Analysis of the Contribution of Intrinsic Disorder in Shaping Potyvirus Genetic Diversity
Abstract
:1. Introduction
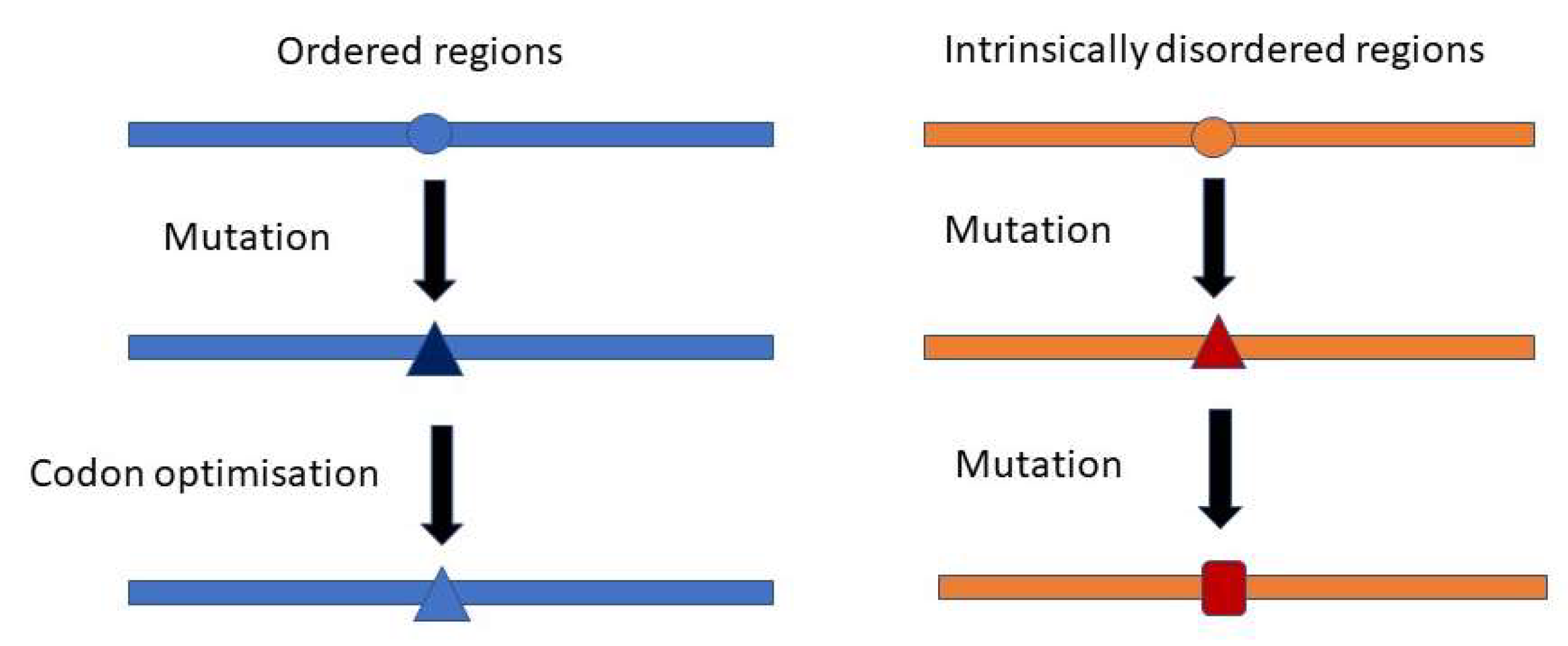
Protein Intrinsic Disorder
2. Material and Methods
Data Sets
- ○
- Disorder prediction
- ○
- Experimental dataset
- ○
- Natural diversity dataset
- ○
- Simulation
- ○
- Adaptive components tested
- ○
- Codon volatility assessment
3. Results
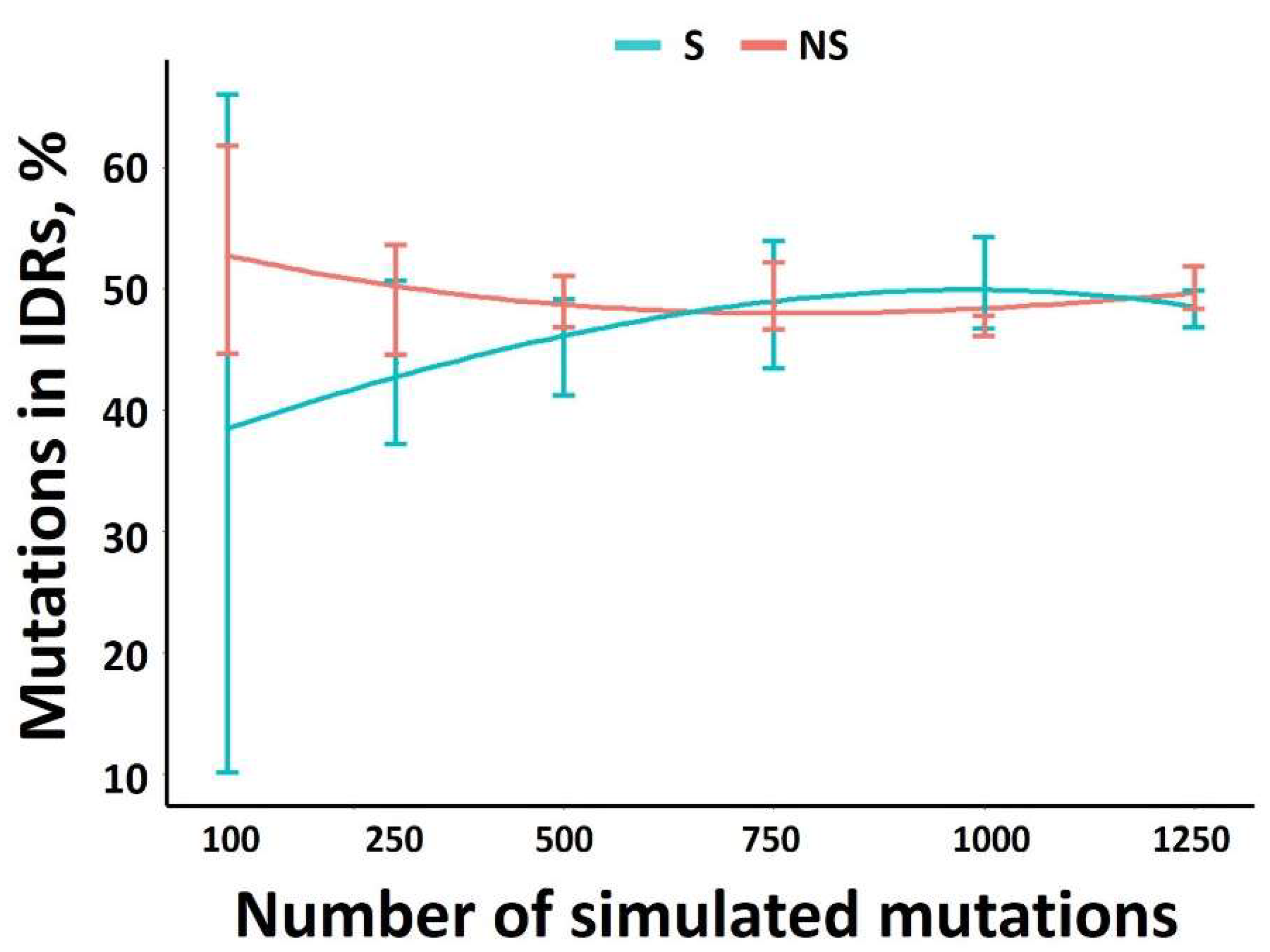
3.1. Theoretical Minimum Number of Mutations Required for an Accurate Estimation of S and NS Distribution in the Genome
3.2. Correlation Assessment between Protein Length and Number of Mutations
3.3. Distribution of NS and S Mutations in IDRs and ORs
3.4. Comparison of the Physicochemical Disturbance of Amino Acid Substitutions in Potyviral Intrinsically Disordered versus Ordered Regions
3.5. Are NS Mutations in IDRs Driven toward the Conservation of Disorder Promoting Amino Acids
3.6. Comparative Assessment of Codon Volatility between IDRs and ORs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Bellay, J.; Michaut, M.; Kim, T.; Han, S.; Colak, R.; Myers, C.L.; Kim, P.M. An omics perspective of protein disorder. Mol. Biosyst. 2012, 8, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Habchi, J.; Tompa, P.; Longhi, S.; Uversky, V.N. Introducing Protein Intrinsic Disorder. Chem. Rev. 2014, 114, 6561–6588. [Google Scholar] [CrossRef] [PubMed]
- Tokuriki, N.; Oldfield, C.J.; Uversky, V.N.; Berezovsky, I.N.; Tawfik, D.S. Do viral proteins possess unique biophysical features? Trends Biochem. Sci. 2009, 34, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Drummond, D.A.; Wilke, C.O. Contact density affects protein evolutionary rate from bacteria to animals. J. Mol. Evol. 2008, 66, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Gitlin, L.; Hagai, T.; LaBarbera, A.; Solovey, M.; Andino, R. Rapid Evolution of Virus Sequences in Intrinsically Disordered Protein Regions. PLoS Pathog. 2014, 10, e1004529. [Google Scholar] [CrossRef]
- Walter, J.; Charon, J.; Hu, Y.; Lachat, J.; Leger, T.; Lafforgue, G.; Barra, A.; Michon, T. Comparative analysis of mutational robustness of the intrinsically disordered viral protein VPg and of its interactor eIF4E. PLoS ONE 2019, 14, e0211725. [Google Scholar] [CrossRef]
- Bershtein, S.; Segal, M.; Bekerman, R.; Tokuriki, N.; Tawfik, D.S. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 2006, 444, 929–932. [Google Scholar] [CrossRef]
- Elena, S.F.; Agudelo-Romero, P.; Carrasco, P.; Codoñer, F.M.; Martín, S.; Torres-Barceló, C.; Sanjuán, R. Experimental evolution of plant RNA viruses. Heredity (Edinb) 2008, 100, 478–483. [Google Scholar] [CrossRef]
- Charon, J.; Theil, S.; Nicaise, V.; Michon, T. Protein intrinsic disorder within the Potyvirus genus: From proteome-wide analysis to functional annotation. Mol. Biosyst. 2016, 12, 634–652. [Google Scholar] [CrossRef]
- Charon, J.; Barra, A.; Walter, J.; Millot, P.; Hebrard, E.; Moury, B.; Michon, T. First Experimental Assessment of Protein Intrinsic Disorder Involvement in an RNA Virus Natural Adaptive Process. Mol. Biol. Evol. 2018, 35, 38–49. [Google Scholar] [CrossRef] [Green Version]
- Kutnjak, D.; Rupar, M.; Gutierrez-Aguirre, I.; Curk, T.; Kreuze, J.F.; Ravnikar, M. Deep Sequencing of Virus-Derived Small Interfering RNAs and RNA from Viral Particles Shows Highly Similar Mutational Landscapes of a Plant Virus Population. J. Virol. 2015, 89, 4760–4769. [Google Scholar] [CrossRef] [PubMed]
- Kutnjak, D.; Elena, S.F.; Ravnikar, M. Time-Sampled Population Sequencing Reveals the Interplay of Selection and Genetic Drift in Experimental Evolution of Potato Virus Y. J. Virol. 2017, 91, e00690-17. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, J.M.; Willemsen, A.; Hillung, J.; Zwart, M.P.; Elena, S.F. Temporal dynamics of intrahost molecular evolution for a plant RNA virus. Mol. Biol. Evol. 2015, 32, 1132–1147. [Google Scholar] [CrossRef]
- Schadt, E.E.; Turner, S.; Kasarskis, A. A Window into Third Generation Sequencing. Hum. Mol. Genet. 2010, 19, 227–240. [Google Scholar] [CrossRef]
- Sickmeier, M.; Hamilton, J.A.; LeGall, T.; Vacic, V.; Cortese, M.S.; Tantos, A.; Szabo, B.; Tompa, P.; Chen, J.; Uversky, V.N.; et al. DisProt: The Database of Disordered Proteins. Nucleic Acids Res. 2007, 35, D786–D793. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Dunbrack, R.L.; Williams, R.W.; Dunker, A.K.; Uversky, V.N. PONDR-FIT: A meta-predictor of intrinsically disordered amino acids. Biochim. Biophys Acta Proteins Proteom. 2010, 1804, 996–1010. [Google Scholar] [CrossRef]
- Tromas, N.; Elena, S.F. The Rate and Spectrum of Spontaneous Mutations in a Plant RNA Virus. Genetics 2010, 185, 983–989. [Google Scholar] [CrossRef]
- Radivojac, P.; Iakoucheva, L.M.; Oldfield, C.J.; Obradovic, Z.; Uversky, V.N.; Dunker, A.K. Intrinsic disorder and functional proteomics. Biophys. J. 2007, 92, 1439–1456. [Google Scholar] [CrossRef]
- Zhang, J. On the evolution of codon volatility. Genetics 2005, 169, 495–501. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef] [Green Version]
- Rudnicki, W.R.; Mroczek, T.; Cudek, P. Amino acid properties conserved in molecular evolution. PLoS ONE 2014, 9, e98983. [Google Scholar] [CrossRef] [PubMed]
- Dunker, A.K.; Obradovic, Z. The protein trinity–linking function and disorder. Nat. Biotechnol. 2001, 19, 805–806. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Romero, P.; Uversky, V.N.; Dunker, A.K. Conservation of intrinsic disorder in protein domains and families: I. A database of conserved predicted disordered regions. J. Proteome Res. 2006, 5, 879–887. Available online: http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=16602695 (accessed on 1 September 2022). [CrossRef] [PubMed]
- Ahrens, J.; Dos Santos, H.G.; Siltberg-Liberles, J. The Nuanced Interplay of Intrinsic Disorder and Other Structural Properties Driving Protein Evolution. Mol. Biol. Evol. 2016, 33, 2248–2256. [Google Scholar] [CrossRef] [PubMed]
- Ahrens, J.B.; Nunez-Castilla, J.; Siltberg-Liberles, J. Evolution of intrinsic disorder in eukaryotic proteins. Cell Mol. Life Sci. 2017, 74, 3163–3174. [Google Scholar] [CrossRef]
- Nicaise, V. Crop immunity against viruses: Outcomes and future challenges. Front. Plant Sci. 2014, 5, 660. [Google Scholar] [CrossRef]
- Scholthof, K.-B.G.; Adkins, S.; Czosnek, H.; Palukaitis, P.; Jacquot, E.; Hohn, T.; Hohn, B.; Saunders, K.; Candresse, T.; Ahlquist, P.; et al. Top 10 plant viruses in molecular plant pathology. Mol. Plant Pathol. 2011, 12, 938–954. [Google Scholar] [CrossRef]
- Moury, B.; Desbiez, C. Host range evolution of potyviruses: A global phylogenetic analysis. Viruses 2020, 12, 111. [Google Scholar] [CrossRef]
- Tokuriki, N.; Tawfik, D.S. Protein dynamism and evolvability. Science 2009, 324, 203–207. [Google Scholar] [CrossRef]
- Jitobaom, K.; Phakaratsakul, S.; Sirihongthong, T.; Chotewutmontri, S.; Suriyaphol, P.; Suptawiwat, O.; Auewarakul, P. Codon usage similarity between viral and some host genes suggests a codon-specific translational regulation. Heliyon 2020, 6, e03915. [Google Scholar] [CrossRef]
- Kumar, N.; Kaushik, R.; Tennakoon, C.; Uversky, V.N.; Longhi, S.; Zhang, K.Y.J.; Bhatia, S. Insights into the evolutionary forces that shape the codon usage in the viral genome segments encoding intrinsically disordered protein regions. Brief Bioinform. 2021, 22, bbab145. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.-H.; Dang, Y.; Zhou, Z.; Wu, C.; Zhao, F.; Sachs, M.S.; Liu, Y. Codon Usage Influences the Local Rate of Translation Elongation to Regulate Co-translational Protein Folding. Mol. Cell. 2015, 59, 744–754. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Yu, C.-H.; Zhao, F.; Dang, Y.; Wu, C.; Xie, P.; Sachs, M.; Liu, Y. ERF1 mediates codon usage effects on mRNA translation efficiency through premature termination at rare codons. Nucleic Acids Res. 2019, 47, 9243–9258. [Google Scholar] [CrossRef] [PubMed]
- Mitarai, N.; Sneppen, K.; Pedersen, S. Ribosome Collisions and Translation Efficiency: Optimization by Codon Usage and mRNA Destabilization. J. Mol. Biol. 2008, 382, 236–245. [Google Scholar] [CrossRef] [PubMed]
- Pechmann, S.; Frydman, J. Interplay between chaperones and protein disorder promotes the evolution of protein networks. PLoS Comput. Biol. 2014, 10, e1003674. [Google Scholar] [CrossRef] [PubMed]
- Tomaszewski, T.; DeVries, R.S.; Dong, M.; Bhatia, G.; Norsworthy, M.D.; Zheng, X.; Caetano-Anollés, G. New Pathways of Mutational Change in SARS-CoV-2 Proteomes Involve Regions of Intrinsic Disorder Important for Virus Replication and Release. Evol. Bioinforma. 2020, 16, 1176934320965149. [Google Scholar] [CrossRef]

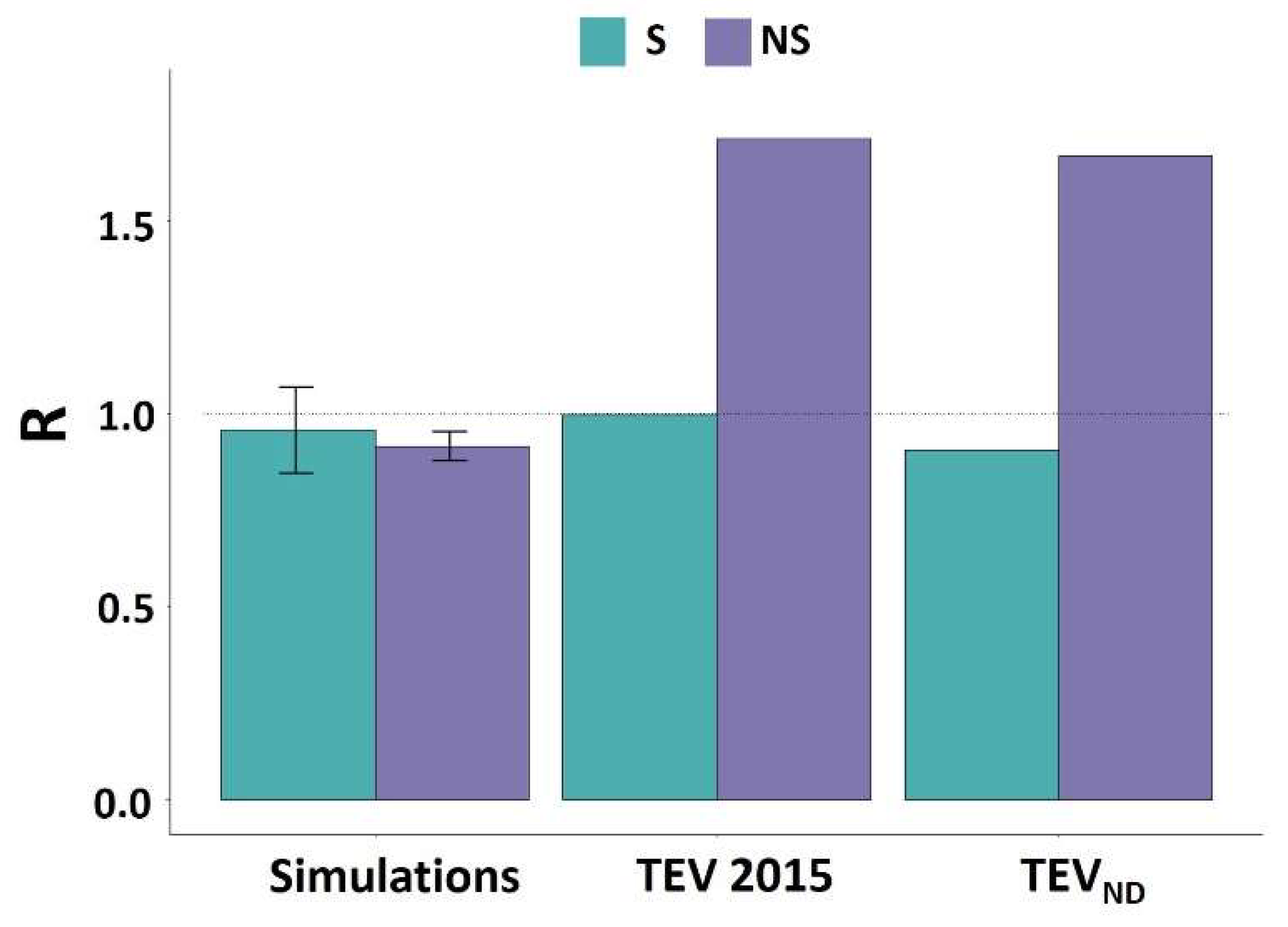
| S | NS | |
| TEV 2015 | 0.63 | 0.19 |
| TEVND | 0.94 | 0.16 |
| Simulations | 1 | 0.98 |
| S | NS | |
| PVY 2015 | 0.93 | 0.12 |
| PVY 2017 | 0.78 | 0.01 |
| PVYND | 0.96 | 0.35 |
| Simulations | 0.96 | 0.97 |
| S | NS | |
| TuMVND | 0.95 | 0.09 |
| Simulations | 0.92 | 0.98 |
| Simulations | TEV 2015 | TEVND | PVY 2015 | PVY 2017 | PVYND | TuMVND |
|---|---|---|---|---|---|---|
| A | 0.014 | 2.10 × 10−6 | 0.52 | 0.04 | 0.0005 | 3.10 × 10−8 |
| B | 0.026 | 1.10 × 10−5 | 0.46 | 0.03 | 0.0001 | 5.10 × 10−8 |
| C | 0.018 | 5.10 × 10−6 | 0.33 | 0.01 | 8.10-6 | 2.10 × 10−8 |
| D | 0.028 | 2.10 × 10−5 | 0.27 | 0.0049 | 1.10-7 | 6.10 × 10−9 |
| A | IDRs | ORs | B | IDRs | ORs | C | IDRs | ORs |
|---|---|---|---|---|---|---|---|---|
| PVY 2015 | abc | ab | TEV 2015 | ab | ab | TuMVND | a | a |
| PVY 2017 | abc | a | TEVND | b | b | Sim A | a | b |
| PVYND | ac | a | Sim A | a | a | Sim B | a | b |
| Sim A | bc | c | Sim B | a | a | Sim C | a | b |
| Sim B | bc | c | Sim C | a | a | Sim D | a | b |
| Sim C | b | bc | Sim D | ab | a | |||
| Sim D | bc | c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lafforgue, G.; Michon, T.; Charon, J. Analysis of the Contribution of Intrinsic Disorder in Shaping Potyvirus Genetic Diversity. Viruses 2022, 14, 1959. https://doi.org/10.3390/v14091959
Lafforgue G, Michon T, Charon J. Analysis of the Contribution of Intrinsic Disorder in Shaping Potyvirus Genetic Diversity. Viruses. 2022; 14(9):1959. https://doi.org/10.3390/v14091959
Chicago/Turabian StyleLafforgue, Guillaume, Thierry Michon, and Justine Charon. 2022. "Analysis of the Contribution of Intrinsic Disorder in Shaping Potyvirus Genetic Diversity" Viruses 14, no. 9: 1959. https://doi.org/10.3390/v14091959





