First Isolation of Punique Virus from Sand Flies Collected in Northern Algeria
Abstract
:1. Introduction
2. Materiel and Methods
2.1. Ethical Considerations
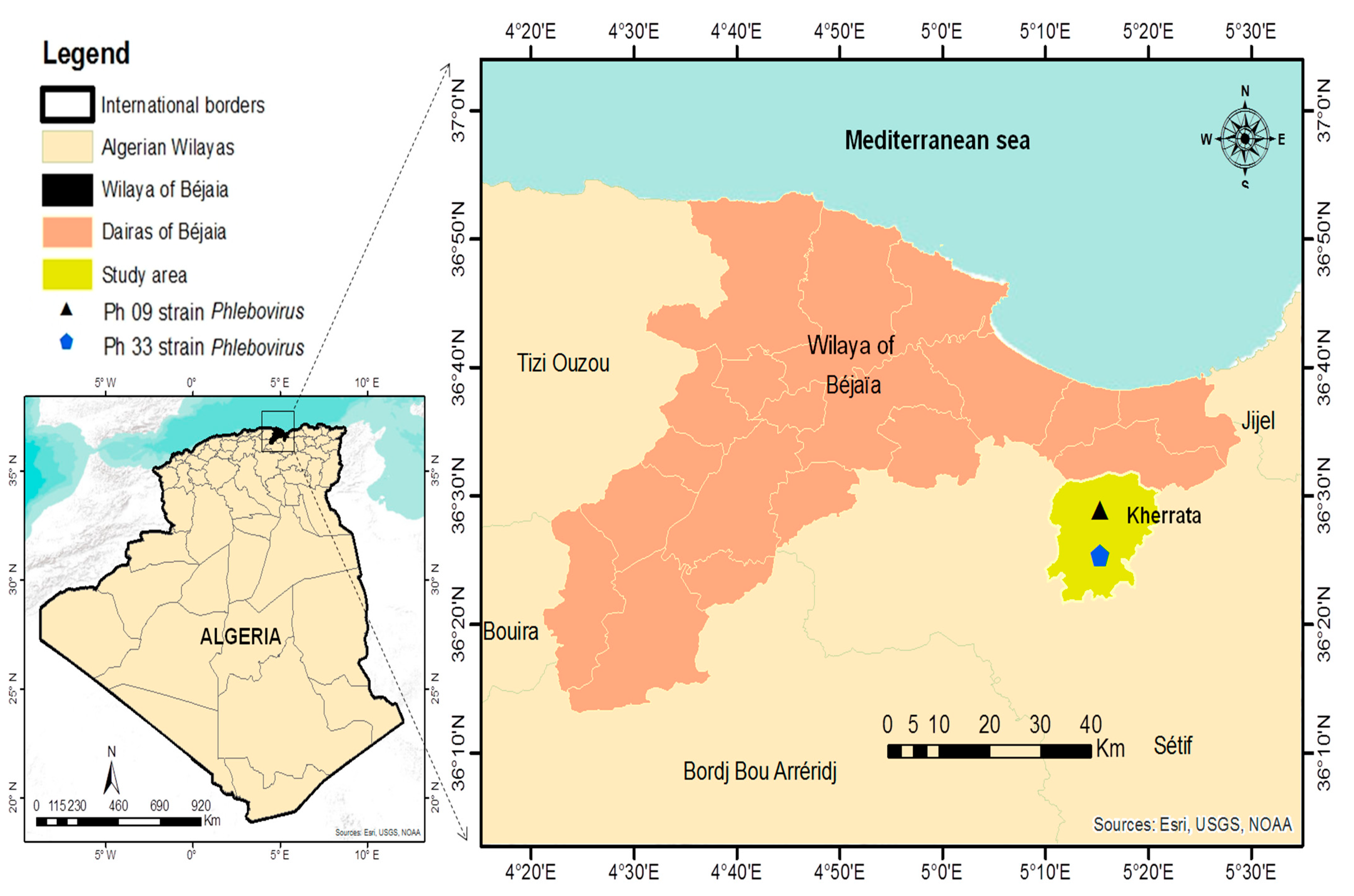
2.2. Sand Flies Sampling
2.3. Phlebovirus Screening
2.3.1. Sand Flies Processing and Viral RNA Extraction
2.3.2. Phlebovirus Detection
2.4. Virus Isolation
2.5. Sequencing and Phylogenetic Analysis
2.6. Sand Flies Identification
2.6.1. Morphological Identification of Sand Flies
2.6.2. Molecular Identification of Positive Pools
3. Results
3.1. Virus Detection and Sand Flies Identification
3.2. Virus Isolation
3.3. Phylogenetic Analysis
3.4. Sand Fly Identification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lafri, I.; Bitam, I. Phlebotomine sandflies and associated pathogens in Algeria: Update and comprehensive overview. Vet. Ital. 2021, 57. [Google Scholar] [CrossRef]
- Benallal, K.E.; Garni, R.; Harrat, Z.; Volf, P.; Dvorak, V. Phlebotomine sand flies (Diptera: Psychodidae) of the Maghreb region: A systematic review of distribution, morphology, and role in the transmission of the pathogens. PLoS Negl. Trop. Dis. 2022, 16, e0009952. [Google Scholar] [CrossRef] [PubMed]
- Moriconi, M.; Rugna, G.; Calzolari, M.; Bellini, R.; Albieri, A.; Angelini, P.; Cagarelli, R.; Landini, M.P.; Charrel, R.N.; Varan, S. Phlebotomine sand fly–borne pathogens in the Mediterranean Basin: Human leishmaniasis and phlebovirus infections. PLoS Negl. Trop. Dis. 2017, 11, e0005660. [Google Scholar] [CrossRef] [PubMed]
- Abudurexiti, A.; Adkins, S.; Alioto, D.; Alkhovsky, S.V.; Avšič-Županc, T.; Ballinger, M.J.; Bente, D.A.; Beer, M.; Bergeron, É.; Blair, C.D.; et al. Taxonomy of the order Bunyavirales: Update 2019. Arch. Virol. 2019, 164, 1949–1965. [Google Scholar] [CrossRef] [PubMed]
- Charrel, R.N.; Gallian, P.; Navarro-Marí, J.M.; Nicoletti, L.; Papa, A.; Sánchez-Seco, M.P.; Tenorio, A.; de Lamballerie, X. Emergence of Toscana virus in Europe. Emerg. Infect. Dis. 2005, 11, 1657. [Google Scholar] [CrossRef]
- Benkő, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarría, M.; Hess, M.; Jones, M.S.; Kaján, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV virus taxonomy profile: Adenoviridae 2022. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef]
- Calisher, C.H.; Calzolari, M. Taxonomy of phleboviruses, emphasizing those that are sandfly-borne. Viruses 2021, 13, 918. [Google Scholar] [CrossRef]
- Ayhan, N.; Charrel, R.N. Of phlebotomines (sandflies) and viruses: A comprehensive perspective on a complex situation. Curr. Opin. Insect. Sci. 2017, 22, 117–124. [Google Scholar] [CrossRef]
- Ayhan, N.; Charrel, R.N. Sandfly-borne viruses of demonstrated/relevant medical importance. In Vectors and Vector-Borne Zoonotic Diseases; Intech Open: London, UK, 2018. [Google Scholar] [CrossRef]
- Ashford, R.W. Phlebotomus fevers. In The Encyclopedia of Arthropod-Transmitted Infections; Service, M.W., Ed.; CABI: Wallingford, UK, 2001. [Google Scholar]
- Amaro, F.; Zé-Zé, L.; Lourenço, J.; Giovanetti, M.; Becker, S.C.; Alves, M.J. Phylogenetic analysis of Massilia phlebovirus in Portugal. Viruses 2021, 13, 1412. [Google Scholar] [CrossRef]
- Ergunay, K.; Tkachev, S.; Kozlova, I.; Růžek, D. A review of methods for detecting tick-borne encephalitis virus infection in tick, animal, and human specimens. Vector Borne Zoonotic Dis. 2016, 16, 4–12. [Google Scholar] [CrossRef]
- Bichaud, L.; Dachraoui, K.; Piorkowski, G.; Chelbi, I.; Moureau, G.; Cherni, S.; de Lamballerie, X.; Sakhria, S.; Charrel, R.N.; Zhioua, E. Toscana virus isolated from sand flies, Tunisia. Emerg. Infect. Dis. 2013, 19, 322. [Google Scholar] [CrossRef] [PubMed]
- Es-Sette, N.; Nourlil, J.; Hamdi, S.; Mellouki, F.; Lemrani, M. First detection of Toscana virus RNA from sand flies in the genus Phlebotomus (Diptera: Phlebotomidae) naturally infected in Morocco. J. Med. Entomol. 2014, 49, 1507–1509. [Google Scholar] [CrossRef] [PubMed]
- Es-sette, N.; Ajaoud, M.; Anga, L.; Mellouki, F.; Lemrani, M. Toscana virus isolated from sand flies, Morocco. Parasit. Vectors 2015, 8, 205. [Google Scholar] [CrossRef] [PubMed]
- Es-Sette, N.; Ajaoud, M.; Charrel, R.N.; Lemrani, M. Épidémiologie moléculaire des Phlebovirus dans quatre provinces du Maroc. Bull. Soc. Pathol. Exot. 2016, 109, 143–150. [Google Scholar] [CrossRef]
- Alkan, C.; Allal-Ikhlef, A.B.; Alwassouf, S.; Baklouti, A.; Piorkowski, G.; de Lamballerie, X.; Izri, A.; Charrel, R.N. Virus isolation, genetic characterization and seroprevalence of Toscana virus in Algeria. Clin. Microbiol. Infect. 2015, 21, 1040-e1. [Google Scholar] [CrossRef]
- Zhioua, E.; Moureau, G.; Chelbi, I.; Ninove, L.; Bichaud, L.; Derbali, M.; Champs, M.; Cherni, S.; Salez, N.; Cook, S.; et al. Punique virus, a novel phlebovirus, related to sand fly fever Naples virus, isolated from sand flies collected in Tunisia. J. Gen. Virol. 2010, 91, 1275. [Google Scholar] [CrossRef]
- Izri, A.; Temmam, S.; Moureau, G.; Hamrioui, B.; De Lamballerie, X.; Charrel, R.N. Sand fly fever Sicilian virus, Algeria. Emerg. Infect. Dis. 2008, 14, 795. [Google Scholar] [CrossRef]
- Moureau, G.; Bichaud, L.; Salez, N.; Ninove, L.; Hamrioui, B.; Belazzoug, S.; de Lamballerie, X.; Izri, A.; Charrel, R.N. Molecular and serological evidence for the presence of novel phleboviruses in sand flies from northern Algeria. Open Virol. J. 2010, 4, 15. [Google Scholar]
- Tahir, D.; Alwassouf, S.; Loudahi, A.; Davoust, B.; Charrel, R.N. Seroprevalence of Toscana virus in dogs from Kabylia (Algeria). Clin. Microbiol. Infect. 2016, 22, e16–e17. [Google Scholar] [CrossRef]
- Dujardin, J.C.; Campino, L.; Cañavate, C.; Dedet, J.P.; Gradoni, L.; Soteriadou, K.; Mazeris, A.; Ozbel, Y.; Boelaer, M. Spread of vector-borne diseases and neglect of Leishmaniasis, Europe. Emerg. Infect. Dis. 2008, 14, 1013. [Google Scholar] [CrossRef]
- Fares, W.; Dachraoui, K.; Barhoumi, W.; Cherni, S.; Chelbi, I.; Zhioua, E. Co-circulation of Toscana virus and Leishmania infantum in a focus of zoonotic visceral leishmaniasis from Central Tunisia. Acta Trop. 2020, 204, 105342. [Google Scholar] [CrossRef] [PubMed]
- Calzolari, M.; Romeo, G.; Callegari, E.; Bonilauri, P.; Chiapponi, C.; Carra, E.; Rugna, G.; Taddei, R.; Lelli, D.; Dottori, M. Co-Circulation of Phleboviruses and Leishmania Parasites in Sand Flies from a Single Site in Italy Monitored between 2017 and 2020. Viruses 2021, 13, 1660. [Google Scholar] [CrossRef]
- Tahir, D. Epidémiologie de la leishmaniose chez le chien et l’homme à Bejaia. Ph.D. Thesis, Institut des Sciences Vétérinaires Université Blida I, Algérie, Africa, 2014. [Google Scholar]
- Ramdane, E.; Berchi, S.; Louad, K. Les phlébotomes (Diptera, Pshycodidae), vecteurs d’agents pathogènes responsables de la leishmaniose humaine dans la région de Constantine (Algérie). Entomofauna 2018, 39, 537–555. [Google Scholar]
- Lafri, I.; Almeras, L.; Bitam, I.; Caputo, A.; Yssouf, A.; Forestier, C.L.; Izri, A.; Raoult, D.; Parola, P. Identification of Algerian field-caught phlebotomine sand fly vectors by MALDI-TOF MS. PLoS Negl. Trop. Dis. 2016, 10, e0004351. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Seco, M.P.; Echevarría, J.M.; Hernández, L.; Estévez, D.; Navarro-Marí, J.M.; Tenorio, A. Detection and identification of Toscana and other phleboviruses by RT-nested-PCR assays with degenerated primers. J. Med. Virol. 2003, 71, 140–149. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Abonnenc, E. Les phlébotomes de la région éthiopienne (Diptera, Psychodidae). Cah. ORSTOM Sér. Ent. Méd. Parasitol. 1972, 55, 1–239. [Google Scholar]
- Leger, N.; Pesson, B.; Madulo-Leblond, G.; Abonnenc, E. Differentiation of females of the subgenus Larroussius Nitzulescu 1931 (Diptera-Phlebotomidae) of the Mediterranean region. Ann. Parasitol. Hum. Comp. 1983, 58, 611–623. [Google Scholar] [CrossRef]
- Dedet, J.P.; Adaddi, K.Y.; Bellazoug, S. Les phlébotomes (Diptera, Psychodidae) d’Algerie. Cah. ORSTOM Sér. Ent. Méd. Parasitol. 1984, 22, 99–127. [Google Scholar]
- Folmer, O.; Hoeh, W.R.; Black, M.B.; Vrijenhoek, R.C. Conserved primers for PCR amplification of mitochondrial DNA from different invertebrate phyla. Mol. Mar. Biol. Biotechnol. 1994, 3, 294–299. [Google Scholar] [PubMed]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Remoli, M.E.; Bongiorno, G.; Fortuna, C.; Marchi, A.; Bianchi, R.; Khoury, C.; Ciufolini, M.G.; Gramiccia, M. Experimental evaluation of sand fly collection and storage methods for the isolation and molecular detection of Phlebotomus-borne viruses. Parasit. Vectors 2015, 8, 576. [Google Scholar] [CrossRef] [PubMed]
- Peyrefitte, C.N.; Grandadam, M.; Bessaud, M.; Andry, P.E.; Fouque, F.; Caro, V.; Diancourt, L.; Schuffenecker, I.; Pagès, F.; Tolou, H.; et al. Diversity of Phlebotomus perniciosus in Provence, southeastern France: Detection of two putative new Phlebovirus sequences. Vector Bor. Zoonotic Dis. 2013, 13, 630–636. [Google Scholar] [CrossRef] [PubMed]
- Remoli, M.E.; Fortuna, C.; Marchi, A.; Bucci, P.; Argentini, C.; Bongiorno, G.; Maroli, M.; Gradoni, L.; Gramiccia, M.; Ciufolini, M.G. Viral isolates of a novel putative Phlebovirus in the Marche Region of Italy. Am. J. Trop. Med. Hyg. 2014, 90, 760. [Google Scholar] [CrossRef]
- Alkan, C.; Alwassouf, S.; Piorkowski, G.; Laurence, B.; Seda, T.; Dincer, E.; Ergunay, K.; Ozbel, Y.; Alten, B.; de Lamballerie, X.; et al. Isolation, genetic characterization, and seroprevalence of Adana virus, a novel Phlebovirus belonging to the Salehabad virus complex, in Turkey. J. Virol. 2015, 89, 4080–4091. [Google Scholar] [CrossRef]
- Fares, W.; Charrel, R.N.; Dachraoui, K.; Bichaud, L.; Barhoumi, W.; Derbali, M.; Cherni, S.; Chelbi, I.; de Lamballerie, X.; Zhioua, E. Infection of sand flies collected from different bio-geographical areas of Tunisia with phleboviruses. Acta Trop. 2015, 141, 1–6. [Google Scholar] [CrossRef]
- Ciufolini, M.G.; Maroli, M.; Verani, P. Growth of two phleboviruses after experimental infection of their suspected sand fly vector, Phlebotomus perniciosus (Diptera: Psychodidae). Am. J. Trop. Med. Hyg. 1985, 34, 174–179. [Google Scholar] [CrossRef]
- Tesh, R.B. The genus Phlebovirus and its vectors. Annu. Rev. Entomol. 1988, 33, 169–181. [Google Scholar] [CrossRef]
- Maroli, M.; Ciufolini, M.G.; Verani, P. Vertical transmission of Toscana virus in the sand fly, Phlebotomus perniciosus, via the second gonotrophic cycle. Med. Vet. Entomol. 1993, 7, 283–286. [Google Scholar] [CrossRef]
- Alkan, C.; Bichaud, L.; de Lamballerie, X.; Alten, B.; Gould, E.A.; Charrel, R.N. Sand fly-borne phleboviruses of Eurasia and Africa: Epidemiology, genetic diversity, geographic range, control measures. Antiviral. Res. 2013, 100, 54–74. [Google Scholar] [CrossRef] [PubMed]
- Sanbonmatsu-Gámez, S.; Pérez-Ruiz, M.; Collao, X.; Sánchez-Seco, M.P.; Morillas-Márquez, F.; De la Rosa-Fraile, M.; Navarro-Marí, J.M.; Tenorio, A. Toscana virus in Spain. Emerg. Infect. Dis. 2005, 11, 1701. [Google Scholar] [CrossRef] [PubMed]
- Bichaud, L.; Izri, A.; de Lamballerie, X.; Moureau, G.; Charrel, R.N. First detection of Toscana virus in Corsica, France. Clin. Microbiol. Infect. 2014, 20, O101–O104. [Google Scholar] [CrossRef]
- Zhioua, E.; Kaabi, B.; Chelbi, I. Entomological investigations following the spread of visceral leishmaniasis in Tunisia. J. Vector Ecol. 2007, 32, 371–374. [Google Scholar] [CrossRef]
- Sakhria, S.; Bichaud, L.; Mensi, M.; Salez, N.; Dachraoui, K.; Thirion, L.; Cherni, S.; Chelbi, I.; De Lamballerie, X.; Zhioua, E.; et al. Co-circulation of Toscana virus and Punique virus in northern Tunisia: A microneutralization-based seroprevalence study. PLoS Negl. Trop. Dis. 2013, 7, e2429. [Google Scholar] [CrossRef]
- Harrat, Z.; Pratlong, F.; Belazzoug, S.; Dereure, J.; Deniau, M.; Rioux, J.A.; Belkaid, M.; Dedet, J.P. Leishmania infantum and L. major in Algeria. Trans. R Soc. Trop. Med. Hyg. 1996, 90, 625–629. [Google Scholar] [CrossRef]
- Verani, P.; Ciufolini, M.G.; Caciolli, S.; Renzi, A.; Nicoletti, L.; Sabatinelli, G.; Bartolozzi, D.; Volpi, G.; Amaducci, L.; Coluzzi, M.; et al. Ecological and epidemiological studies of Toscana virus, an arbovirus isolated from Phlebotomus. Ann. Ist. Super. Sanita. 1982, 18, 397–399. [Google Scholar]
- Ehrnst, A.; Peters, C.J.; Niklasson, B.; Svedmyr, A.; Holmgren, B. Neurovirulent Toscana virus (a sand fly fever virus) in Swedish man after visit to Portugal. Lancet 1985, 325, 1212–1213. [Google Scholar] [CrossRef]

| Male | Female | Total | |
|---|---|---|---|
| Number of sand flies | 940 | 2111 | 3051 |
| Number of pools | 19 | 43 | 62 |
| Number of positive pools | 1(Ph33) | 1(Ph9) | 2 |
| Infection rate (%) | 0.1% | 0.05% | 0.06% |
| N° Sequences | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Amino Acids Distances | |||||||
| 1. ON524174 PUNV strain Ph9 Kherrata Algeria | 0.000 | 0.0061 | 0.0061 | 0.0381 | 0.0121 | 0.0121 | |
| 2. AB905362.1 PUNV strain 14 Tunisia | 0.0327 | 0.0061 | 0.0118 | 0.0381 | 0.0121 | 0.0236 | |
| 3. ON524173 PUNV strain Ph33 Kherrata Algeria | 0.0500 | 0.0412 | 0.000 | 0.0305 | 0.0061 | 0.0182 | |
| 4. MT250046.1 PUNV strain Pu0518.0.2 2018 Algeria | 0.0476 | 0.0515 | 0.0348 | 0.0305 | 0.0061 | 0.0287 | |
| 5. OM362899.1 PUNV strain2010T114 Tunisia | 0.0590 | 0.0561 | 0.0478 | 0.0505 | 0.0229 | 0.0229 | |
| 6. OM362900.1 PUNV strain 2010T122 | 0.0412 | 0.0369 | 0.0328 | 0.0348 | 0.0128 | 0.0121 | |
| 7. NC 055302.1 PUNV straiPI-B4-2008 Tunisia | 0.0632 | 0.0770 | 0.0676 | 0.0763 | 0.0705 | 0.0542 | |
| Nucleotides distances | |||||||
| Sex | ||||
|---|---|---|---|---|
| Species | Male | Female | (%) | Total |
| Ph. perniciosus | 152 | 144 | 98.97% | 296 |
| Ph. longicuspis | 0 | 2 | 0.67% | 2 |
| Ph. perfiliewi | 1 | 1 | 0.67% | 2 |
| Total | 153 | 147 | 100% | 300 |
| Positive Pools | Phlebovirus | Sand Flies Species |
|---|---|---|
| Ph 9 | Punique virus | Ph. perniciosus |
| Ph 33 | Punique virus | Ph. perniciosus |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manseur, H.; Hachid, A.; Khardine, A.F.; BENALLAL, K.E.; Bia, T.; Temani, M.; HAKEM, A.; Sánchez-Seco, M.P.; Bitam, I.; Vázquez, A.; et al. First Isolation of Punique Virus from Sand Flies Collected in Northern Algeria. Viruses 2022, 14, 1796. https://doi.org/10.3390/v14081796
Manseur H, Hachid A, Khardine AF, BENALLAL KE, Bia T, Temani M, HAKEM A, Sánchez-Seco MP, Bitam I, Vázquez A, et al. First Isolation of Punique Virus from Sand Flies Collected in Northern Algeria. Viruses. 2022; 14(8):1796. https://doi.org/10.3390/v14081796
Chicago/Turabian StyleManseur, Hemza, Aissam Hachid, Ahmed Fayez Khardine, Kamal Eddine BENALLAL, Taha Bia, Merbouha Temani, Ahcene HAKEM, Maria Paz Sánchez-Seco, Idir Bitam, Ana Vázquez, and et al. 2022. "First Isolation of Punique Virus from Sand Flies Collected in Northern Algeria" Viruses 14, no. 8: 1796. https://doi.org/10.3390/v14081796
APA StyleManseur, H., Hachid, A., Khardine, A. F., BENALLAL, K. E., Bia, T., Temani, M., HAKEM, A., Sánchez-Seco, M. P., Bitam, I., Vázquez, A., & LAFRI, I. (2022). First Isolation of Punique Virus from Sand Flies Collected in Northern Algeria. Viruses, 14(8), 1796. https://doi.org/10.3390/v14081796






