A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections
Abstract
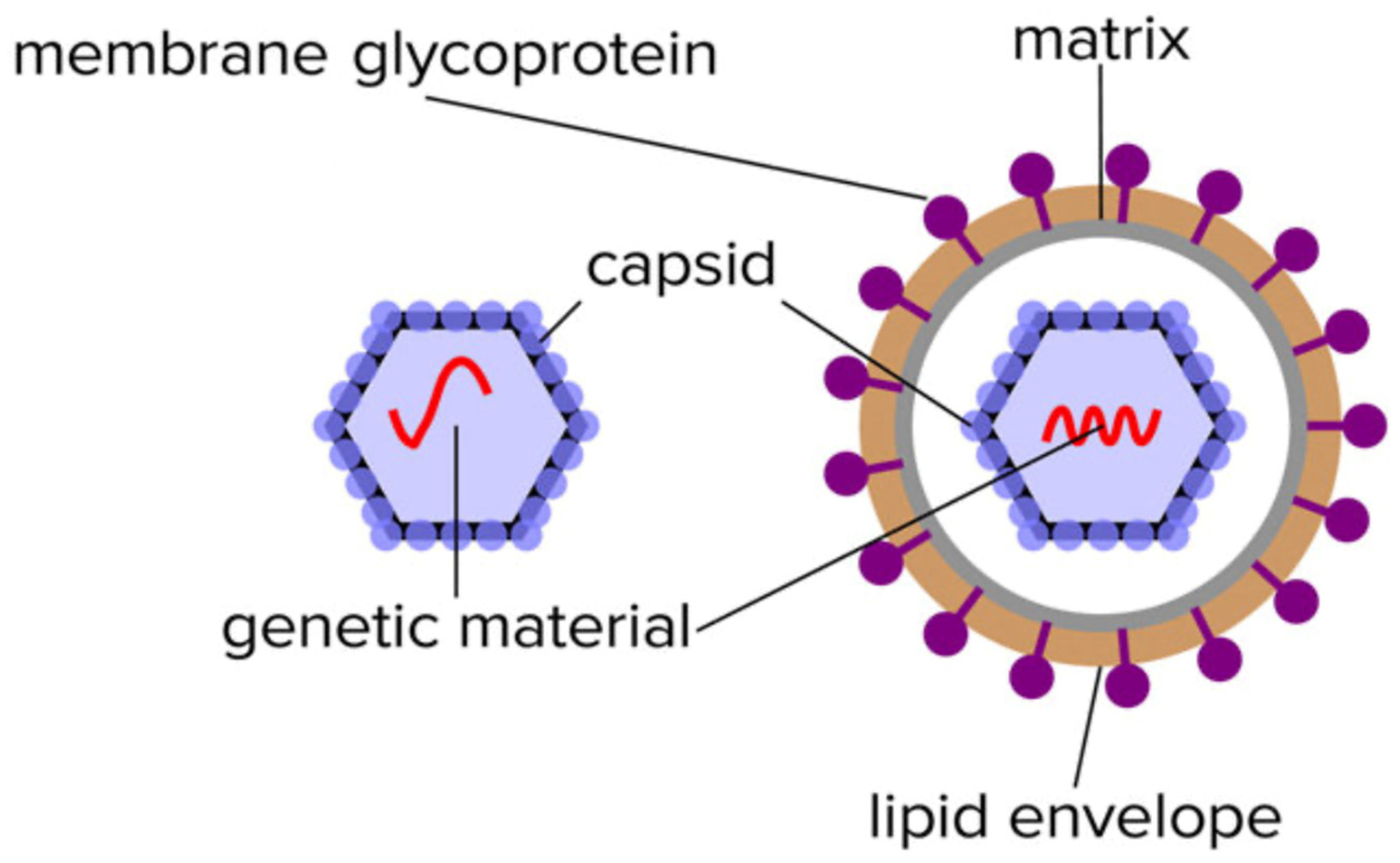
:1. Introduction
1.1. Understanding the Systems Biology of the Virus and Viral Infections
1.2. Understanding the Nature of Viral Proteins and Molecular Biology

2. Sampling and Virion Enrichment
2.1. Virion Protein Solubilisation and Extraction
2.2. Accelerating MALDI-ToF Assay Protocol Development Using Pseudotypes/Pseudoviruses
2.3. Understanding the Operational Parameters of Your MALDI-ToF Mass Spectrometer
2.4. Peak Identification–Quantification and Bioinformatics
2.5. Promise and Pitfalls of Machine Learning Bioinformatics
3. Clinical Testing Laboratory, Validation and International Accreditation
Limitation and Advantages of CLIA-Laboratory Developed Tests (LDT)
4. MALDI–ToF Mass Spectrometry Screening Test for SARS-CoV-2
- Prepare positive control: Take 300 μL of ultra-pure water and add to the freeze-dried positive control SARS-CoV-2 pseudo-virus tube Vortex until there are no visible clumps in the tube. Spike 150 μL into 4.85 mL of saline. Process as for samples.
- Prepare gargle-saliva samples: Irradiate collected gargle-saliva samples with UV light at 280 nm for 15 min Collect the liquid into a lure lock syringe attached with a 21 G, 1.5″ needle. Once the liquid has been collected into the syringe, create an air bubble to keep the liquid in place whilst removing the needle from the syringe. Add the plastic cover and dispose it into a sharps box.
- Apply the 0.45 μL filter onto syringe and push 5 mL of sample/spiked saline through a 0.45 μL filter into a clean 10 mL centrifuge tube.
- Viral particle enrichment: Add ice-cold acetone (4 °C) to match the amount of sample in the tube (1:1); it should be 5 mL, giving a total volume of 10 mL. Centrifuge and spin the samples at 16,000 RCF at 4 °C, for 30 min. Discard the supernatant completely into a suitable container. Note: No visible pellet can be observed.
- Dissolution of virions and solubilization of viral proteins: Add 50 μL of the buffer LBSD-X working solution made up with 20 mM TCEP. Manually, with the pipette tip, scrape it off the side of the container and pump mix with a pipette. Vortex the sample well, and incubate for 15 min at a room temperature.
- MALDI–ToF mass spectrometry: After 15 min of sample incubation with the buffer LBSD-X and TCEP, vortex the sample again to ensure homogeneity and that 1 μL of the sample is ready to plate following the sample plating procedures using 15 mg/mL Sinapinic Acid matrices.
5. CLIA LDT Validation of a MALDI-ToF Mass Spectrometry Test for SARS-CoV-2
- Limit of detection (LoD);
- Interfering substances and specificity;
- Clinical performance evaluation;
- Reproducibility;
- Stability.
5.1. Limit of Detection (LoD)
5.2. Interfering Substances and Specificity
5.3. Clinical Performance Evaluation
- Clinical evaluation was specific to this purpose.
- ○
- No other test for SARS-CoV-2 testing with emergency use authorization has been evaluated for this purpose in the clinical performance evaluations.
- ○
- All previous EUA IVD tests have been evaluated on hospitalized, clinically confirmed, patient samples only.
5.3.1. Establishing Operational Cut-Off Values
5.3.2. Direct Comparison with an rtPCR SARS-CoV-2 Test
5.3.3. Internal Sampling QC
5.3.4. Daily System QCs
5.4. Reproducibility
5.5. Stability
6. Conclusion—Validation Disposition
Conclusion—Global Biosecurity
7. Patents
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Leber, W.; Lammel, O.; Siebenhofer, A.; Redlberger-Fritz, M.; Panovska-Griffiths, J.; Czypionka, T. Comparing the diagnostic accuracy of point-of-care lateral flow antigen testing for SARS-CoV-2 with RT-PCR in primary care (REAP-2). EClinicalMedicine 2021, 38, 101011. [Google Scholar] [CrossRef] [PubMed]
- Deeks, J.J.; Dinnes, J.; Davenport, C.; Takwoingi, Y.; McInnes, M.; Mg Leeflang, M.; Cunningham, J. Letter to the Editor regarding Peto T; UK COVID-19 Lateral Flow Oversight Team: COVID-19: Rapid antigen detection for SARS-CoV-2 by lateral flow assay. EClinicalMedicine 2021, 38, 101037. [Google Scholar] [CrossRef] [PubMed]
- Lamb, G.; Heskin, J.; Randell, P.; Mughal, N.; Moore, L.S.; Jones, R.; Davies, G.W.; Rayment, M. Real-world evaluation of COVID-19 lateral flow device (LFD) mass-testing in healthcare workers at a London hospital; a prospective cohort analysis. J. Infect. 2021, 83, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, J.; Dunn, S.; Best, A.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T.; Moles-Garcia, E.; et al. Validation testing to determine the sensitivity of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings: Testing frequency and public health messaging is key. PLoS Biol. 2021, 19, e3001216. [Google Scholar] [CrossRef]
- De la Matta, M.; Delgado-Sánchez, J.M.; Gutiérrez, G.M.; Romero, J.L.; Gómez, M.M.; Blanco, A.D. Utility of preoperative polymerase chain reaction testing during SARS-CoV-2 pandemic: The challenge of evolving incidence. Rev. Española De Anestesiol. Y Reanim. (Engl. Ed.) 2021, 68, 346–352. [Google Scholar] [CrossRef]
- Tanriover, M.D.; Fathi, A.; Raspe, M.; Novais, A.G.; Uyaroğlu, O.A. Characteristics and management of asymptomatic SARS-CoV-2 infections. J. Basic Clin. Physiol. Pharmacol. 2022, 33, 1–7. [Google Scholar] [CrossRef]
- Subudhi, P.D.; Rooge, S.; Thangriyal, S.; Aggarwal, R.; Gupta, E.; Baweja, S. Prolonged Detection of SARS CoV2 RNA in Extracellular Vesicles in Nasal Swab RT-PCR Negative Patients. medRxiv 2021. [Google Scholar]
- Lefebvre, D.; Blanco-Valle, K.; Feraudet-Tarisse, C.; Merda, D.; Simon, S.; Fenaille, F.; Hennekinne, J.-A.; Nia, Y.; Becher, F. Quantitative Determination of Staphylococcus aureus Enterotoxins Types A to I and Variants in Dairy Food Products by Multiplex Immuno-LC-MS/MS. J. Agric. Food Chem. 2021, 69, 2603–2610. [Google Scholar] [CrossRef]
- Kang, L.; Weng, N.; Jian, W. LC–MS bioanalysis of intact proteins and peptides. Biomed. Chromatogr. 2020, 34, e4633. [Google Scholar] [CrossRef]
- Donnelly, D.P.; Rawlins, C.M.; DeHart, C.J.; Fornelli, L.; Schachner, L.F.; Lin, Z.; Lippens, J.L.; Aluri, K.C.; Sarin, R.; Chen, B.; et al. Best practices and benchmarks for intact protein analysis for top-down mass spectrometry. Nat. Methods 2019, 16, 587–594. [Google Scholar] [CrossRef]
- Sogawa, K.; Watanabe, M.; Sato, K.; Segawa, S.; Ishii, C.; Miyabe, A.; Murata, S.; Saito, T.; Nomura, F. Use of the MALDI BioTyper system with MALDI–TOF mass spectrometry for rapid identification of microorganisms. Anal. Bioanal. Chem. 2011, 400, 1905–1911. [Google Scholar] [CrossRef] [PubMed]
- Monopoli, A.; Nacci, A.; Cataldi, T.R.; Calvano, C.D. Synthesis and Matrix Properties of α-Cyano-5-phenyl-2, 4-pentadienic Acid (CPPA) for Intact Proteins Analysis by Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry. Molecules 2020, 25, 6054. [Google Scholar] [CrossRef] [PubMed]
- Cubuk, J.; Alston, J.J.; Incicco, J.J.; Singh, S.; Stuchell-Brereton, M.D.; Ward, M.D.; Zimmerman, M.I.; Vithani, N.; Griffith, D.; Wagoner, J.A.; et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Fung, T.S.; Liu, T.X. Post-translational modifications of coronavirus proteins: Roles and function. Future Virol. 2018, 13, 405–430. [Google Scholar] [CrossRef]
- Iles, R.K.; Zmuidinaite, R.; Iles, J.K.; Carnell, G.; Sampson, A.; Heeney, J.L. Development of a clinical MALDI-ToF mass spectrometry assay for SARS-CoV-2: Rational design and multi-disciplinary team work. Diagnostics 2020, 10, 746. [Google Scholar] [CrossRef]
- Depfenhart, M.; de Villiers, D.; Lemperle, G.; Meyer, M.; Di Somma, S. Potential new treatment strategies for COVID-19: Is there a role for bromhexine as add-on therapy? Intern. Emerg. Med. 2020, 15, 801–812. [Google Scholar] [CrossRef]
- Kupferschmidt, K.; Cohen, J. Race to find COVID-19 treatments accelerates. Science 2020, 367, 1412–1413. [Google Scholar] [CrossRef]
- Mason, R.J. Pathogenesis of COVID-19 from a cell biology perspective. Eur. Respir. J. 2020, 55, 2000607. [Google Scholar] [CrossRef]
- Nachtigall, F.M.; Pereira, A.; Trofymchuk, O.S.; Trofymchuk, O.S.; Santos, L.S. Detection of SARS-CoV-2 in nasal swabs using MALDI-MS. Nat. Biotechnol. 2020, 38, 1168–1173. [Google Scholar] [CrossRef]
- Tran, N.K.; Howard, T.; Walsh, R.; Pepper, J.; Loegering, J.; Phinney, B.; Salemi, M.R.; Rashidi, H.H. Novel application of automated machine learning with MALDI-TOF-MS for rapid high-throughput screening of COVID-19: A proof of concept. Sci. Rep. 2021, 11, 8219. [Google Scholar] [CrossRef]
- Armengaud, J. Unleashing immuno-mass spectrometry superpowers to detect SARS-CoV-2. EBioMedicine 2021, 69, 103480. [Google Scholar] [CrossRef] [PubMed]
- Chavan, S.; Mangalaparthi, K.K.; Singh, S.; Renuse, S.; Vanderboom, P.M.; Madugundu, A.K.; Budhraja, R.; McAulay, K.; Grys, T.E.; Rule, A.D.; et al. Mass spectrometric analysis of urine from COVID-19 patients for detection of SARS-CoV-2 viral antigen and to study host response. J. Proteome Res. 2021, 20, 3404–3413. [Google Scholar] [CrossRef] [PubMed]
- Bentley, E.M.; Mather, S.T.; Temperton, N.J. The use of pseudotypes to study viruses, virus sero-epidemiology and vaccination. Vaccine 2015, 33, 2955–2962. [Google Scholar] [CrossRef] [PubMed]
- Carnell, G.; Grehan, K.; Ferrara, F.; Molesti, E.; Temperton, N. An optimized method for the production using PEI, titration and neutralization of SARS-CoV spike luciferase pseudotypes. Bio. Protoc. 2017, 7, e2514. [Google Scholar] [CrossRef]
- Anderson, M. The ultraviolet offense: Germicidal UV lamps destroy vicious viruses. New tech might put them many more places without harming humans. IEEE Spectr. 2020, 57, 50–55. [Google Scholar] [CrossRef]
- Ruetalo, N.; Businger, R.; Schindler, M. Rapid, dose-dependent and efficient inactivation of surface dried SARS-CoV-2 by 254 nm UV-C irradiation. Eurosurveillance 2021, 26, 2001718. [Google Scholar] [CrossRef]
- Chen, C.H.; Lin, J.L.; Chu, M.L.; Chen, C.H. MALDI ion trap mass spectrometer with charge detector for large biomolecule detection. Anal. Chem. 2010, 82, 10125–10128. [Google Scholar] [CrossRef]
- Sage, E.; Brenac, A.; Alava, T.; Morel, R.; Dupre, C.; Hanay, M.S.; Roukes, M.L.; Duraffourg, L.; Masselon, C.; Hentz, S. Neutral particle mass spectrometry with nanomechanical systems. Nat. Commun. 2015, 6, 6482. [Google Scholar] [CrossRef]
- Witze, E.; Old, W.; Resing, K.; Ahn, N.G. Mapping protein post-translational modifications with mass spectrometry. Nat. Methods 2007, 4, 798–806. [Google Scholar] [CrossRef]
- Nishikaze, T. Sialic acid derivatization for glycan analysis by mass spectrometry. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2019, 95, 523–537. [Google Scholar] [CrossRef]
- Peng, Y.; Gu, B.; Sun, Z.; Li, Y.; Zhang, Y.; Lu, H. Linkage-selective derivatization for glycosylation site-and glycoform-specific characterization of sialic acid isomers using mass spectrometry. Chem. Commun. 2021, 57, 9590–9593. [Google Scholar] [CrossRef] [PubMed]
- Bouwman, K.M.; Tomris, I.; Turner, H.L.; van der Woude, R.; Shamorkina, T.M.; Bosman, G.P.; Rockx, B.; Herfst, S.; Snijder, J.; Haagmans, B.L.; et al. Multimerization-and glycosylation-dependent receptor binding of SARS-CoV-2 spike proteins. PLoS Pathog. 2021, 17, e1009282. [Google Scholar] [CrossRef]
- Pikora, C.A. Glycosylation of the ENV spike of primate immunodeficiency viruses and antibody neutralization. Curr. HIV Res. 2004, 2, 243–254. [Google Scholar] [CrossRef]
- Zhang, M.; Gaschen, B.; Blay, W.; Foley, B.; Haigwood, N.; Kuiken, C.; Korber, B. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 2004, 14, 1229–1246. [Google Scholar] [PubMed]
- Pais, R.J.; Iles, R.K.; Zmuidinaite, R. MALDI-ToF Mass Spectra Phenomic Analysis for Human Disease Diagnosis Enabled by Cutting-Edge Data Processing Pipelines and Bioinformatics Tools. Curr. Med. Chem. 2021, 28, 6532–6547. [Google Scholar] [CrossRef] [PubMed]
- Seethi, V.D.R.; LaCasse, Z.; Chivte, P.; Gaillard, E.R.; Bharti, P. An Explainable-AI approach for Diagnosis of COVID-19 using MALDI-ToF Mass Spectrometry. arXiv 2021, preprint. arXiv:2109.14099. [Google Scholar]
- Kononenko, I. Inductive and Bayesian learning in medical diagnosis. Appl. Artif. Intell. Int. J. 1993, 7, 317–337. [Google Scholar] [CrossRef]
- Butler-Laporte, G.; Lawandi, A.; Schiller, I.; Yao, M.C.; Dendukuri, N.; McDonald, E.G.; Lee, T.C. Comparison of Saliva and Nasopharyngeal Swab Nucleic Acid Amplification Testing for Detection of SARS-CoV-2: A Systematic Review and Meta-analysis. JAMA Intern. Med. 2021, 181, 353–360. [Google Scholar] [CrossRef]
- Xu, X.; Sun, J.; Nie, S.; Li, H.; Kong, Y.; Liang, M.; Hou, J.; Huang, X.; Li, D.; Ma, T.; et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 2020, 26, 1193–1195. [Google Scholar] [CrossRef]
- Meyerowitz, E.A.; Richterman, A.; Isaac, I.; Bogoch, I.I.; Low, N.; Cevik, M. Towards an accurate and systematic characterisation of persistently asymptomatic infection with SARS-CoV-2. Lancet Infect. Dis. 2020, 21, e163–e169. [Google Scholar] [CrossRef]
- Buitrago-Garcia, D.; Egli-Gany, D.; Counotte, M.J. Occurrence and transmission potential of asymptomatic and presymptomatic SARS-CoV-2 infections: A living systematic review and meta-analysis. PLoS Med. 2020, 17, e1003346. [Google Scholar]
- Byambasuren, O.; Cardona, M.; Bell, K.; Clark, J.; McLaws, M.-L.; Glasziou, P. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: Systematic review and meta-analysis. JAMMI 2020, 5, 223–234. [Google Scholar] [CrossRef]
- Dietterich, T. Overfitting and Undercomputing in Machine Learning. ACM Computing Surveys (CSUR). 1995. Available online: https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.55.2069&rep=rep1&type=pdf (accessed on 5 July 2022).
- Peng, Y.; Nagata, M.H. An empirical overview of nonlinearity and overfitting in machine learning using COVID-19 data. Chaos Solitons Fractals 2020, 139, 110055. [Google Scholar] [CrossRef] [PubMed]
- van Belkum, A.; Chatellier, S.; Girard, V.; Pincus, D.; Deol, P.; Dunne, W.M., Jr. Progress in proteomics for clinical microbiology: MALDI-TOF MS for microbial species identification and more. Expert Rev. Proteom. 2015, 12, 595–605. [Google Scholar] [CrossRef]
- Lefferts, J.A.; Gutmann, E.J.; Martin, I.W.; Wells, W.A.; Tsongalis, G.J. Implementation of an Emergency Use Authorization test during an impending national crisis. J. Mol. Diagn. 2020, 22, 844–846. [Google Scholar] [CrossRef]
- Chan, D.W.; Semmes, O.J.; Petricoin, E.F.; Liotta, L.A.; van der Merwe, D.; Diamandis, E.P. National Academy of Clinical Biochemistry Guidelines: The Use of MALDI-TOF Mass Spectrometry Profiling to Diagnose Cancer; American Association for Clinical Chemistry: Washington, DC, USA, 2006. [Google Scholar]
- Chivte, P.; LaCasse, Z.; Seethi, V.D.R.; Bharti, P.; Bland, J.; Kadkol, S.S.; Gaillard, E.R. MALDI-ToF protein profiling as a potential rapid diagnostic platform for COVID-19. J. Mass Spectrom. Adv. Clin. Lab 2021, 21, 31–41. [Google Scholar] [CrossRef]
- Ferguson, J.; Dunn, S.; Best, A.; Mirza, J.; Percival, B.; Mayhew, M.; Megram, O.; Ashford, F.; White, T.; Moles-Garcia, E.; et al. Validation testing to determine the effectiveness of lateral flow testing for asymptomatic SARS-CoV-2 detection in low prevalence settings. medRxiv 2020. [Google Scholar] [CrossRef]
- Meldrum, O.W.; Yakubov, G.E.; Bonilla, M.R.; Deshmukh, O.; McGuckin, M.A.; Gidley, M.J. Mucin gel assembly is controlled by a collective action of non-mucin proteins, disulfide bridges, Ca2+-mediated links, and hydrogen bonding. Sci. Rep. 2018, 8, 5802. [Google Scholar] [CrossRef]
- Iles, R.K.; Iles, J.K. US 2021/0356475 A1: Virus and Exosome Sample Preparation and Analysis Methods. Available online: https://patents.google.com/patent/US20210356475A1/en (accessed on 25 August 2022).
- Griffin, J.H.; Downard, K.M. Mass spectrometry analytical responses to the SARS-CoV2 coronavirus in review. TrAC Trends Anal. Chem. 2021, 142, 116328. [Google Scholar] [CrossRef]
- Sivanesan, I.; Gopal, J.; Vinay, R.S.; Hanna, L.E.; Oh, J.W.; Muthu, M. Consolidating the potency of Matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) in viral diagnosis: Extrapolating its applicability for COVID diagnosis? TrAC Trends Anal. Chem. 2022, 150, 1165. [Google Scholar] [CrossRef]







| (A) PFU | ||
| Conc. | POS/NEG | % Positive |
| 1000 PFU (200 per mL) | 10/10 | 100% |
| 750 PFU (150 per mL) | 10/10 | 100% |
| 500 PFU (100 per mL) | 5/10 | 50% |
| 10 PFU (2 per mL) | 0/10 | 0% |
| (B) Copy Number | ||
| Conc. | POS/NEG | % Positive |
| 2304 copies per mL | 10/10 | 100% |
| 1152 copies per mL | 10/10 | 100% |
| 576 copies per mL | 10/10 | 100% |
| 288 copies per mL | 9/10 | 90% |
| 144 copies per mL | 5/10 | 50% |
| 77 copies per mL | 0/10 | 0% |
| Positive Samples Detected | Negative Samples Detected | ||||||
|---|---|---|---|---|---|---|---|
| Potential Interfering Substance | Concentration of Substance | Pool 1 | Pool 2 | Pool 3 | Pool 1 | Pool 2 | Pool 3 |
| Toothpaste | NOT EVALUATED | ||||||
| Mouthwash * | 0.1% v/v Benzalkonium chloride And/or 0.1% Chlorohexidine | neg | neg | neg | neg | neg | neg |
| Nicotine | NOT EVALUATED | ||||||
| Benzocaine/Menthol | |||||||
| Caffeine | |||||||
| No Interfering substance (controls) | Pseudotype SARCOV-2 103 FU/μL | Pos | Pos | Pos | neg | neg | neg |
| SARS Virus | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | neg | neg | neg |
| Rhino Virus | Infected Individuals Gargle (concn unknown) | Pos | Pos | Pos | neg | neg | neg |
| Human Coronavirus MERS | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | neg | neg | neg |
| Human Coronavirus OC43 ** | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | Pos | Pos | neg |
| Human Coronavirus 229E ** | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | neg | neg | Pos |
| Human Coronavirus NL63 | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | neg | neg | neg |
| Human Coronavirus HKU1 | Pseudotypes at 103 FU/μL | Pos | Pos | Pos | neg | neg | neg |
| Influenza A virus (H1N1 Swine Flu) | 1000 PFU | Pos | Pos | Pos | neg | neg | neg |
| Influenza A virus (H1N21 Pueto Rico) | 1000 PFU | Pos | Pos | Pos | neg | neg | neg |
| (A) High Cut-Off Threshold | Abbott SARS-CoV-2 rtPCR Test ** | |||
| Pre- Symptomatic Positive (3) | Asymptomatic Positive (57) | Negative (92) | ||
| MALDI–ToF Gargle sample test | Pre-Symptomatic Positive (3) | 3/3 | N/A | 0 |
| Asymptomatic Positive (47) | N/A | 29/57 | 18 | |
| Negative (102) | 0 | 28 | 74/92 | |
| Positive Agreement | Pre symptomatic cases 100% Asymptomatic cases 51% | |||
| Negative Agreement | 80% agreement on negatives | |||
| (B) Low Cut-Off Threshold | Abbott SARS-CoV-2 rtPCR Test ** | |||
| Pre- Symptomatic Positive (3) | Asymptomatic Positive (57) | Negative (92) | ||
| MALDI–ToF Gargle sample test | Pre-Symptomatic Positive (3) | 3/3 | N/A | 0 |
| Asymptomatic Positive (47) | N/A | 45/57 | 18 | |
| Negative (102) | 0 | 5 | 102/92 | |
| Positive Agreement | Pre symptomatic cases 100% Asymptomatic cases 75% | |||
| Negative Agreement | 65% agreement on negatives | |||
| Week 1 POS/NEG | Week 2 POS/NEG | Week 3 POS/NEG | Week 4 POS/NEG | Total POS/NEG | |
|---|---|---|---|---|---|
| Positive (PCR) | 4/0 | 4/0 | 4/0 | 4/0 | 16/0 |
| Negative (PCR) | 0/4 | 0/4 | 0/4 | 0/4 | 0/16 |
| Extract Stability | |||||
| (A) CDC Inactivated Virus | Week 1 POS/NEG | Week 2 POS/NEG | Week 3 POS/NEG | Week 4 POS/NEG | Total POS/NEG |
| Positive (PCR) | 5/0 | 5/0 | 5/0 | - | 15/0 |
| Negative (PCR) | 0/5 | 0/5 | 0/5 | - | 0/15 |
| (B) Pseudo-Type Virus | Week 1 POS/NEG | Week 2 POS/NEG | Week 3 POS/NEG | Week 4 POS/NEG | Total POS/NEG |
| Positive (PCR) | 10/0 | 10/0 | 10/0 | 10/0 | 40/0 |
| Negative (PCR) | 0/10 | 0/10 | 0/10 | 0/10 | 0/40 |
| Sample Stability | |||||
| (C) CDC Inactivated Virus | Week 1 POS/NEG | Week 2 POS/NEG | Week 3 POS/NEG | Week 4 POS/NEG | Total POS/NEG |
| Positive (PCR) | 5/0 | 5/0 | 5/0 | 5/0 | 20/0 |
| Negative (PCR) | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 |
| (D) Pseudo-Type Virus | Week 1 POS/NEG | Week 2 POS/NEG | Week 3 POS/NEG | Week 4 POS/NEG | Total POS/NEG |
| Positive (PCR) | 5/0 | 5/0 | 5/0 | 5/0 | 20/0 |
| Negative (PCR) | 0/5 | 0/5 | 0/5 | 0/5 | 0/20 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iles, R.K.; Iles, J.K.; Zmuidinaite, R.; Roberts, M. A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections. Viruses 2022, 14, 1958. https://doi.org/10.3390/v14091958
Iles RK, Iles JK, Zmuidinaite R, Roberts M. A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections. Viruses. 2022; 14(9):1958. https://doi.org/10.3390/v14091958
Chicago/Turabian StyleIles, Ray K., Jason K. Iles, Raminta Zmuidinaite, and Michael Roberts. 2022. "A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections" Viruses 14, no. 9: 1958. https://doi.org/10.3390/v14091958
APA StyleIles, R. K., Iles, J. K., Zmuidinaite, R., & Roberts, M. (2022). A How to Guide: Clinical Population Test Development and Authorization of MALDI-ToF Mass Spectrometry-Based Screening Tests for Viral Infections. Viruses, 14(9), 1958. https://doi.org/10.3390/v14091958





