Memon, D.; Meyer, B.; White, K.M.; Rezelj, V.V.; Correa Marrero, M.; Polacco, B.J.; Melnyk, J.E.; Ulferts, S.; Kaake, R.M.; et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell 2020, 182, 685–712.e19. [Google Scholar] [CrossRef]
- Broeckel, R.; Sarkar, S.; May, N.A.; Totonchy, J.; Kreklywich, C.N.; Smith, P.; Graves, L.; DeFilippis, V.R.; Heise, M.T.; Morrison, T.E.; et al. Src Family Kinase Inhibitors Block Translation of Alphavirus Subgenomic MRNAs. Antimicrob. Agents Chemother. 2019, 63, e02325-18. [Google Scholar] [CrossRef]
- De Campos, R.M.; Ferreira, D.F.; Da Veiga, V.F.; Rebello, M.A.; Rebello, M.C.S. Effect of Monensin on Mayaro Virus Replication in Monkey Kidney and Aedes Albopictus Cells. Acta Virol. 2003, 47, 113–119. [Google Scholar] [PubMed]
- Gall, B.; Pryke, K.; Abraham, J.; Mizuno, N.; Botto, S.; Sali, T.M.; Broeckel, R.; Haese, N.; Nilsen, A.; Placzek, A.; et al. Emerging Alphaviruses Are Sensitive to Cellular States Induced by a Novel Small-Molecule Agonist of the STING Pathway. J. Virol. 2018, 92, e01913-17. [Google Scholar] [CrossRef] [PubMed]
- Da Costa, L.J.; Rebello, M.A. Effect of Brefeldin A on Mayaro Virus Replication in Aedes Albopictus and Vero Cells. Acta Virol. 1999, 43, 357–360. [Google Scholar] [PubMed]
- Burlandy, F.M.; Meneses, M.D.F. De Prostaglandin A 1 Inhibits the Replication of Sindbis Virus in Monkey Kidney and Mosquito Cells. Rev. Ciênc. Médicas E Biológicas 2008, 7, 169–174. [Google Scholar]
- Santoro, M.G.; Jaffe, B.M.; Garaci, E.; Esteban, M. Antiviral Effect of Prostaglandins of the A Series: Inhibition of Vaccinia Virus Replication in Cultured Cells. J. Gen. Virol. 1982, 63 2, 435–440. [Google Scholar] [CrossRef]
- Santoro, M.G.; Fukushima, M.; Benedetto, A.; Amici, C. PGJ2, a New Antiviral Prostaglandin: Inhibition of Sendai Virus Replication and Alteration of Virus Protein Synthesis. J. Gen. Virol. 1987, 68 Pt 4, 1153–1158. [Google Scholar] [CrossRef]
- Burlandy, F.M.; Rebello, M.A. Inhibition of Mayaro Virus Replication by Prostaglandin A1 in Vero Cells. Intervirology 2001, 44, 344–349. [Google Scholar] [CrossRef]
- Sivropoulou, A.; Nikolaou, C.; Papanikolaou, E.; Kokkini, S.; Lanaras, T.; Arsenakis, M. Antimicrobial, Cytotoxic, and Antiviral Activities of Salvia Fructicosa Essential Oil. J. Agric. Food Chem. 1997, 45, 3197–3201. [Google Scholar] [CrossRef]
- Shagufta; Ahmad, I. Recent Insight into the Biological Activities of Synthetic Xanthone Derivatives. Eur. J. Med. Chem. 2016, 116, 267–280. [Google Scholar] [CrossRef]
- da Silva, Í.E.P.; Lopes da Silva, M.; Dias, R.S.; Santos, E.G.; Brangioni de Paula, M.C.; Silva de Oliveira, A.; Costa da Silveira Oliveira, A.F.; Marques de Oliveira, F.; Canedo da Silva, C.; Teixeira, R.R.; et al. Xanthenedione (and Intermediates Involved in Their Synthesis) Inhibit Zika Virus Migration to the Central Nervous System in Murine Neonatal Models. Microbes Infect. 2020, 22, 489–499. [Google Scholar] [CrossRef]
- Fernandes, L.S.; da Silva, M.L.; Dias, R.S.; Lucindo, M.S.d.S.; da Silva, Í.E.P.; Silva, C.C.; Teixeira, R.R.; de Paula, S.O. Evaluation of Antiviral Activity of Cyclic Ketones against Mayaro Virus. Viruses 2021, 13, 2123. [Google Scholar] [CrossRef] [PubMed]
- Bakhache, W.; Neyret, A.; McKellar, J.; Clop, C.; Bernard, E.; Weger-Lucarelli, J.; Briant, L. Fatty Acid Synthase and Stearoyl-CoA Desaturase-1 Are Conserved Druggable Cofactors of Old World Alphavirus Genome Replication. Antivir. Res. 2019, 172, 104642. [Google Scholar] [CrossRef]
- Pereira, H.S.; Rebello, M.A. Inhibition of Mayaro Virus Replication by Cerulenin in Aedes Albopictus Cells. Acta Virol. 1998, 42, 383–388. [Google Scholar] [PubMed]
- Karpe, Y.A.; Pingale, K.D.; Kanade, G.D. Activities of Proteasome and M-Calpain Are Essential for Chikungunya Virus Replication. Virus Genes 2016, 52, 716–721. [Google Scholar] [CrossRef]
- Contin, R.; Arnoldi, F.; Mano, M.; Burrone, O.R. Rotavirus Replication Requires a Functional Proteasome for Effective Assembly of Viroplasms. J. Virol. 2011, 85, 2781–2792. [Google Scholar] [CrossRef] [PubMed]
- Satheshkumar, P.S.; Anton, L.C.; Sanz, P.; Moss, B. Inhibition of the Ubiquitin-Proteasome System Prevents Vaccinia Virus DNA Replication and Expression of Intermediate and Late Genes. J. Virol. 2009, 83, 2469–2479. [Google Scholar] [CrossRef] [PubMed]
- Isaacson, M.K.; Ploegh, H.L. Ubiquitination, Ubiquitin-like Modifiers, and Deubiquitination in Viral Infection. Cell Host Microbe 2009, 5, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin Modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef]
- Viswanathan, K.; Früh, K.; DeFilippis, V. Viral Hijacking of the Host Ubiquitin System to Evade Interferon Responses. Curr. Opin. Microbiol. 2010, 13, 517–523. [Google Scholar] [CrossRef]
- La Frazla, S.; Amici, C.; Santoro, M.G. Antiviral Activity of Proteasome Inhibitors in Herpes Simplex Virus-1 Infection: Role of Nuclear Factor-ΚB. Antivir. Ther. 2006, 11, 995–1004. [Google Scholar] [CrossRef]
- Watanabe, H.; Tanaka, Y.; Shimazu, Y.; Sugahara, F.; Kuwayama, M.; Hiramatsu, A.; Kiyotani, K.; Yoshida, T.; Sakaguchi, T. Cell-Specific Inhibition of Paramyxovirus Maturation by Proteasome Inhibitors. Microbiol. Immunol. 2005, 49, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Harty, R.N.; Brown, M.E.; McGettigan, J.P.; Wang, G.; Jayakar, H.R.; Huibregtse, J.M.; Whitt, M.A.; Schnell, M.J. Rhabdoviruses and the Cellular Ubiquitin-Proteasome System: A Budding Interaction. J. Virol. 2001, 75, 10623–10629. [Google Scholar] [CrossRef] [PubMed]
- Schubert, U.; Ott, D.E.; Chertova, E.N.; Welker, R.; Tessmer, U.; Princiotta, M.F.; Bennink, J.R.; Kräusslich, H.G.; Yewdell, J.W. Proteasome Inhibition Interferes with Gag Polyprotein Processing, Release, and Maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 2000, 97, 13057–13062. [Google Scholar] [CrossRef] [PubMed]
- Amaya, M.; Keck, F.; Lindquist, M.; Voss, K.; Scavone, L.; Kehn-Hall, K.; Roberts, B.; Bailey, C.; Schmaljohn, C.; Narayanan, A. The Ubiquitin Proteasome System Plays a Role in Venezuelan Equine Encephalitis Virus Infection. PLoS ONE 2015, 10, e0124792. [Google Scholar] [CrossRef]
- Kisselev, A.F.; Goldberg, A.L. Proteasome Inhibitors: From Research Tools to Drug Candidates. Chem. Biol. 2001, 8, 739–758. [Google Scholar] [CrossRef]
- Camini, F.C.; da Silva, T.F.; da Silva Caetano, C.C.; Almeida, L.T.; Ferraz, A.C.; Alves Vitoreti, V.M.; de Mello Silva, B.; de Queiroz Silva, S.; de Magalhães, J.C.; de Brito Magalhães, C.L. Antiviral Activity of Silymarin against Mayaro Virus and Protective Effect in Virus-Induced Oxidative Stress. Antivir. Res. 2018, 158, 8–12. [Google Scholar] [CrossRef]
- Caetano, C.C.d.S.; Camini, F.C.; Almeida, L.T.; Ferraz, A.C.; da Silva, T.F.; Lima, R.L.S.; de Freitas Carvalho, M.M.; de Freitas Castro, T.; Carneiro, C.M.; de Mello Silva, B.; et al. Mayaro Virus Induction of Oxidative Stress Is Associated With Liver Pathology in a Non-Lethal Mouse Model. Sci. Rep. 2019, 9, 15289. [Google Scholar] [CrossRef]
- Ferraz, A.C.; Almeida, L.T.; da Silva Caetano, C.C.; da Silva Menegatto, M.B.; Souza Lima, R.L.; de Senna, J.P.N.; de Oliveira Cardoso, J.M.; Perucci, L.O.; Talvani, A.; Geraldo de Lima, W.; et al. Hepatoprotective, Antioxidant, Anti-Inflammatory, and Antiviral Activities of Silymarin against Mayaro Virus Infection. Antivir. Res. 2021, 194, 105168. [Google Scholar] [CrossRef]
- Tappe, D.; Pérez-Girón, J.V.; Just-Nübling, G.; Schuster, G.; Gómez-Medina, S.; Günther, S.; Muñoz-Fontela, C.; Schmidt-Chanasit, J. Sustained Elevated Cytokine Levels during Recovery Phase of Mayaro Virus Infection. Emerg. Infect. Dis. 2016, 22, 750–752. [Google Scholar] [CrossRef]
- Santiago, F.W.; Halsey, E.S.; Siles, C.; Vilcarromero, S.; Guevara, C.; Silvas, J.A.; Ramal, C.; Ampuero, J.S.; Aguilar, P.V. Long-Term Arthralgia after Mayaro Virus Infection Correlates with Sustained Pro-Inflammatory Cytokine Response. PLoS Negl. Trop. Dis. 2015, 9, e0004104. [Google Scholar] [CrossRef]
- Camini, F.C.; da Silva Caetano, C.C.; Almeida, L.T.; da Costa Guerra, J.F.; de Mello Silva, B.; de Queiroz Silva, S.; de Magalhães, J.C.; de Brito Magalhães, C.L. Oxidative Stress in Mayaro Virus Infection. Virus Res. 2017, 236, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sugasti-Salazar, M.; Llamas-González, Y.Y.; Campos, D.; González-Santamaría, J. Inhibition of P38 Mitogen-Activated Protein Kinase Impairs Mayaro Virus Replication in Human Dermal Fibroblasts and Hela Cells. Viruses 2021, 13, 1156. [Google Scholar] [CrossRef] [PubMed]
- Plotnikov, A.; Zehorai, E.; Procaccia, S.; Seger, R. The MAPK Cascades: Signaling Components, Nuclear Roles and Mechanisms of Nuclear Translocation. Biochim. Biophys. Acta-Mol. Cell Res. 2011, 1813, 1619–1633. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Shi, M.; Zhang, L.; Li, Y.; Sun, J.; Zhang, L.; Wang, X.; Xu, X.; Zhang, X.; Mao, Y.; et al. Activation of JNK1/2 and P38 MAPK Signaling Pathways Promotes Enterovirus 71 Infection in Immature Dendritic Cells. BMC Microbiol. 2014, 14, 147. [Google Scholar] [CrossRef] [PubMed]
- Santio, N.M.; Koskinen, P.J. PIM Kinases: From Survival Factors to Regulators of Cell Motility. Int. J. Biochem. Cell Biol. 2017, 93, 74–85. [Google Scholar] [CrossRef]
- Eerola, S.K.; Kohvakka, A.; Tammela, T.L.J.; Koskinen, P.J.; Latonen, L.; Visakorpi, T. Expression and ERG Regulation of PIM Kinases in Prostate Cancer. Cancer Med. 2021, 10, 3427–3436. [Google Scholar] [CrossRef]
- Wu, J.; Chu, E.; Kang, Y. Pim Kinases in Multiple Myeloma. Cancers 2021, 13, 4304. [Google Scholar] [CrossRef]
- Zhang, X.; Song, M.; Kundu, J.K.; Lee, M.-H.; Liu, Z.-Z. PIM Kinase as an Executional Target in Cancer. J. Cancer Prev. 2018, 23, 109–116. [Google Scholar] [CrossRef]
- Keeton, E.K.; McEachern, K.; Dillman, K.S.; Palakurthi, S.; Cao, Y.; Grondine, M.R.; Kaur, S.; Wang, S.; Chen, Y.; Wu, A.; et al. AZD1208, a Potent and Selective Pan-Pim Kinase Inhibitor, Demonstrates Efficacy in Preclinical Models of Acute Myeloid Leukemia. Blood 2014, 123, 905–913. [Google Scholar] [CrossRef]
- Sugasti-Salazar, M.; Campos, D.; Valdés-Torres, P.; González-Santamaría, J. Targeting Host PIM Protein Kinases Reduces MayaroVirus Replication. Viruses 2022, 14, 422. [Google Scholar] [CrossRef]
- Zhou, F.; Wan, Q.; Chen, Y.; Chen, S.; He, M. liang PIM1 Kinase Facilitates Zika Virus Replication by Suppressing Host Cells’ Natural Immunity. Signal Transduct. Target. Ther. 2021, 6, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Mutricy, R.; Matheus, S.; Mosnier, É.; Martinez-Lorenzi, E.; De Laval, F.; Nacher, M.; Niemetzky, F.; Naudion, P.; Djossou, F.; Rousset, D.; et al. Mayaro Virus Infection in French Guiana, a Cross Sectional Study 2003–2019. Infect. Genet. Evol. 2022, 99, 105243. [Google Scholar] [CrossRef] [PubMed]
- de Souza Costa, M.C.; Siqueira Maia, L.M.; Costa de Souza, V.; Gonzaga, A.M.; Correa de Azevedo, V.; Ramos Martins, L.; Chavez Pavoni, J.H.; Gomes Naveca, F.; Dezengrini Slhessarenko, R. Arbovirus Investigation in Patients from Mato Grosso during Zika and Chikungunya Virus Introdution in Brazil, 2015–2016. Acta Trop. 2019, 190, 395–402. [Google Scholar] [CrossRef] [PubMed]
- Salgado, B.B.; de Jesus Maués, F.C.; Pereira, R.L.; Chiang, J.O.; de Oliveira Freitas, M.N.; Ferreira, M.S.; Martins, L.C.; da Costa Vasconcelos, P.F.; Ganoza, C.; Lalwani, P. Prevalence of Arbovirus Antibodies in Young Healthy Adult Population in Brazil. Parasites Vectors 2021, 14, 403. [Google Scholar] [CrossRef] [PubMed]
- Caicedo, E.Y.; Charniga, K.; Rueda, A.; Dorigatti, I.; Mendez, Y.; Hamlet, A.; Carrera, J.P.; Cucunubá, Z.M. The Epidemiology of Mayaro Virus in the Americas: A Systematic Review and Key Parameter Estimates for Outbreak Modelling. PLoS Negl. Trop. Dis. 2021, 15, e0009418. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andreolla, A.P.; Borges, A.A.; Bordignon, J.; Duarte dos Santos, C.N. Mayaro Virus: The State-of-the-Art for Antiviral Drug Development. Viruses 2022, 14, 1787. https://doi.org/10.3390/v14081787
Andreolla AP, Borges AA, Bordignon J, Duarte dos Santos CN. Mayaro Virus: The State-of-the-Art for Antiviral Drug Development. Viruses. 2022; 14(8):1787. https://doi.org/10.3390/v14081787
Chicago/Turabian StyleAndreolla, Ana Paula, Alessandra Abel Borges, Juliano Bordignon, and Claudia Nunes Duarte dos Santos. 2022. "Mayaro Virus: The State-of-the-Art for Antiviral Drug Development" Viruses 14, no. 8: 1787. https://doi.org/10.3390/v14081787
APA StyleAndreolla, A. P., Borges, A. A., Bordignon, J., & Duarte dos Santos, C. N. (2022). Mayaro Virus: The State-of-the-Art for Antiviral Drug Development. Viruses, 14(8), 1787. https://doi.org/10.3390/v14081787





