The FlagT4G Vaccine Confers a Strong and Regulated Immunity and Early Virological Protection against Classical Swine Fever
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Viruses
2.2. Experimental Design
2.3. Antibody Detection by ELISA and Virus Neutralization Assay
2.4. Evaluation of the Cytokine Response by ELISA and Luminex Assays
2.5. Detection of CSFV RNA
2.6. PBMC Collection
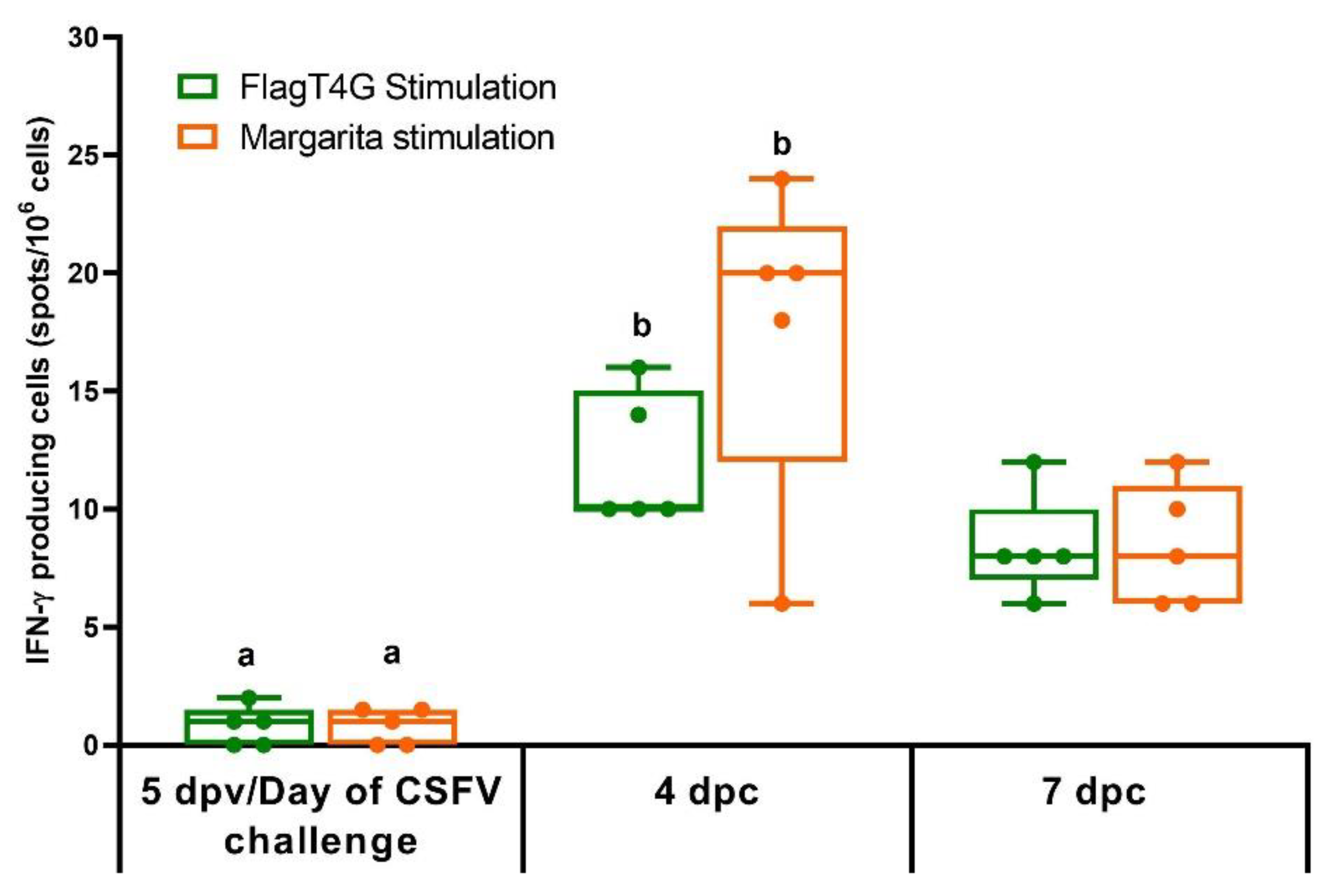
2.7. Assessment of Cellular Immune Response by IFN-γ ELISPOT
2.8. Phenotypical Profile Evaluation by Flow Cytometry
2.9. Statistical Analysis
3. Results
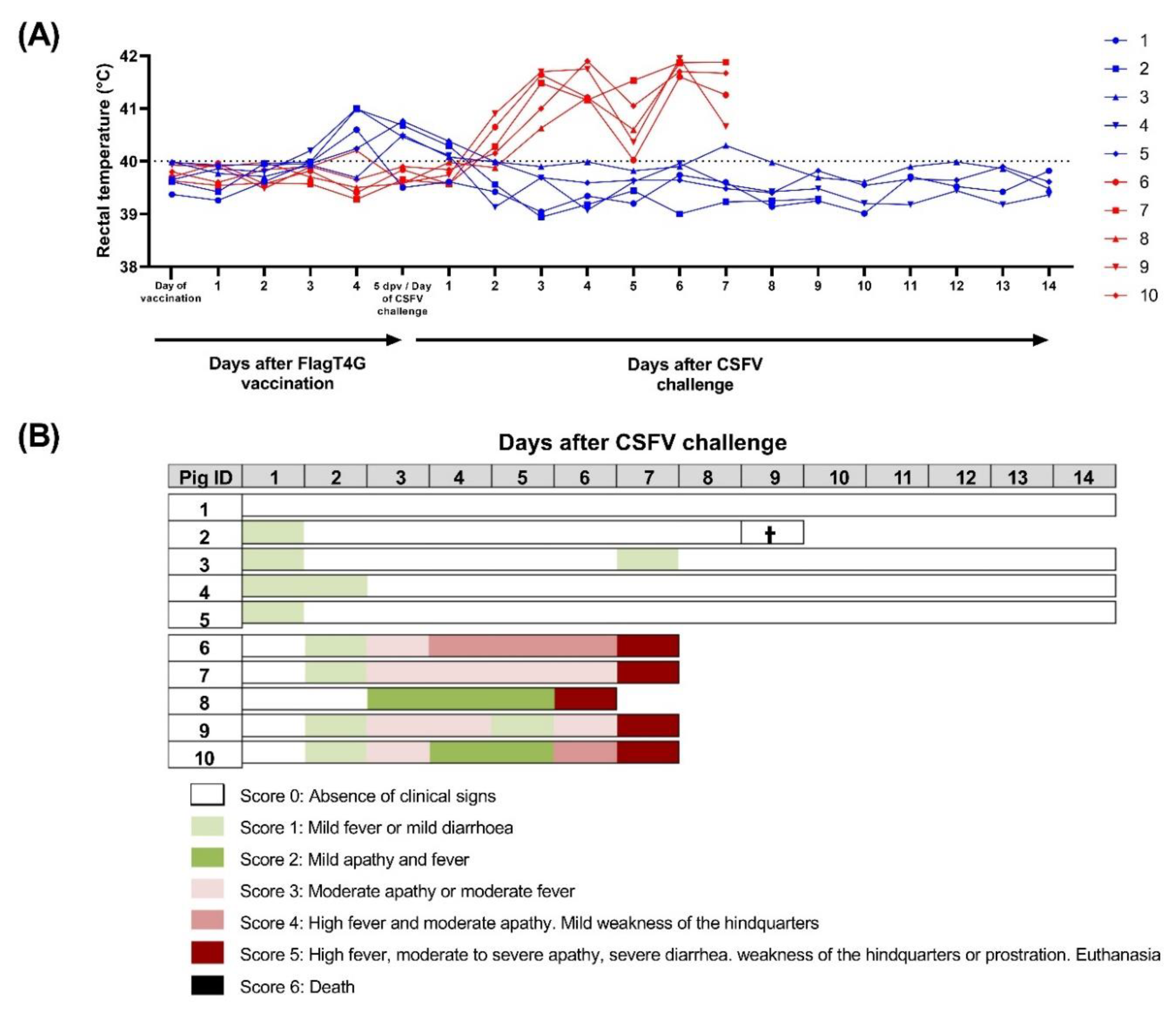
3.1. FlagT4G Affords Early Clinical Protection against CSFV Challenge in Pigs
3.2. FlagT4G Induced Fast Protection after Five Days of Vaccination in the Absence of Antibody Response
3.3. Cytokine Response after FlagT4G Vaccination and CSFV Challenge
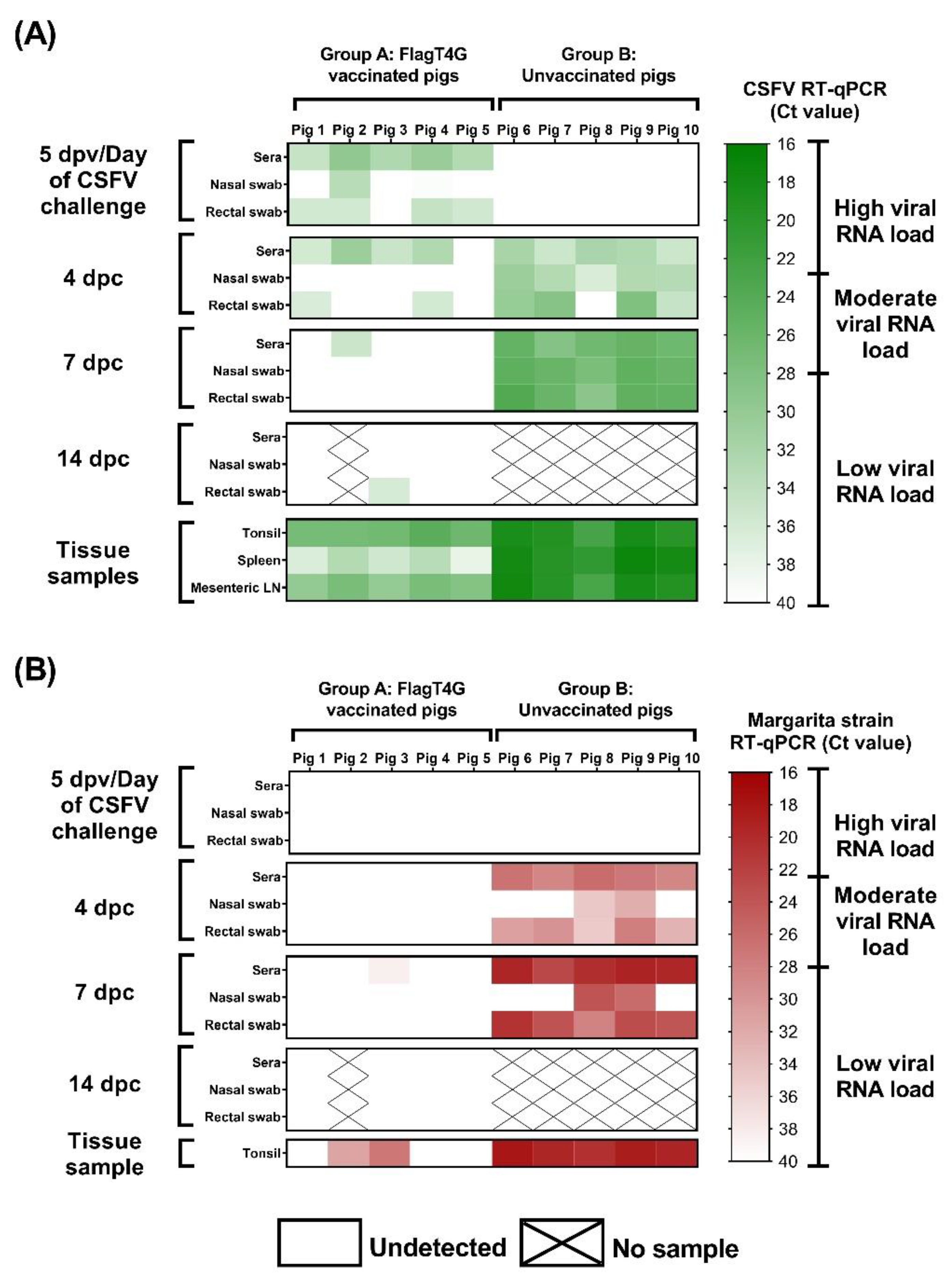
3.4. The FlagT4G Vaccine Induces Rapid Protection against Systemic Viral Replication
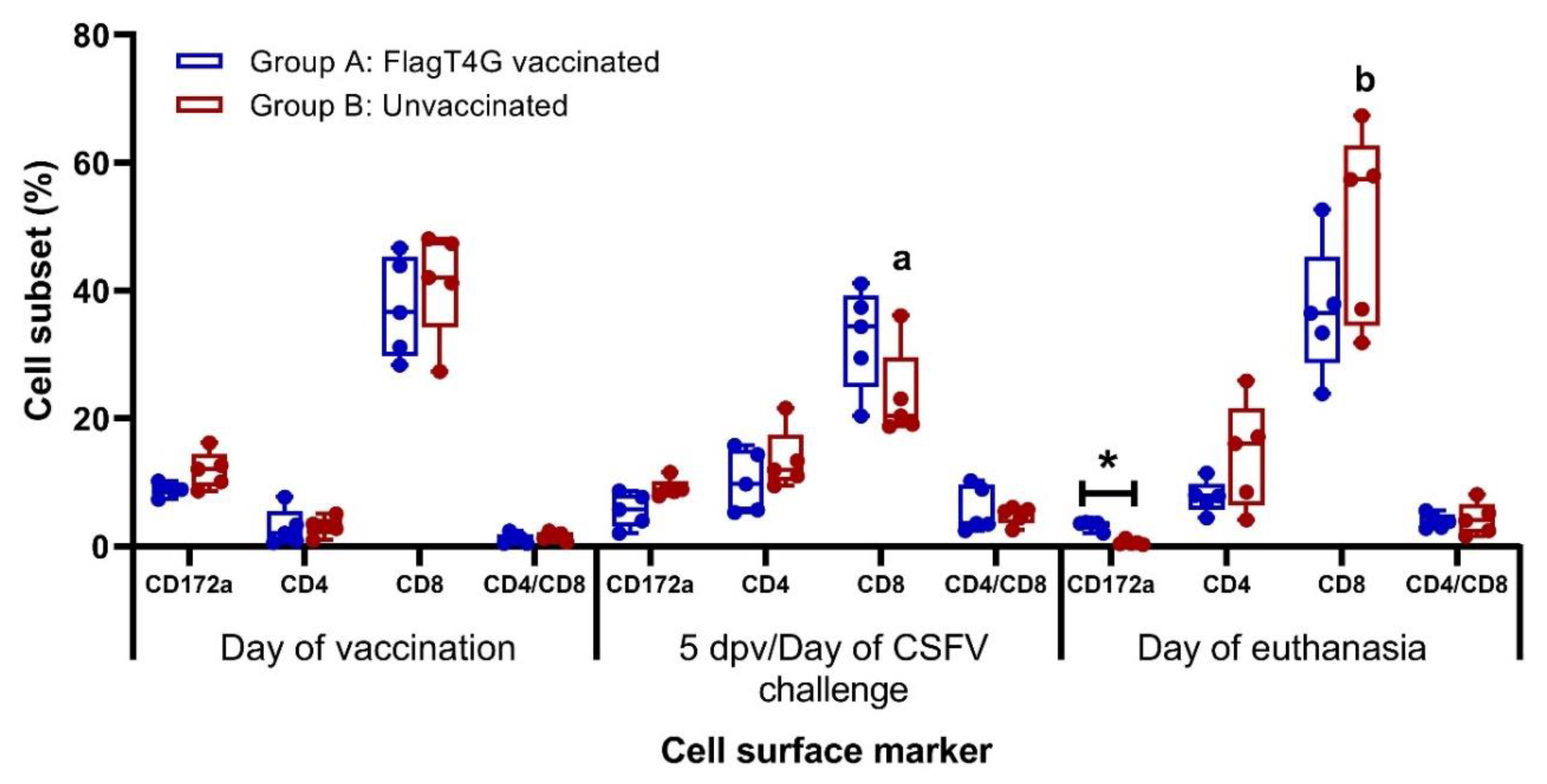
3.5. FlagT4G Vaccination Improves Cellular Immune Response in Swine
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ganges, L.; Crooke, H.R.; Bohórquez, J.A.; Postel, A.; Sakoda, Y.; Becher, P.; Ruggli, N. Classical swine fever virus: The past, present and future. Virus Res. 2020, 289, 198151. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Staubach, C.; Henke, J.; Carlson, J.; Beer, M. Classical swine fever—An updated review. Viruses 2017, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.; Peterhans, E. Pestiviruses. Annu. Rev. Anim. Biosci. 2014, 2, 141–163. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (OIE). Chapter 15.2 Infection with Classical Swine fever virus. In Terrestrial Animal Health Code; World Organisation for Animal Health (OIE), Ed.; World Organisation for Animal Health (OIE): Paris, France, 2019. [Google Scholar]
- Coronado, L.; Perera, C.L.; Rios, L.; Frías, M.T.; Pérez, L.J. A Critical Review about Different Vaccines against Classical Swine Fever Virus and Their Repercussions in Endemic Regions. Vaccines 2021, 9, 154. [Google Scholar] [CrossRef]
- Ganges, L.; Núñez, J.I.; Sobrino, F.; Borrego, B.; Fernández-Borges, N.; Frías-Lepoureau, M.T.; Rodríguez, F. Recent advances in the development of recombinant vaccines against classical swine fever virus: Cellular responses also play a role in protection. Vet. J. 2008, 177, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Lamothe-Reyes, Y.; Bohórquez, J.A.; Wang, M.; Alberch, M.; Pérez-Simó, M.; Rosell, R.; Ganges, L. Early and solid protection afforded by the thiverval vaccine provides novel vaccination alternatives against classical swine fever virus. Vaccines 2021, 9, 464. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (OIE). OIE World Animal Health Information System. Available online: https://www.oie.int/wahis_2/public/wahid.php/Diseaseinformation/Diseaseoutbreakmaps?disease_type_hidden=&disease_id_hidden=&selected_disease_name_hidden=&disease_type=0&disease_id_terrestrial=13&disease_id_aquatic=-999&selected_start_day=1&selected_start_ (accessed on 22 February 2020).
- Postel, A.; Nishi, T.; Kameyama, K.; Meyer, D.; Suckstorff, O.; Fukai, K.; Becher, P. Reemergence of Classical Swine Fever, Japan, 2018. Emerg. Infect. Dis. 2019, 25, 1228–1231. [Google Scholar] [CrossRef]
- Isoda, N.; Baba, K.; Ito, S.; Ito, M.; Sakoda, Y.; Makita, K. Dynamics of Classical Swine Fever Spread in Wild Boar in 2018–2019, Japan. Pathogens 2020, 9, 119. [Google Scholar] [CrossRef]
- World Organisation for Animal Health (WOAH). Resolution No. 20. Recognition of the Classical Swine Fever Status of Members; World Organisation for Animal Health: Paris, France, 2021. [Google Scholar]
- Coronado, L.; Rios, L.; Frías, M.T.; Amarán, L.; Naranjo, P.; Percedo, M.I.; Perera, C.L.; Prieto, F.; Fonseca-Rodriguez, O.; Pérez, L.J. Positive selection pressure on E2 protein of classical swine fever virus drives variations in virulence, pathogenesis and antigenicity: Implication for epidemiological surveillance in endemic areas. Transbound. Emerg. Dis. 2019, 66, 2362–2382. [Google Scholar] [CrossRef]
- Ji, W.; Niu, D.-D.D.; Si, H.-L.L.; Ding, N.-Z.Z.; He, C.-Q.Q. Vaccination influences the evolution of classical swine fever virus. Infect. Genet. Evol. 2014, 25, 69–77. [Google Scholar] [CrossRef]
- Hu, D.; Lv, L.; Gu, J.; Chen, T.; Xiao, Y.; Liu, S. Genetic Diversity and Positive Selection Analysis of Classical Swine Fever Virus Envelope Protein Gene E2 in East China under C-Strain Vaccination. Front. Microbiol. 2016, 7, 85. [Google Scholar] [CrossRef] [PubMed]
- Jang, G.; Kim, J.A.; Kang, W.M.; Yang, H.S.; Park, C.; Jeong, K.; Moon, S.U.; Park, C.K.; Lyoo, Y.S.; Lee, C. Endemic outbreaks due to the re-emergence of classical swine fever after accidental introduction of modified live LOM vaccine on Jeju Island, South Korea. Transbound. Emerg. Dis. 2019, 66, 634–639. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.; Pei, J.; Bai, J.; Zhao, M.; Ju, C.; Yi, L.; Kang, Y.; Zhang, X.; Chen, L.; Li, Y.; et al. Genetic diversity and positive selection analysis of classical swine fever virus isolates in south China. Virus Genes 2011, 43, 234–242. [Google Scholar] [CrossRef] [PubMed]
- World Organisation for Animal Health (OIE). Chapter 3.9.3. Classical swine fever (Infection with Classical swine fever virus). In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 2022; World Organisation for Animal Health (OIE), Ed.; World Organisation for Animal Health: Paris, France, 2018. [Google Scholar]
- EMEA/V/C/002757—R/0006; Suvaxyn CSF Marker. European Medicines Agency (EMA): Amsterdam, The Netherlands, 2019.
- EMEA/V/C/000046—R/0011; Porcilis Pesti. European Medicines Agency (EMA): Amsterdam, The Netherlands, 2011.
- Xia, H.; Harimoorthy, R.; Vijayaraghavan, B.; Blome, S.; Widén, F.; Beer, M.; Belák, S.; Liu, L. Differentiation of classical swine fever virus infection from CP7-E2alf marker vaccination by a multiplex microsphere immunoassay. Clin. Vaccine Immunol. 2015, 22, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Aebischer, A.; Müller, M.; Hofmann, M.A. Two newly developed Erns-based ELISAs allow the differentiation of Classical Swine Fever virus-infected from marker-vaccinated animals and the discrimination of pestivirus antibodies. Vet. Microbiol. 2013, 161, 274–285. [Google Scholar] [CrossRef]
- Postel, A.; Schmeiser, S.; Oguzoglu, T.C.; Indenbirken, D.; Alawi, M.; Fischer, N.; Grundhoff, A.; Becher, P. Close relationship of ruminant pestiviruses and classical swine fever virus. Emerg. Infect. Dis. 2015, 21, 668–672. [Google Scholar] [CrossRef]
- Wang, M.; Sozzi, E.; Bohórquez, J.A.; Alberch, M.; Pujols, J.; Cantero, G.; Gaffuri, A.; Lelli, D.; Rosell, R.; Bensaid, A.; et al. Decrypting the Origin and Pathogenesis in Pregnant Ewes of a New Ovine Pestivirus Closely Related to Classical Swine Fever Virus. Viruses 2020, 12, 775. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Sozzi, E.; Wang, M.; Alberch, M.; Abad, X.; Gaffuri, A.; Lelli, D.; Rosell, R.; Pérez, L.J.; Moreno, A.; et al. The new emerging ovine pestivirus can infect pigs and confers strong protection against classical swine fever virus. Transbound. Emerg. Dis. 2022, 69, 1539–1555. [Google Scholar] [CrossRef]
- Holinka, L.G.; Fernandez-Sainz, I.J.; Sanford, B.; O’Donnell, V.; Gladue, D.P.; Carlson, J.; Lu, Z.; Risatti, G.R.; Borca, M.V. Development of an improved live attenuated antigenic marker CSF vaccine strain candidate with an increased genetic stability. Virology 2014, 471, 13–18. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Defaus, S.; Rosell, R.; Pérez-Simó, M.; Alberch, M.; Gladue, D.P.; Borca, M.V.; Andreu, D.; Ganges, L. Development of a Dendrimeric Peptide-Based Approach for the Differentiation of Animals Vaccinated with FlagT4G against Classical Swine Fever from Infected Pigs. Viruses 2021, 13, 1980. [Google Scholar] [CrossRef]
- Holinka, L.G.; O’Donnell, V.; Risatti, G.R.; Azzinaro, P.; Arzt, J.; Stenfeldt, C.; Velazquez-Salinas, L.; Carlson, J.; Gladue, D.P.; Borca, M.V. Early protection events in swine immunized with an experimental live attenuated classical swine fever marker vaccine, FlagT4G. PLoS ONE 2017, 12, e0177433. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Wensvoort, G.; Terpstra, C.; Boonstra, J.; Bloemraad, M.; Zaane, D. Van Production of monoclonal antibodies against swine fever virus and their use in laboratory diagnosis. Vet. Microbiol. 1986, 12, 101–108. [Google Scholar] [CrossRef]
- Terpstra, C.; Bloemraad, M.; Gielkens, A.L. The neutralizing peroxidase-linked assay for detection of antibody against swine fever virus. Vet. Microbiol. 1984, 9, 113–120. [Google Scholar] [CrossRef]
- Wang, M.; Bohórquez, J.A.; Hinojosa, Y.; Muñoz-González, S.; Gerber, M.; Coronado, L.; Perera, C.L.; Liniger, M.; Ruggli, N.; Ganges, L. Abrogation of the RNase activity of Erns in a low virulence classical swine fever virus enhances the humoral immune response and reduces virulence, transmissibility, and persistence in pigs. Virulence 2021, 12, 2037–2049. [Google Scholar] [CrossRef] [PubMed]
- Diaz de Arce, H.; Artursson, K.; L’Haridon, R.; Perers, A.; La Bonnardiere, C.; Alm, G.V. A sensitive immunoassay for porcine interferon-α. Vet. Immunol. Immunopathol. 1992, 30, 319–327. [Google Scholar] [CrossRef]
- Hoffmann, B.; Beer, M.; Schelp, C.; Schirrmeier, H.; Depner, K. Validation of a real-time RT-PCR assay for sensitive and specific detection of classical swine fever. J. Virol. Methods 2005, 130, 36–44. [Google Scholar] [CrossRef]
- Muñoz-González, S.; Pérez-Simó, M.; Colom-Cadena, A.; Cabezón, O.; Bohórquez, J.A.; Rosell, R.; Pérez, L.J.; Marco, I.; Lavín, S.; Domingo, M.; et al. Classical swine fever virus vs. Classical swine fever virus: The superinfection exclusion phenomenon in experimentally infected wild boar. PLoS ONE 2016, 11, e0149469. [Google Scholar] [CrossRef]
- Tarradas, J.; Monsó, M.; Fraile, L.; de la Torre, B.G.; Muñoz, M.; Rosell, R.; Riquelme, C.; Pérez, L.J.; Nofrarías, M.; Domingo, M.; et al. A T-cell epitope on NS3 non-structural protein enhances the B and T cell responses elicited by dendrimeric constructions against CSFV in domestic pigs. Vet. Immunol. Immunopathol. 2012, 150, 36–46. [Google Scholar] [CrossRef]
- Li, Y.; Mateu, E.; Díaz, I. Impact of Cryopreservation on Viability, Phenotype, and Functionality of Porcine PBMC. Front. Immunol. 2021, 12, 5011. [Google Scholar] [CrossRef]
- Coronado, L.; Bohórquez, J.A.; Muñoz-González, S.; Pérez, L.J.; Rosell, R.; Fonseca, O.; Delgado, L.; Perera, C.L.; Frías, M.T.; Ganges, L. Investigation of chronic and persistent classical swine fever infections under field conditions and their impact on vaccine efficacy. BMC Vet. Res. 2019, 15, 247. [Google Scholar] [CrossRef] [PubMed]
- Blome, S.; Moß, C.; Reimann, I.; König, P.; Beer, M. Classical swine fever vaccines—State-of-the-art. Vet. Microbiol. 2017, 206, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Meuwissen, M.P.M.; Horst, S.H.; Huirne, R.B.M.; Dijkhuizen, A.A. A model to estimate the financial consequences of classical swine fever outbreaks: Principles and outcomes. Prev. Vet. Med. 1999, 42, 249–270. [Google Scholar] [CrossRef]
- Graham, S.P.; Everett, H.E.; Haines, F.J.; Johns, H.L.; Sosan, O.A.; Salguero, F.J.; Clifford, D.J.; Steinbach, F.; Drew, T.W.; Crooke, H.R. Challenge of pigs with classical swine fever viruses after C-strain vaccination reveals remarkably rapid protection and insights into early immunity. PLoS ONE 2012, 7, e29310. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, R.R.; Everett, H.E.; Graham, S.P.; Steinbach, F.; Crooke, H.R. Head Start Immunity: Characterizing the Early Protection of C Strain Vaccine Against Subsequent Classical Swine Fever Virus Infection. Front. Immunol. 2019, 10, 1584. [Google Scholar] [CrossRef]
- Ganges, L.; Barrera, M.; Núñez, J.I.; Blanco, I.; Frías, M.T.; Rodríguez, F.; Sobrino, F. A DNA vaccine expressing the E2 protein of classical swine fever virus elicits T cell responses that can prime for rapid antibody production and confer total protection upon viral challenge. Vaccine 2005, 23, 3741–3752. [Google Scholar] [CrossRef]
- Tarradas, J.; Argilaguet, J.M.; Rosell, R.; Nofrarías, M.; Crisci, E.; Córdoba, L.; Pérez-Martín, E.; Díaz, I.; Rodríguez, F.; Domingo, M.; et al. Interferon-gamma induction correlates with protection by DNA vaccine expressing E2 glycoprotein against classical swine fever virus infection in domestic pigs. Vet. Microbiol. 2010, 142, 51–58. [Google Scholar] [CrossRef]
- Wang, M.; Liniger, M.; Muñoz-González, S.; Bohórquez, J.A.; Hinojosa, Y.; Gerber, M.; López-Soria, S.; Rosell, R.; Ruggli, N.; Ganges, L. A Polyuridine Insertion in the 3′ Untranslated Region of Classical Swine Fever Virus Activates Immunity and Reduces Viral Virulence in Piglets. J. Virol. 2019, 94. [Google Scholar] [CrossRef]
- Suradhat, S.; Intrakamhaeng, M.; Damrongwatanapokin, S. The correlation of virus-specific interferon-gamma production and protection against classical swine fever virus infection. Vet. Immunol. Immunopathol. 2001, 83, 177–189. [Google Scholar] [CrossRef]
- Suradhat, S.; Damrongwatanapokin, S.; Thanawongnuwech, R. Factors critical for successful vaccination against classical swine fever in endemic areas. Vet. Microbiol. 2007, 119, 1–9. [Google Scholar] [CrossRef]
- Summerfield, A.; Ruggli, N. Immune responses against classical swine fever virus: Between ignorance and lunacy. Front. Vet. Sci. 2015, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sun, Y.; Meng, X.Y.; Li, L.F.; Li, Y.; Luo, Y.; Wang, W.; Yu, S.; Yin, C.; Li, S.; et al. Comprehensive evaluation of the host responses to infection with differentially virulent classical swine fever virus strains in pigs. Virus Res. 2018, 255, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Gou, H.; Zhao, M.; Fan, S.; Yuan, J.; Liao, J.; He, W.; Xu, H.; Chen, J. Autophagy induces apoptosis and death of T lymphocytes in the spleen of pigs infected with CSFV. Sci. Rep. 2017, 7, 13577. [Google Scholar] [CrossRef]
- Bohórquez, J.A.; Wang, M.; Pérez-Simó, M.; Vidal, E.; Rosell, R.; Ganges, L. Low CD4/CD8 ratio in classical swine fever postnatal persistent infection generated at 3 weeks after birth. Transbound. Emerg. Dis. 2019, 66, 752–762. [Google Scholar] [CrossRef]
- Appleyard, G.D.; Furesz, S.E.; Wilkie, B.N. Blood lymphocyte subsets in pigs vaccinated and challenged with Actinobacillus pleuropneumoniae. Vet. Immunol. Immunopathol. 2002, 86, 221–228. [Google Scholar] [CrossRef]
- Agrati, C.; Castilletti, C.; Casetti, R.; Sacchi, A.; Falasca, L.; Turchi, F.; Tumino, N.; Bordoni, V.; Cimini, E.; Viola, D.; et al. Longitudinal characterization of dysfunctional T cell-activation during human acute Ebola infection. Cell Death Dis. 2016, 7, e2164. [Google Scholar] [CrossRef]
- Serrano-Villar, S.; Moreno, S.; Fuentes-Ferrer, M.; Sánchez-Marcos, C.; Ávila, M.; Sainz, T.; de Villar, N.G.P.; Fernández-Cruz, A.; Estrada, V. The CD4:CD8 ratio is associated with markers of age-associated disease in virally suppressed HIV-infected patients with immunological recovery. HIV Med. 2014, 15, 40–49. [Google Scholar] [CrossRef]
- Postel, A.; Becher, P. Genetically distinct pestiviruses pave the way to improved classical swine fever marker vaccine candidates based on the chimeric pestivirus concept. Emerg. Microbes Infect. 2020, 9, 2180–2189. [Google Scholar] [CrossRef]


| Pig ID | FlagT4G | CSFV Margarita Strain | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 5 DPV/Day of CSFV Challenge | 4 dpi | 7 dpi | 14 dpi | 5 DPV/Day of CSFV Challenge | 4 dpi | 7 dpi | 14 dpi | ||
| FlagT4G vaccinated pigs | 1 | (-) | (-) | 1:40 | 1:120 | (-) | (-) | (-) | 1:15 |
| 2 † | (-) | 1:15 | 1:80 | N/S | (-) | (-) | (-) | N/S | |
| 3 | (-) | 1:20 | 1:80 | 1:160 | (-) | (-) | 1:15 | 1:80 | |
| 4 | (-) | 1:15 | 1:30 | 1:60 | (-) | 1:10 | 1:20 | 1:30 | |
| 5 | (-) | 1:10 | 1:30 | 1:240 | (-) | (-) | (-) | 1:10 | |
| Unvaccinated pigs | 6 * | (-) | (-) | (-) | N/S | (-) | (-) | (-) | N/S |
| 7 * | (-) | (-) | (-) | N/S | (-) | (-) | (-) | N/S | |
| 8 * | (-) | (-) | (-) | N/S | (-) | (-) | (-) | N/S | |
| 9 * | (-) | (-) | (-) | N/S | (-) | (-) | (-) | N/S | |
| 10 * | (-) | (-) | (-) | N/S | (-) | (-) | (-) | N/S | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bohórquez, J.A.; Wang, M.; Díaz, I.; Alberch, M.; Pérez-Simó, M.; Rosell, R.; Gladue, D.P.; Borca, M.V.; Ganges, L. The FlagT4G Vaccine Confers a Strong and Regulated Immunity and Early Virological Protection against Classical Swine Fever. Viruses 2022, 14, 1954. https://doi.org/10.3390/v14091954
Bohórquez JA, Wang M, Díaz I, Alberch M, Pérez-Simó M, Rosell R, Gladue DP, Borca MV, Ganges L. The FlagT4G Vaccine Confers a Strong and Regulated Immunity and Early Virological Protection against Classical Swine Fever. Viruses. 2022; 14(9):1954. https://doi.org/10.3390/v14091954
Chicago/Turabian StyleBohórquez, José Alejandro, Miaomiao Wang, Ivan Díaz, Mònica Alberch, Marta Pérez-Simó, Rosa Rosell, Douglas P. Gladue, Manuel V. Borca, and Llilianne Ganges. 2022. "The FlagT4G Vaccine Confers a Strong and Regulated Immunity and Early Virological Protection against Classical Swine Fever" Viruses 14, no. 9: 1954. https://doi.org/10.3390/v14091954






