Modeling HPV-Associated Disease and Cancer Using the Cottontail Rabbit Papillomavirus
Abstract
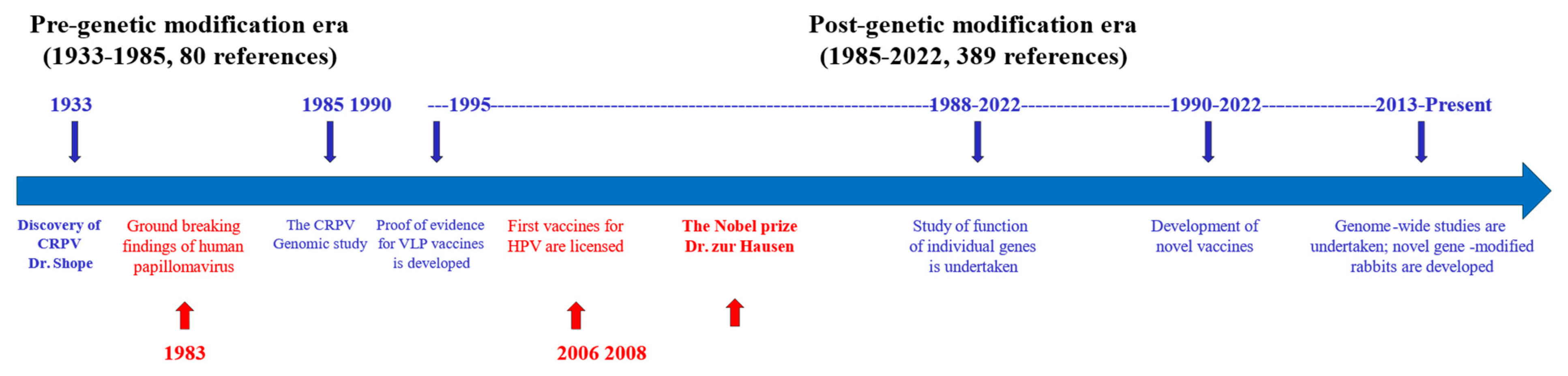
1. Introduction
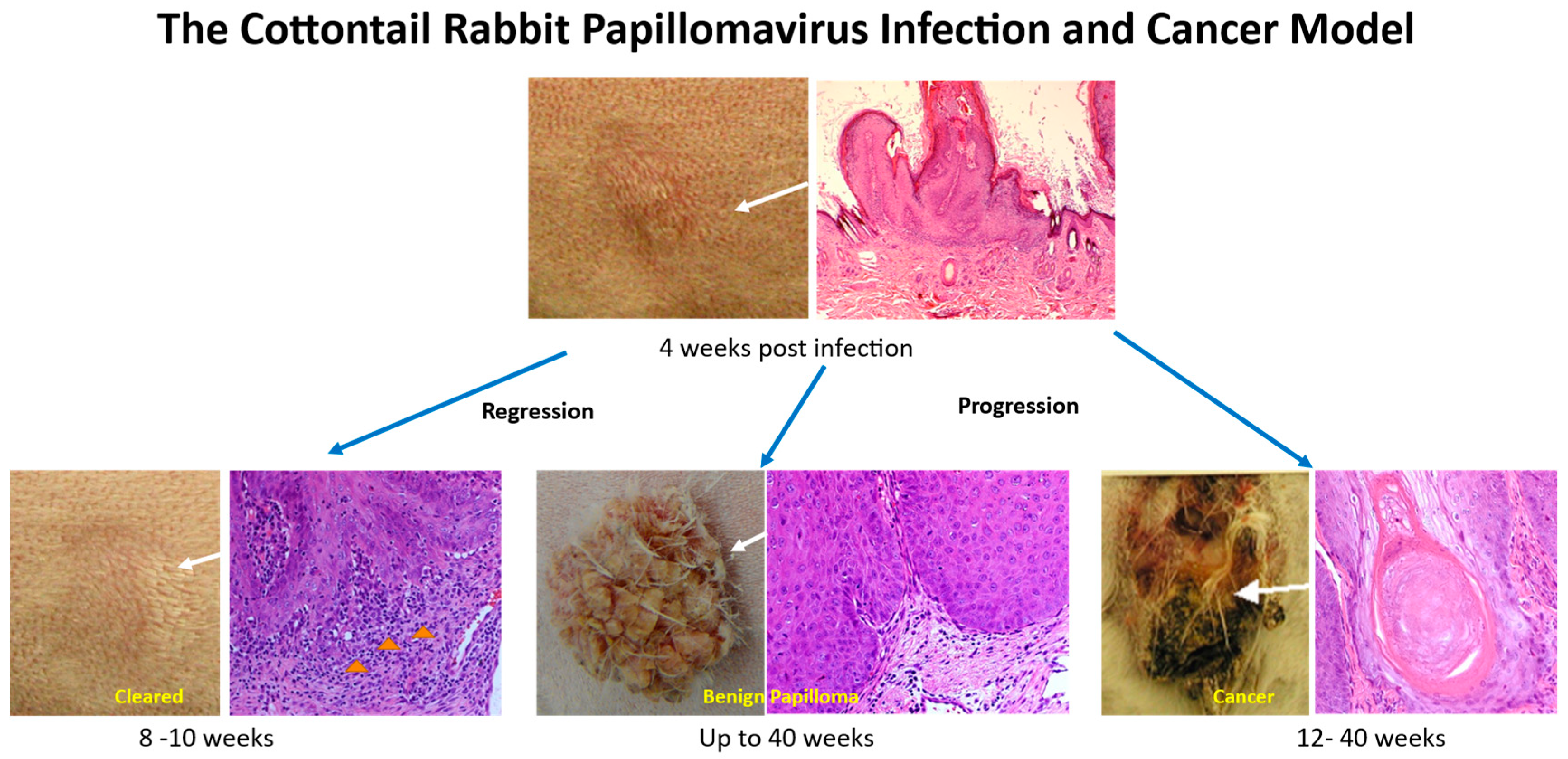
2. Cottontail Rabbit Papillomavirus (CRPV)-Associated Pathogenesis
2.1. Increased Viral Infection and Tumor Growth Using a Pre-Wounding Strategy
2.2. The Use of Mutant CRPV Genomes to Understand the Viral Life Cycle In Vivo
2.2.1. Early Genes Play a Crucial Role in Viral Life Cycle and Tumor Growth
2.2.2. Synonymous Codon Optimization Increases Oncogenicity and Immunogenicity of the Virus
2.2.3. Early Gene E6 Is Important for Tumor Regression
3. Genetic Analyses of Changes during CRPV Infections
3.1. Host Changes during Viral Infection
3.2. In Situ Analysis of Tissues at Different Disease Stages
4. Rabbits for Studying Viral–Host Interactions during Tumor Progression
4.1. Inbred and Outbred Rabbits
4.2. Transgenic Rabbits
4.3. Novel Genetically Modified Rabbits
5. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roden, R.B.S.; Stern, P.L. Opportunities and challenges for human papillomavirus vaccination in cancer. Nat. Rev. Cancer 2018, 18, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J. Host control of human papillomavirus infection and disease. Best Pract. Res. Clin. Obstet. Gynaecol. 2018, 47, 27–41. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D.; Budgeon, L.R.; Cladel, N.M.; Hu, J. Recent advances in preclinical model systems for papillomaviruses. Virus Res. 2017, 231, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Shope, R.E.; Hurst, E.W. Infectious papillomatosis of rabbits; with a note on the histopathology. J. Exp. Med. 1933, 58, 607–624. [Google Scholar] [CrossRef]
- Escudero Duch, C.; Williams, R.A.; Timm, R.M.; Perez-Tris, J.; Benitez, L. A Century of Shope Papillomavirus in Museum Rabbit Specimens. PLoS ONE 2015, 10, e0132172. [Google Scholar] [CrossRef]
- Brandsma, J.L. The cottontail rabbit papillomavirus model of high-risk HPV-induced disease. Methods Mol. Med. 2005, 119, 217–235. [Google Scholar]
- Breitburd, F.; Salmon, J.; Orth, G. The rabbit viral skin papillomas and carcinomas: A model for the immunogenetics of HPV-associated carcinogenesis. Clin. Dermatol. 1997, 15, 237–247. [Google Scholar] [CrossRef]
- Cladel, N.M.; Peng, X.; Christensen, N.; Hu, J. The rabbit papillomavirus model: A valuable tool to study viral-host interactions. Philos. Trans. R. Soc. B 2019, 374, 20180294. [Google Scholar] [CrossRef]
- Christensen, N.D.; Pickel, M.D.; Budgeon, L.R.; Kreider, J.W. In vivo anti-papillomavirus activity of nucleoside analogues including cidofovir on CRPV-induced rabbit papillomas. Antivir. Res. 2000, 48, 131–142. [Google Scholar] [CrossRef]
- Morse, M.A.; Balogh, K.K.; Brendle, S.A.; Campbell, C.A.; Chen, M.X.; Furze, R.C.; Harada, I.L.; Holyer, I.D.; Kumar, U.; Lee, K.; et al. BET bromodomain inhibitors show anti-papillomavirus activity in vitro and block CRPV wart growth in vivo. Antivir. Res. 2018, 154, 158–165. [Google Scholar] [CrossRef]
- Bolhassani, A.; Mohit, E.; Rafati, S. Different spectra of therapeutic vaccine development against HPV infections. Hum. Vaccin. 2009, 5, 671–689. [Google Scholar] [CrossRef] [PubMed]
- Fausch, S.C.; Da Silva, D.M.; Eiben, G.L.; Le Poole, I.C.; Kast, W.M. HPV protein/peptide vaccines: From animal models to clinical trials. Front. Biosci. 2003, 8, s81–s91. [Google Scholar] [PubMed]
- Amella, C.A.; Lofgren, L.A.; Ronn, A.M.; Nouri, M.; Shikowitz, M.J.; Steinberg, B.M. Latent infection induced with cottontail rabbit papillomavirus. A model for human papillomavirus latency. Am. J. Pathol. 1994, 144, 1167–1171. [Google Scholar] [PubMed]
- Duan, J.; Paris, W.; De Marte, J.; Roopchand, D.; Fleet, T.; Cordingley, M.G. Topical effects of cidofovir on cutaneous rabbit warts: Treatment regimen and inoculum dependence. Antivir. Res. 2000, 46, 135–144. [Google Scholar] [CrossRef]
- Christensen, N.D.; Cladel, N.M.; Hu, J.; Balogh, K.K. Formulation of cidofovir improves the anti-papillomaviral activity of topical treatments in the CRPV/rabbit model. Antivir. Res. 2014, 108, 148–155. [Google Scholar] [CrossRef]
- Breitburd, F.; Kirnbauer, R.; Hubbert, N.L.; Nonnenmacher, B.; Trin-Dinh-Desmarquet, C.; Orth, G.; Schiller, J.T.; Lowy, D.R. Immunization with viruslike particles from cottontail rabbit papillomavirus (CRPV) can protect against experimental CRPV infection. J. Virol. 1995, 69, 3959–3963. [Google Scholar] [CrossRef]
- Christensen, N.D.; Reed, C.A.; Cladel, N.M.; Han, R.; Kreider, J.W. Immunization with viruslike particles induces long-term protection of rabbits against challenge with cottontail rabbit papillomavirus. J. Virol. 1996, 70, 960–965. [Google Scholar] [CrossRef]
- Giri, I.; Danos, O.; Yaniv, M. Genomic structure of the cottontail rabbit (Shope) papillomavirus. Proc. Natl. Acad. Sci. USA 1985, 82, 1580–1584. [Google Scholar] [CrossRef]
- Brandsma, J.L.; Xiao, W. Some human papillomavirus (HPV) E7 DNA sequences can replace wild-type E7 sequences in cottontail rabbit papillomavirus (CRPV) to induce papillomas in rabbits. Abstract book. In Proceedings of the 14th International Papillomavirus Conference, Abstract book. Quebec City, QC, Canada, 23–28 July 1995; p. 92. [Google Scholar]
- Brandsma, J.L.; Yang, Z.-H.; Barthold, S.W.; Johnson, E.A. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc. Natl. Acad. Sci. USA 1991, 88, 4816–4820. [Google Scholar] [CrossRef]
- Wu, X.; Xiao, W.; Brandsma, J. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J. Virol. 1994, 68, 6097–6102. [Google Scholar] [CrossRef]
- Jeckel, S.; Loetzsch, E.; Huber, E.; Stubenrauch, F.; Iftner, T. Identification of the E9/E2C cDNA and functional characterization of the gene product reveal a new repressor of transcription and replication in cottontail rabbit papillomavirus. J. Virol. 2003, 77, 8736–8744. [Google Scholar] [CrossRef] [PubMed]
- Jeckel, S.; Huber, E.; Stubenrauch, F.; Iftner, T. A transactivator function of cottontail rabbit papillomavirus e2 is essential for tumor induction in rabbits. J. Virol. 2002, 76, 11209–11215. [Google Scholar] [CrossRef] [PubMed]
- Probst, S.; Notz, E.; Wolff, M.; Buehlmann, J.; Stubenrauch, F.; Iftner, T. A recombinant cottontail rabbit papillomavirus genome for ectopic expression of genes in cells infected with virus in vivo. J. Virol. Methods 2013, 187, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Hu, J.; Balogh, K.K.; Christensen, N.D. CRPV genomes with synonymous codon optimizations in the CRPV E7 gene show phenotypic differences in growth and altered immunity upon E7 vaccination. PLoS ONE 2008, 3, e2947. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cladel, N.M.; Budgeon, L.; Balogh, K.K.; Christensen, N.D. Papillomavirus DNA complementation in vivo. Virus Res. 2009, 144, 117–122. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.M.; Balogh, K.; Budgeon, L.; Christensen, N.D. Impact of genetic changes to the CRPV genome and their application to the study of pathogenesis in vivo. Virology 2007, 358, 384–390. [Google Scholar] [CrossRef][Green Version]
- Egawa, N.; Egawa, K.; Griffin, H.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef]
- Brandsma, J.L.; Shylankevich, M.; Su, Y.; Roberts, A.; Rose, J.K.; Zelterman, D.; Buonocore, L. Vesicular stomatitis virus-based therapeutic vaccination targeted to the E1, E2, E6, and E7 proteins of cottontail rabbit papillomavirus. J. Virol. 2007, 81, 5749–5758. [Google Scholar] [CrossRef]
- Han, R.; Reed, C.A.; Cladel, N.M.; Christensen, N.D. Immunization of rabbits with cottontail rabbit papillomavirus E1 and E2 genes: Protective immunity induced by gene gun-mediated intracutaneous delivery but not by intramuscular injection. Vaccine 2000, 18, 2937–2944. [Google Scholar] [CrossRef]
- Schneider, M.; Yigitliler, A.; Stubenrauch, F.; Iftner, T. Cottontail Rabbit Papillomavirus E1 and E2 Proteins Mutually Influence Their Subcellular Localizations. J. Virol. 2018, 92, E00704–E00718. [Google Scholar] [CrossRef]
- Christensen, N.D.; Han, R.; Cladel, N.M.; Pickel, M.D. Combination treatment with intralesional cidofovir and viral-DNA vaccination cures large cottontail rabbit papillomavirus-induced papillomas and reduces recurrences. Antimicrob. Agents Chemother. 2001, 45, 1201–1209. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hu, J.; Han, R.; Cladel, N.M.; Pickel, M.D.; Christensen, N.D. Intracutaneous DNA vaccination with the E8 gene of cottontail rabbit papillomavirus induces protective immunity against virus challenge in rabbits. J. Virol. 2002, 76, 6453–6459. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cladel, N.M.; Wang, Z.; Han, R.; Pickel, M.D.; Christensen, N.D. GM-CSF enhances protective immunity to cottontail rabbit papillomavirus E8 genetic vaccination in rabbits. Vaccine 2004, 22, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cladel, N.M.; Budgeon, L.R.; Reed, C.A.; Pickel, M.D.; Christensen, N.D. Protective cell-mediated immunity by DNA vaccination against Papillomavirus L1 capsid protein in the Cottontail Rabbit Papillomavirus model. Viral Immunol. 2006, 19, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Embers, M.E.; Budgeon, L.R.; Pickel, M.; Christensen, N.D. Protective immunity to rabbit oral and cutaneous papillomaviruses by immunization with short peptides of l2, the minor capsid protein. J. Virol. 2002, 76, 9798–9805. [Google Scholar] [CrossRef] [PubMed]
- Palmer, K.E.; Benko, A.; Doucette, S.A.; Cameron, T.I.; Foster, T.; Hanley, K.M.; McCormick, A.A.; McCulloch, M.; Pogue, G.P.; Smith, M.L.; et al. Protection of rabbits against cutaneous papillomavirus infection using recombinant tobacco mosaic virus containing L2 capsid epitopes. Vaccine 2006, 24, 5516–5525. [Google Scholar] [CrossRef]
- Lin, Y.L.; Borenstein, L.A.; Selvakumar, R.; Ahmed, R.; Wettstein, F.O. Effective vaccination against papilloma development by immunization with L1 or L2 structural protein of cottontail rabbit papillomavirus. Virology 1992, 187, 612–619. [Google Scholar] [CrossRef]
- Kalnin, K.; Tibbitts, T.; Yan, Y.; Stegalkina, S.; Shen, L.; Costa, V.; Sabharwal, R.; Anderson, S.F.; Day, P.M.; Christensen, N.; et al. Low doses of flagellin-L2 multimer vaccines protect against challenge with diverse papillomavirus genotypes. Vaccine 2014, 32, 3540–3547. [Google Scholar] [CrossRef]
- Vambutas, A.; DeVoti, J.; Nouri, M.; Drijfhout, J.W.; Lipford, G.B.; Bonagura, V.R.; Van der Burg, S.H.; Melief, C.J. Therapeutic vaccination with papillomavirus E6 and E7 long peptides results in the control of both established virus-induced lesions and latently infected sites in a pre-clinical cottontail rabbit papillomavirus model. Vaccine 2005, 23, 5271–5280. [Google Scholar] [CrossRef]
- Hu, J.; Budgeon, L.R.; Balogh, K.K.; Peng, X.; Cladel, N.M.; Christensen, N.D. Long-peptide therapeutic vaccination against CRPV-induced papillomas in HLA-A2.1 transgenic rabbits. Trials Vaccinol. 2014, 3, 134–142. [Google Scholar] [CrossRef]
- Mejia, A.F.; Culp, T.D.; Cladel, N.M.; Balogh, K.K.; Budgeon, L.R.; Buck, C.B.; Christensen, N.D. Preclinical Model To Test Human Papillomavirus Virus (HPV) Capsid Vaccines In Vivo Using Infectious HPV/Cottontail Rabbit Papillomavirus Chimeric Papillomavirus Particles. J. Virol. 2006, 80, 12393–12397. [Google Scholar] [CrossRef] [PubMed]
- Gambhira, R.; Jagu, S.; Karanam, B.; Gravitt, P.E.; Culp, T.D.; Christensen, N.D.; Roden, R.B. Protection of rabbits against challenge with rabbit papillomaviruses by immunization with the N terminus of human papillomavirus type 16 minor capsid antigen L2. J. Virol. 2007, 81, 11585–11592. [Google Scholar] [CrossRef] [PubMed]
- Jagu, S.; Karanam, B.; Wang, J.W.; Zayed, H.; Weghofer, M.; Brendle, S.A.; Balogh, K.K.; Tossi, K.P.; Roden, R.B.; Christensen, N.D. Durable immunity to oncogenic human papillomaviruses elicited by adjuvanted recombinant Adeno-associated virus-like particle immunogen displaying L2 17–36 epitopes. Vaccine 2015, 33, 5553–5563. [Google Scholar] [CrossRef]
- Olczak, P.; Matsui, K.; Wong, M.; Alvarez, J.; Lambert, P.; Christensen, N.D.; Hu, J.; Huber, B.; Kirnbauer, R.; Wang, J.W.; et al. RG2-VLP: A Vaccine Designed to Broadly Protect against Anogenital and Skin Human Papillomaviruses Causing Human Cancer. J. Virol. 2022, 96, e00566-22. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Griffith, J.W.; Han, R.; Lang, C.M.; Kreider, J.W. Development of keratoacanthomas and squamous cell carcinomas in transgenic rabbits with targeted expression of EJras oncogene in epidermis. Am. J. Pathol. 1999, 155, 315–324. [Google Scholar] [CrossRef]
- Peng, X.; Lang, C.M.; Kreider, J.W. Immortalization of inbred rabbit keratinocytes from a Shope papilloma and tumorigenic transformation of the cells by EJ-ras. Cancer Lett. 1996, 108, 101–109. [Google Scholar] [CrossRef]
- Peng, X.; Olson, R.O.; Christian, C.B.; Lang, C.M.; Kreider, J.W. Papillomas and carcinomas in transgenic rabbits carrying EJ-ras DNA and cottontail rabbit papillomavirus DNA. J. Virol. 1993, 67, 1698–1701. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.M.; Christensen, N.D. Increased immunity to cottontail rabbit papillomavirus infection in EIII/JC inbred rabbits after vaccination with a mutant E6 that correlates with spontaneous regression. Viral Immunol. 2007, 20, 320–325. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.M.; Pickel, M.D.; Christensen, N.D. Amino acid residues in the carboxy-terminal region of cottontail rabbit papillomavirus E6 influence spontaneous regression of cutaneous papillomas. J. Virol. 2002, 76, 11801–11808. [Google Scholar] [CrossRef]
- Hu, J.; Peng, X.; Schell, T.D.; Budgeon, L.R.; Cladel, N.M.; Christensen, N.D. An HLA-A2.1-transgenic rabbit model to study immunity to papillomavirus infection. J. Immunol. 2006, 177, 8037–8045. [Google Scholar] [CrossRef]
- Hu, J.; Schell, T.D.; Peng, X.; Cladel, N.M.; Balogh, K.K.; Christensen, N.D. Using HLA-A2.1 Transgenic Rabbit Model to Screen and Characterize New HLA-A2.1 Restricted Epitope DNA Vaccines. J. Vaccines Vaccin. 2010, 1. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.; Nonnenmacher, M.; Caze, S.; Flamant, P.; Croissant, O.; Orth, G.; Breitburd, F. Variation in the nucleotide sequence of cottontail rabbit papillomavirus a and b subtypes affects wart regression and malignant transformation and level of viral replication in domestic rabbits. J. Virol. 2000, 74, 10766–10777. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Breitburd, F.; Marche, P.N.; Orth, G. Linkage of Regression and Malignant Conversion of Rabbit Viral Papillomas to MHC Class II Genes. Nature 1992, 356, 66–68. [Google Scholar] [PubMed]
- Xu, J.; Zhang, J.; Yang, D.; Song, J.; Pallas, B.; Zhang, C.; Hu, J.; Peng, X.; Christensen, N.D.; Han, R.; et al. Gene Editing in Rabbits: Unique Opportunities for Translational Biomedical Research. Front. Genet. 2021, 12, 642444. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Budgeon, L.R.; Cladel, N.M.; Culp, T.D.; Balogh, K.K.; Christensen, N.D. Detection of L1, infectious virions and anti-L1 antibody in domestic rabbits infected with cottontail rabbit papillomavirus. J. Gen. Virol. 2007, 88, 3286–3293. [Google Scholar] [CrossRef]
- Han, R.; Cladel, N.M.; Reed, C.A.; Christensen, N.D. Characterization of transformation function of cottontail rabbit papillomavirus E5 and E8 genes. Virology 1998, 251, 253–263. [Google Scholar] [CrossRef][Green Version]
- Nonnenmacher, M.; Salmon, J.; Jacob, Y.; Orth, G.; Breitburd, F. Cottontail rabbit papillomavirus E8 protein is essential for wart formation and provides new insights into viral pathogenesis. J. Virol. 2006, 80, 4890–4900. [Google Scholar] [CrossRef]
- Du, M.; Fan, X.; Hanada, T.; Gao, H.; Lutchman, M.; Brandsma, J.L.; Chishti, A.H.; Chen, J.J. Association of cottontail rabbit papillomavirus E6 oncoproteins with the hDlg/SAP97 tumor suppressor. J. Cell. Biochem. 2005, 94, 1038–1045. [Google Scholar] [CrossRef]
- Meyers, C.; Harry, J.; Lin, Y.-L.; Wettstein, F.O. Identification of three transforming proteins encoded by cottontail rabbit papillomavirus. J. Virol. 1992, 66, 1655–1664. [Google Scholar] [CrossRef]
- Hu, J.; Peng, X.; Cladel, N.M.; Pickel, M.D.; Christensen, N.D. Large cutaneous rabbit papillomas that persist during cyclosporin A treatment can regress spontaneously after cessation of immunosuppression. J. Gen. Virol. 2005, 86, 55–63. [Google Scholar] [CrossRef]
- Nasseri, M.; Meyers, C.; Wettstein, F.O. Genetic analysis of CRPV pathogenesis: The L1 open reading frame is dispensable for cellular transformation but is required for papilloma formation. Virology 1989, 170, 321–325. [Google Scholar] [CrossRef]
- Defeo-Jones, D.; Vuocolo, G.A.; Haskell, K.M.; Hanobik, M.G.; Kiefer, D.M.; McAvoy, E.M.; Ivey-Hoyle, M.; Brandsma, J.L.; Oliff, A.; Jones, R.E. Papillomavirus E7 protein binding to the retinoblastoma protein is not required for viral induction of warts. J. Virol. 1993, 67, 716–725. [Google Scholar] [CrossRef] [PubMed]
- Cladel, N.M.; Hu, J.; Balogh, K.K.; Christensen, N.D. Differences in methodology, but not differences in viral strain, account for variable experimental outcomes in laboratories utilizing the cottontail rabbit papillomavirus model. J. Virol. Methods 2010, 165, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Salmon, J.; Ramoz, N.; Cassonnet, P.; Orth, G.; Breitburd, F. A cottontail rabbit papillomavirus strain (CRPVb) with strikingly divergent E6 and E7 oncoproteins: An insight in the evolution of papillomaviruses. Virology 1997, 235, 228–234. [Google Scholar] [CrossRef][Green Version]
- Reuter, J.D.; Gomez, D.; Brandsma, J.L.; Rose, J.K.; Roberts, A. Optimization of cottontail rabbit papilloma virus challenge technique. J. Virol. Methods 2001, 98, 127–134. [Google Scholar] [CrossRef]
- Kreider, J.W.; Cladel, N.M.; Patrick, S.D.; Welsh, P.A.; DiAngelo, S.L.; Bower, J.M.; Christensen, N.D. High efficiency induction of papillomas in vivo using recombinant cottontail rabbit papillomavirus DNA. J. Virol. Methods 1995, 55, 233–244. [Google Scholar] [CrossRef]
- Brandsma, J.L.; Xiao, W. Infectious virus replication in papillomas induced by molecularly cloned cottontail rabbit papillomavirus DNA. J. Virol. 1993, 67, 567–571. [Google Scholar] [CrossRef]
- Cladel, N.M.; Hu, J.; Balogh, K.; Mejia, A.; Christensen, N.D. Wounding prior to challenge substantially improves infectivity of cottontail rabbit papillomavirus and allows for standardization of infection. J. Virol. Methods 2008, 148, 34–39. [Google Scholar] [CrossRef][Green Version]
- Peh, W.L.; Middleton, K.; Christensen, N.; Nicholls, P.; Egawa, K.; Sotlar, K.; Brandsma, J.; Percival, A.; Lewis, J.; Liu, W.J.; et al. Life cycle heterogeneity in animal models of human papillomavirus-associated disease. J. Virol. 2002, 76, 10401–10416. [Google Scholar] [CrossRef]
- Peh, W.L.; Brandsma, J.L.; Christensen, N.D.; Cladel, N.M.; Wu, X.; Doorbar, J. The viral E4 protein is required for the completion of the cottontail rabbit papillomavirus productive cycle in vivo. J. Virol. 2004, 78, 2142–2151. [Google Scholar] [CrossRef]
- Cladel, N.M.; Budgeon, L.R.; Hu, J.; Balogh, K.K.; Christensen, N.D. Synonymous codon changes in the oncogenes of the cottontail rabbit papillomavirus lead to increased oncogenicity and immunogenicity of the virus. Virology 2013, 438, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Aleem, S.A.; Abdelwahab, S.; Am-Sherief, H.; Sayed, A. Cellular and physiological upregulation of inducible nitric oxide synthase, arginase, and inducible cyclooxygenase in wound healing. J. Cell. Physiol. 2019, 234, 23618–23632. [Google Scholar] [CrossRef] [PubMed]
- Theilgaard-Monch, K.; Knudsen, S.; Follin, P.; Borregaard, N. The transcriptional activation program of human neutrophils in skin lesions supports their important role in wound healing. J. Immunol. 2004, 172, 7684–7693. [Google Scholar] [CrossRef] [PubMed]
- Sistigu, A.; Di Modugno, F.; Manic, G.; Nistico, P. Deciphering the loop of epithelial-mesenchymal transition, inflammatory cytokines and cancer immunoediting. Cytokine Growth Factor Rev. 2017, 36, 67–77. [Google Scholar] [CrossRef]
- Woodworth, C.D. HPV innate immunity. Front. Biosci. 2002, 7, d2058–d2071. [Google Scholar] [CrossRef]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Cerqueira, C.; Samperio Ventayol, P.; Vogeley, C.; Schelhaas, M. Kallikrein-8 Proteolytically Processes Human Papillomaviruses in the Extracellular Space To Facilitate Entry into Host Cells. J. Virol. 2015, 89, 7038–7052. [Google Scholar] [CrossRef]
- Hung, V.C.; Lee, J.Y.; Zitelli, J.A.; Hebda, P.A. Topical tretinoin and epithelial wound healing. Arch. Dermatol. 1989, 125, 65–69. [Google Scholar] [CrossRef]
- Szondi, D.C.; Wong, J.K.; Vardy, L.A.; Cruickshank, S.M. Arginase Signalling as a Key Player in Chronic Wound Pathophysiology and Healing. Front. Mol. Biosci. 2021, 8, 773866. [Google Scholar] [CrossRef]
- Crompton, R.A.; Williams, H.; Campbell, L.; Hui Kheng, L.; Saville, C.; Ansell, D.M.; Reid, A.; Wong, J.; Vardy, L.A.; Hardman, M.J.; et al. An Epidermal-Specific Role for Arginase1 during Cutaneous Wound Repair. J. Investig. Dermatol. 2022, 142, 1206–1216.e8. [Google Scholar] [CrossRef]
- Cladel, N.M.; Jiang, P.; Li, J.J.; Peng, X.; Cooper, T.K.; Majerciak, V.; Balogh, K.K.; Meyer, T.J.; Brendle, S.A.; Budgeon, L.R.; et al. Papillomavirus can be transmitted through the blood and produce infections in blood recipients: Evidence from two animal models. Emerg. Microbes. Infect. 2019, 8, 1108–1121. [Google Scholar] [CrossRef] [PubMed]
- Bodaghi, S.; Wood, L.V.; Roby, G.; Ryder, C.; Steinberg, S.M.; Zheng, Z.M. Could human papillomaviruses be spread through blood? J. Clin. Microbiol. 2005, 43, 5428–5434. [Google Scholar] [CrossRef] [PubMed]
- Bounds, C.E.; Hu, J.; Cladel, N.M.; Balogh, K.; Christensen, N.D. Vaccine generated immunity targets an HPV16 E7 HLA-A2.1-restricted CD8(+) T cell epitope relocated to an early gene or a late gene of the cottontail rabbit papillomavirus (CRPV) genome in HLA-A2.1 transgenic rabbits. Vaccine 2011, 29, 1194–1200. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Leder, C.; Kleinschmidt, J.A.; Wiethe, C.; Muller, M. Enhancement of capsid gene expression: Preparing the human papillomavirus type 16 major structural gene L1 for DNA vaccination purposes. J. Virol. 2001, 75, 9201–9209. [Google Scholar] [CrossRef]
- Liu, W.J.; Gao, F.; Zhao, K.N.; Zhao, W.; Fernando, G.J.; Thomas, R.; Frazer, I.H. Codon modified human papillomavirus type 16 E7 DNA vaccine enhances cytotoxic T-lymphocyte induction and anti-tumour activity. Virology 2002, 301, 43–52. [Google Scholar] [CrossRef][Green Version]
- Schaeffer, A.J.; Nguyen, M.; Liem, A.; Lee, D.; Montagna, C.; Lambert, P.F.; Ried, T.; Difilippantonio, M.J. E6 and E7 oncoproteins induce distinct patterns of chromosomal aneuploidy in skin tumors from transgenic mice. Cancer Res. 2004, 64, 538–546. [Google Scholar] [CrossRef]
- Duensing, S.; Duensing, A.; Crum, C.P.; Munger, K. Human papillomavirus type 16 E7 oncoprotein-induced abnormal centrosome synthesis is an early event in the evolving malignant phenotype. Cancer Res. 2001, 61, 2356–2360. [Google Scholar]
- Nasseri, M.; Wettstein, F.O. Differences exist between viral transcripts in cottontail rabbit papillomavirus-induced benign and malignant tumors as well as non-virus-producing and virus-producing tumors. J. Virol. 1984, 51, 706–712. [Google Scholar] [CrossRef]
- Huber, E.; Vlasny, D.; Jeckel, S.; Stubenrauch, F.; Iftner, T. Gene profiling of cottontail rabbit papillomavirus-induced carcinomas identifies upregulated genes directly Involved in stroma invasion as shown by small interfering RNA-mediated gene silencing. J. Virol. 2004, 78, 7478–7489. [Google Scholar] [CrossRef]
- Probst-Hunczek, S.; Jager, G.; Schneider, M.; Notz, E.; Stubenrauch, F.; Iftner, T. RNA sequencing analysis identifies novel spliced transcripts but does not indicate quantitative or qualitative changes of viral transcripts during progression of cottontail rabbit papillomavirus-induced tumours. J. Gen. Virol. 2015, 96, 3083–3089. [Google Scholar] [CrossRef]
- Zhang, P.; Nouri, M.; Brandsma, J.L.; Iftner, T.; Steinberg, B.M. Induction of E6/E7 expression in cottontail rabbit papillomavirus latency following UV activation. Virology 1999, 263, 388–394. [Google Scholar] [CrossRef] [PubMed]
- zur Hausen, H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J. Natl. Cancer Inst. 2000, 92, 690–698. [Google Scholar]
- Doorbar, J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006, 110, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Tewari, K.S.; Monk, B.J. New strategies in advanced cervical cancer: From angiogenesis blockade to immunotherapy. Clin. Cancer Res. 2014, 20, 5349–5358. [Google Scholar] [CrossRef] [PubMed]
- Christensen, N.D.; Cladel, N.M.; Reed, C.A.; Han, R. Rabbit oral papillomavirus complete genome sequence and immunity following genital infection. Virology 2000, 269, 451–461. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Evans, C.A.; Gorman, L.R.; Ito, Y.; Weiser, R.S. Antitumor immunity in the Shope papilloma-carcinoma complex of rabbits. I. Papilloma regression induced by homologous and autologous tissue vaccines. JNCI 1962, 29, 277–285. [Google Scholar] [PubMed]
- Tagami, H. Regression phenomenon of numerous flat wart—An experiment on the nature of tumor immunity in man. Int. J. Dermatol. 1983, 22, 570–571. [Google Scholar] [CrossRef]
- Nicholls, P.K.; Stanley, M.A. The immunology of animal papillomaviruses. Vet. Immunol. Immunopathol. 2000, 73, 101–127. [Google Scholar] [CrossRef]
- Selvakumar, R.; Ahmed, R.; Wettstein, F.O. Tumor regression is associated with a specific immune response to the E2 protein of cottontail rabbit papillomavirus. Virology 1995, 208, 298–302. [Google Scholar] [CrossRef][Green Version]
- Okabayashi, M.; Angell, M.G.; Christensen, N.D.; Kreider, J.W. Morphometric analysis and identification of infiltrating leucocytes in regressing and progressing Shope rabbit papillomas. Int. J. Cancer 1991, 49, 919–923. [Google Scholar] [CrossRef]
- Okabayashi, M.; Pickel, M.D.; Budgeon, L.R.; Cladel, N.M.; Kreider, J.W. Podofilox-induced regression of Shope papillomas may be independent of host immunity. J. Investig. Dermatol. 1993, 101, 852–857. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Selvakumar, R.; Schmitt, A.; Iftner, T.; Ahmed, R.; Wettstein, F.O. Regression of papillomas induced by cottontail rabbit papillomavirus is associated with infiltration of CD8+ cells and persistence of viral DNA after regression. J. Virol. 1997, 71, 5540–5548. [Google Scholar] [CrossRef] [PubMed]
- Höpfl, R.M.; Christensen, N.D.; Angell, M.G.; Kreider, J.W. Skin test to assess immunity against cottontail rabbit papillomavirus antigens in rabbits with progressing papillomas or after papilloma regression. J. Investig. Dermatol. 1993, 101, 227–231. [Google Scholar] [CrossRef][Green Version]
- Hopfl, R.M.; Christensen, N.D.; Heim, K.; Kreider, J.W. Skin test reactivity to papilloma cells is long lasting in domestic rabbits after regression of cottontail rabbit papillomavirus induced papillomas. In Immunology of Human Papillomavirus; Stanley, M.A., Ed.; Plenum Press: New York, NY, USA; London, UK, 1994; p. 259. [Google Scholar]
- Hopfl, R.; Christensen, N.D.; Angell, M.G.; Kreider, J.W. Leukocyte proliferation in vitro against cottontail rabbit papillomavirus in rabbits with persisting papillomas/cancer or after regression. Arch. Dermatol. Res. 1995, 287, 652–658. [Google Scholar] [CrossRef]
- Hibma, M.H. The immune response to papillomavirus during infection persistence and regression. Open Virol. J. 2012, 6, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Coleman, N.; Birley, H.D.; Renton, A.M.; Hanna, N.F.; Ryait, B.K.; Byrne, M.; Taylor-Robinson, D.; Stanley, M.A. Immunological events in regressing genital warts. Am. J. Clin. Pathol. 1994, 102, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Cladel, N.M.; Budgeon, L.R.; Christensen, N.D. Characterization of three rabbit oral papillomavirus oncogenes. Virology 2004, 325, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Wilgenburg, B.J.; Budgeon, L.R.; Lang, C.M.; Griffith, J.W.; Christensen, N.D. Characterization of immune responses during regression of rabbit oral papillomavirus infections. Comp. Med. 2005, 55, 431–439. [Google Scholar]
- Maglennon, G.A.; McIntosh, P.; Doorbar, J. Persistence of viral DNA in the epithelial basal layer suggests a model for papillomavirus latency following immune regression. Virology 2011, 414, 153–163. [Google Scholar] [CrossRef]
- Muench, P.; Probst, S.; Schuetz, J.; Leiprecht, N.; Busch, M.; Wesselborg, S.; Stubenrauch, F.; Iftner, T. Cutaneous papillomavirus E6 proteins must interact with p300 and block p53-mediated apoptosis for cellular immortalization and tumorigenesis. Cancer Res. 2010, 70, 6913–6924. [Google Scholar] [CrossRef]
- Wurdak, M.; Schneider, M.; Iftner, T.; Stubenrauch, F. The contribution of SP100 to cottontail rabbit papillomavirus transcription and replication. J. Gen. Virol. 2018, 99, 344–354. [Google Scholar] [CrossRef] [PubMed]
- Zeltner, R.; Borenstein, L.A.; Wettstein, F.O.; Iftner, T. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J. Virol. 1994, 68, 3620–3630. [Google Scholar] [CrossRef] [PubMed]
- Biesaga, B.; Mucha-Malecka, A.; Janecka-Widla, A.; Kolodziej-Rzepa, M.; Szostek, S.; Slonina, D.; Kowalczyk, A.; Halaszka, K.; Przewoznik, M. Differences in the prognosis of HPV16-positive patients with squamous cell carcinoma of head and neck according to viral load and expression of P16. J. Cancer Res. Clin. Oncol. 2018, 144, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Jelen, M.M.; Chen, Z.; Kocjan, B.J.; Burt, F.J.; Chan, P.K.; Chouhy, D.; Combrinck, C.E.; Coutlee, F.; Estrade, C.; Ferenczy, A.; et al. Global genomic diversity of human papillomavirus 6 based on 724 isolates and 190 complete genome sequences. J. Virol. 2014, 88, 7307–7316. [Google Scholar] [CrossRef]
- Jelen, M.M.; Chen, Z.; Kocjan, B.J.; Hosnjak, L.; Burt, F.J.; Chan, P.K.; Chouhy, D.; Combrinck, C.E.; Estrade, C.; Fiander, A.; et al. Global Genomic Diversity of Human Papillomavirus 11 Based on 433 Isolates and 78 Complete Genome Sequences. J. Virol 2016, 90, 5503–5513. [Google Scholar] [CrossRef]
- Chen, L.; Qiu, X.; Zhang, N.; Wang, Y.; Wang, M.; Li, D.; Wang, L.; Du, Y. APOBEC-mediated genomic alterations link immunity and viral infection during human papillomavirus-driven cervical carcinogenesis. Biosci. Trends 2017, 11, 383–388. [Google Scholar] [CrossRef]
- Yang-Chun, F.; Sen-Yu, W.; Yuan, Z.; Yan-Chun, H. Genome-Wide Profiling of Human Papillomavirus DNA Integration into Human Genome and Its Influence on PD-L1 Expression in Chinese Uygur Cervical Cancer Women. J. Immunol. Res. 2020, 2020, 6284960. [Google Scholar] [CrossRef]
- von Knebel, D.M. New markers for cervical dysplasia to visualise the genomic chaos created by aberrant oncogenic papillomavirus infections. Eur. J. Cancer 2002, 38, 2229–2242. [Google Scholar] [CrossRef]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.L.; Zhao, Z.K.; Wang, L.; Li, B.; Li, G.B.; Dean, M.; Yu, Q.C.; Wang, Y.H.; Lin, X.X.; et al. Full-length single-cell RNA-seq applied to a viral human cancer: Applications to HPV expression and splicing analysis in HeLa S3 cells. Gigascience 2015, 4, s13742-015. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Argyris, P.P.; Slama, Z.M.; Ross, K.F.; Khammanivong, A.; Herzberg, M.C. Calprotectin and the Initiation and Progression of Head and Neck Cancer. J. Dent. Res. 2018, 97, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Khammanivong, A.; Sorenson, B.S.; Ross, K.F.; Dickerson, E.B.; Hasina, R.; Lingen, M.W.; Herzberg, M.C. Involvement of calprotectin (S100A8/A9) in molecular pathways associated with HNSCC. Oncotarget 2016, 7, 14029–14047. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Christensen, N.D.; Kreider, J.W. Neutralization of CRPV infectivity by monoclonal antibodies that identify conformational epitopes on intact virions. Virus Res. 1991, 21, 169–179. [Google Scholar] [CrossRef]
- Esteves, P.J.; Abrantes, J.; Baldauf, H.M.; BenMohamed, L.; Chen, Y.; Christensen, N.; Gonzalez-Gallego, J.; Giacani, L.; Hu, J.; Kaplan, G.; et al. The wide utility of rabbits as models of human diseases. Exp. Mol. Med. 2018, 50, 66. [Google Scholar] [CrossRef] [PubMed]
- Mehta, A.M.; Mooij, M.; Brankovic, I.; Ouburg, S.; Morre, S.A.; Jordanova, E.S. Cervical Carcinogenesis and Immune Response Gene Polymorphisms: A Review. J. Immunol. Res. 2017, 2017, 8913860. [Google Scholar] [CrossRef]
- Breitburd, F.; Ramoz, N.; Salmon, J.; Orth, G. HLA control in the progression of human papillomavirus infections. Sem. Cancer Biol. 1997, 7, 359. [Google Scholar] [CrossRef]
- Allen, M.; Kalantari, M.; Ylitalo, N.; Pettersson, B.; Hagmar, B.; Scheibenpflug, L.; Johansson, B.; Petterson, U.; Gyllensten, U. HLA DQ-DR haplotype and susceptibility to cervical carcinoma: Indications of increased risk for development of cervical carcinoma in individuals infected with HPV 18. Tissue Antigens 1996, 48, 32–37. [Google Scholar] [CrossRef]
- Beskow, A.H.; Josefsson, A.M.; Gyllensten, U.B. HLA class II alleles associated with infection by HPV16 in cervical cancer in situ. Int. J. Cancer 2001, 93, 817–822. [Google Scholar] [CrossRef]
- Lin, P.; Koutsky, L.A.; Critchlow, C.W.; Apple, R.J.; Hawes, S.E.; Hughes, J.P.; Toure, P.; Dembele, A.; Kiviat, N.B. HLA class II DR-DQ and increased risk of cervical cancer among Senegalese women. Cancer Epidemiol. Biomarkers Prev. 2001, 10, 1037–1045. [Google Scholar] [PubMed]
- Hu, J.; Peng, X.; Budgeon, L.R.; Cladel, N.M.; Balogh, K.K.; Christensen, N.D. Establishment of a Cottontail Rabbit Papillomavirus/HLA-A2.1 Transgenic Rabbit Model. J. Virol. 2007, 81, 7171–7177. [Google Scholar] [CrossRef] [PubMed]
- Yan, Q.; Zhang, Q.; Yang, H.; Zou, Q.; Tang, C.; Fan, N.; Lai, L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen. 2014, 3, 12. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, D.; Xu, J.; Chen, Y.E. Generation of Rabbit Models by Gene Editing Nucleases. Methods Mol. Biol. 2019, 1874, 327–345. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, D.; Ruan, J.; Zhang, J.; Chen, Y.E.; Xu, J. Production of immunodeficient rabbits by multiplex embryo transfer and multiplex gene targeting. Sci. Rep. 2017, 7, 12202. [Google Scholar] [CrossRef]
- Christensen, N.D.; Kreider, J.W.; Kan, N.C.; DiAngelo, S.L. The open reading frame L2 of cottontail rabbit papillomavirus contains antibody-inducing neutralizing epitopes. Virology 1991, 181, 572–579. [Google Scholar] [CrossRef]
- Peng, X.; Griffith, J.W.; Lang, C.M. Reinitiated expression of EJras transgene in targeted epidermal cells of transgenic rabbits by cottontail rabbit papillomavirus infection. Cancer Lett. 2001, 171, 193–200. [Google Scholar] [CrossRef]
- Zhao, K.N.; Liu, W.J.; Frazer, I.H. Codon usage bias and A+T content variation in human papillomavirus genomes. Virus Res. 2003, 98, 95–104. [Google Scholar] [CrossRef]
- Hu, J.; Cladel, N.; Balogh, K.; Christensen, N. Mucosally delivered peptides prime strong immunity in HLA-A2.1 transgenic rabbits. Vaccine 2010, 28, 3706–3713. [Google Scholar] [CrossRef]
- Chesne, P.; Adenot, P.G.; Viglietta, C.; Baratte, M.; Boulanger, L.; Renard, J.P. Cloned rabbits produced by nuclear transfer from adult somatic cells. Nat. Biotechnol. 2002, 20, 366–369. [Google Scholar] [CrossRef]
- Wolf, D.P.; Mitalipov, S.; Norgren, R.B., Jr. Nuclear transfer technology in mammalian cloning. Arch. Med. Res. 2001, 32, 609–613. [Google Scholar] [CrossRef]
- Flisikowska, T.; Thorey, I.S.; Offner, S.; Ros, F.; Lifke, V.; Zeitler, B.; Rottmann, O.; Vincent, A.; Zhang, L.; Jenkins, S.; et al. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS ONE 2011, 6, e21045. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Yang, D.; Xu, J.; Zhu, T.; Chen, Y.E.; Zhang, J. RS-1 enhances CRISPR/Cas9- and TALEN-mediated knock-in efficiency. Nat. Commun. 2016, 7, 10548. [Google Scholar] [CrossRef]
- Bosticardo, M.; Yamazaki, Y.; Cowan, J.; Giardino, G.; Corsino, C.; Scalia, G.; Prencipe, R.; Ruffner, M.; Hill, D.A.; Sakovich, I.; et al. Heterozygous FOXN1 Variants Cause Low TRECs and Severe T Cell Lymphopenia, Revealing a Crucial Role of FOXN1 in Supporting Early Thymopoiesis. Am. J. Hum. Genet. 2019, 105, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Wang, G.; Hoenerhoff, M.J.; Ruan, J.; Yang, D.; Zhang, J.; Yang, J.; Lester, P.A.; Sigler, R.; Bradley, M.; et al. Bacterial and Pneumocystis Infections in the Lungs of Gene-Knockout Rabbits with Severe Combined Immunodeficiency. Front. Immunol. 2018, 9, 429. [Google Scholar] [CrossRef]
- Vonsky, M.; Shabaeva, M.; Runov, A.; Lebedeva, N.; Chowdhury, S.; Palefsky, J.M.; Isaguliants, M. Carcinogenesis Associated with Human Papillomavirus Infection. Mechanisms and Potential for Immunotherapy. Biochemistry 2019, 84, 782–799. [Google Scholar] [CrossRef]
- Nunes, R.A.L.; Morale, M.G.; Silva, G.A.F.; Villa, L.L.; Termini, L. Innate immunity and HPV: Friends or foes. Clinics 2018, 73, e549s. [Google Scholar] [CrossRef]
- Yang, X.; Cheng, Y.; Li, C. The role of TLRs in cervical cancer with HPV infection: A review. Signal Transduct. Target. Ther. 2017, 2, 17055. [Google Scholar] [CrossRef]
- Cattani, P.; Zannoni, G.F.; Ricci, C.; D’Onghia, S.; Trivellizzi, I.N.; Di Franco, A.; Vellone, V.G.; Durante, M.; Fadda, G.; Scambia, G.; et al. Clinical performance of human papillomavirus E6 and E7 mRNA testing for high-grade lesions of the cervix. J. Clin. Microbiol. 2009, 47, 3895–3901. [Google Scholar] [CrossRef]
| Constructs (>300) | Tumor Phenotype | Cancer |
|---|---|---|
| Wild type (>3) | Latent, persistent, cancer | Yes, >12 months (Hu et al., 2002, 2005, 2009; Cladel et al., 2009, 2013) |
| Regressive strain (>5) | Regressive | No (Hu et al., 2002, 2005, 2009) |
| Hybrid, epitope etc., mutants (>200) | Varies | Maybe (Hu et al., 2002, 2005, 2009; Cladel et al., 2009, 2013; Bounds, 2010) and unpublished |
| E8 and SE6 mutants (>10) | Persistent, benign, and small | Maybe, >12 months (Hu et al., 2002, 2005, 2009; Cladel et al., 2009, 2013) |
| E7 mutant genomes (>5) | Persistent and benign | No (unpublished observations) |
| E6 and E7 codon optimized genomes (>20) | Regressive or Cancer | Yes, >3 months (Cladel et al., 2009, 2013) |
| Genes | Changes in CRPV-Infected Tumors | Pathways |
|---|---|---|
| Krt1, 2, 3, 4, 7, 10, 14, 16, 78; Krt13, 75 | UP/Down | Cytokeratin |
| KLF3, 10; KLF 1, 9, 11, 15 | UP/Down | Keratinocyte proliferation |
| BRCA1, BRCA2, FANCD2, PCNA; DDR2 | UP/Down | DNA damage |
| MAPK6, 13; MAPK12 | UP/Down | p38 MAPKs |
| PCNA, CDK2, CASP8, ERBB3, PDCD5,6; TGFBR2, PDCD4 | UP/Down | Cell growth and death |
| TP53I3, CDKN2A | UP | Tumor suppressor |
| CTLA-4, RNF149, Cblb, Rel, PD-L1; Gata3, NFATC1, 4, CD34, NR4A1, Foxp1, CD8b | UP/Down | T cell function |
| CXCL8, IFNgR1, STAT4; Cox-2, CX3CL1 | UP/Down | Cytokines, chemokines, and ligands |
| IL1A, IL4R, IL10RA, IL13, IL17F, IL23A, IL36A, IL36g; IL6R, 11RA, IL13, IL16 | UP/Down | Interleukins |
| Rabbit Strain | Phenotype after Infection | References |
|---|---|---|
| Outbred | Persistent and cancer (wild-type CRPV) Regressive (regressive CRPV) | Hu et al., 2002, 2005, 2009; Cladel et al., 2009, 2013, 2019 |
| EIII/JC inbred | Higher regression rate for wild-type CRPV | Hu et al., 2002, 2005, 2006, 2007, 2009 |
| HLA-A2.1 outbred | Persistent and cancer (wild-type CRPV) with higher regression rates Regressive (regressive CRPV) | Hu et al., 2006, 2007, Bounds et al., 2009, Cladel et al., 2019 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cladel, N.M.; Xu, J.; Peng, X.; Jiang, P.; Christensen, N.D.; Zheng, Z.-M.; Hu, J. Modeling HPV-Associated Disease and Cancer Using the Cottontail Rabbit Papillomavirus. Viruses 2022, 14, 1964. https://doi.org/10.3390/v14091964
Cladel NM, Xu J, Peng X, Jiang P, Christensen ND, Zheng Z-M, Hu J. Modeling HPV-Associated Disease and Cancer Using the Cottontail Rabbit Papillomavirus. Viruses. 2022; 14(9):1964. https://doi.org/10.3390/v14091964
Chicago/Turabian StyleCladel, Nancy M., Jie Xu, Xuwen Peng, Pengfei Jiang, Neil D. Christensen, Zhi-Ming Zheng, and Jiafen Hu. 2022. "Modeling HPV-Associated Disease and Cancer Using the Cottontail Rabbit Papillomavirus" Viruses 14, no. 9: 1964. https://doi.org/10.3390/v14091964
APA StyleCladel, N. M., Xu, J., Peng, X., Jiang, P., Christensen, N. D., Zheng, Z.-M., & Hu, J. (2022). Modeling HPV-Associated Disease and Cancer Using the Cottontail Rabbit Papillomavirus. Viruses, 14(9), 1964. https://doi.org/10.3390/v14091964









