Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review
Abstract
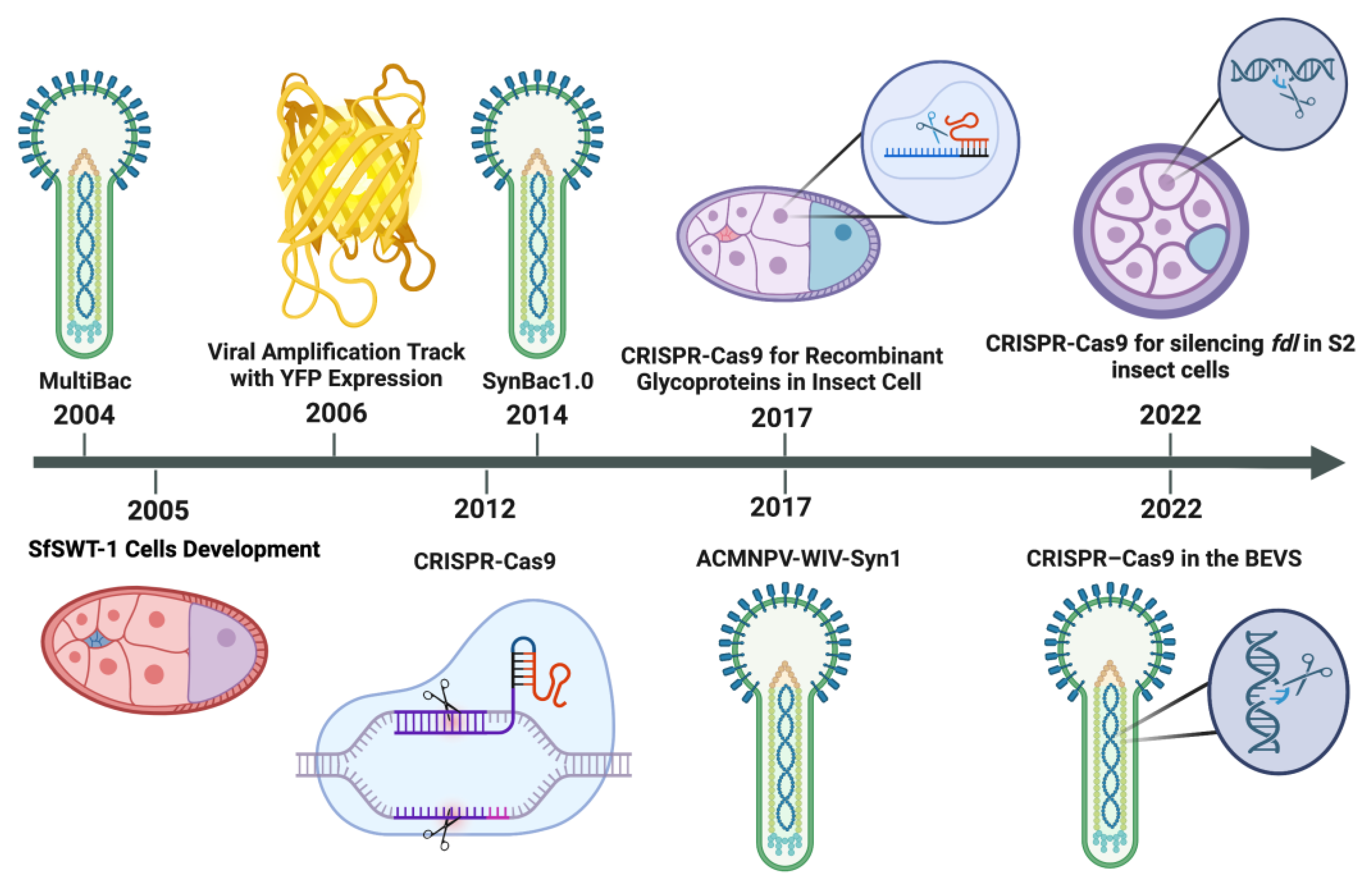
:1. Introduction
2. Baculovirus Expression Vector System (BEVS)
3. Development of MultiBac
4. CRISPR/Cas9 Technology
5. CRISPR/dCas9 Technology
6. Insect Cell Line Modification Using CRISPR/Cas9
7. Glycosylation
8. Glycosylation in BICS
9. Transgenic Insect Cells for BICS
10. The Future of CRISPR/Cas9 to Complex Glycosylation in BICS
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Possee, R.D.; Griffiths, C.M.; Hitchman, R.B.; Chambers, A.; Murguia-Meca, F.; Danquah, J.; Jeshtadi, A.; King, L.A. Baculoviruses: Biology, replication and exploitation. In Insect Virology; Asgari, S., Johnson, K.N., Eds.; Caister Academic Press: Wymondham, UK, 2010; pp. 35–58. [Google Scholar]
- Jehle, J.A.; Lange, M.; Wang, H.; Hu, Z.; Wang, Y.; Hauschild, R. Molecular Identification and Phylogenetic Analysis of Baculoviruses from Lepidoptera. Virology 2006, 346, 180–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohrmann, G.F. Baculovirus Molecular Biology, 3rd ed.; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2013. [Google Scholar]
- Slack, J.; Arif, B.M. The Baculoviruses Occlusion-Derived Virus: Virion Structure and Function. Adv. Virus Res. 2006, 69, 99–165. [Google Scholar] [CrossRef]
- Summers, M.D. Milestones Leading to the Genetic Engineering of Baculoviruses as Expression Vector Systems and Viral Pesticides. Adv. Virus Res. 2006, 68, 3–73. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.E.; Summers, M.D.; Fraser, M.J. Production of Human Beta Interferon in Insect Cells Infected with a Baculovirus Expression Vector. Mol. Cell. Biol. 1983, 3, 2156–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pennock, G.D.; Shoemaker, C.; Miller, L.K. Strong and Regulated Expression of Escherichia coli Beta-Galactosidase in Insect Cells with a Baculovirus Vector. Mol. Cell. Biol. 1984, 4, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Kost, T.A.; Condreay, J.P.; Jarvis, D.L. Baculovirus as Versatile Vectors for Protein Expression in Insect and Mammalian Cells. Nat. Biotechnol. 2005, 23, 567–575. [Google Scholar] [CrossRef]
- Noad, R.; Roy, P. Bluetongue Vaccines. Vaccine 2009, 27, D86–D89. [Google Scholar] [CrossRef]
- Pérez de Diego, A.C.; Athmaram, T.N.; Stewart, M.; Rodríguez-Sánchez, B.; Sánchez-Vizcaíno, J.M.; Noad, R.; Roy, P. Characterization of Protection Afforded by a Bivalent Virus-Like Particle Vaccine against Bluetongue Virus Serotypes 1 and 4 in Sheep. PLoS ONE 2011, 6, e26666. [Google Scholar] [CrossRef] [Green Version]
- Vicente, T.; Roldão, A.; Peixoto, C.; Carrondo, M.J.T.; Alves, P.M. Large-Scale Production and Purification of VLP-Based Vaccines. J. Invertebr. Pathol. 2011, 107, S42–S48. [Google Scholar] [CrossRef]
- Felberbaum, R.S. The Baculovirus Expression Vector System: A Commercial Manufacturing Platform for Viral Vaccines and Gene Therapy Vectors. Biotechnol. J. 2015, 10, 702–714. [Google Scholar] [CrossRef]
- Sandro, Q.; Relizani, K.; Benchaouir, R. AAV Production Using Baculovirus Expression Vector System. Viral Vectors Gene Ther. 2019, 91–99. [Google Scholar] [CrossRef]
- Hong, M.; Li, T.; Xue, W.; Zhang, S.; Cui, L.; Wang, H.; Zhang, Y.; Zhou, L.; Gu, Y.; Xia, N.; et al. Genetic Engineering of Baculovirus-Insect Cell System to Improve Protein Production. Front. Bioeng. Biotechnol. 2022, 10. [Google Scholar] [CrossRef] [PubMed]
- Krammer, F.; Schinko, T.; Palmberger, D.; Tauer, C.; Messner, P.; Grabherr, R. Trichoplusia Ni Cells (High FiveTM) Are Highly Efficient for the Production of Influenza A Virus-like Particles: A Comparison of Two Insect Cell Lines as Production Platforms for Influenza Vaccines. Mol. Biotechnol. 2010, 45, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Palomares, L.A.; Joosten, C.E.; Hughes, P.R.; Granados, R.R.; Shuler, M.L. Novel Insect Cell Line Capable of Complex N-Glycosylation and Sialylation of Recombinant Proteins. Biotechnol. Prog. 2003, 19, 185–192. [Google Scholar] [CrossRef]
- Carbonell, L.F.; Klowden, M.J.; Miller, L.K. Baculovirus-Mediated Expression of Bacterial Genes in Dipteran and Mammalian Cells. J. Virol. 1985, 56, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, C.; Sandig, V.; Jennings, G.; Rudolpht, M.; Schlag, P.; Strauss, M. Efficient Gene Transfer into Human Hepatocytes by Baculovirus Vectors. Proc. Natl. Acad. Sci. USA 1995, 92, 10099–10103. [Google Scholar] [CrossRef] [Green Version]
- Boyce, F.M.; Bucher, N.L. Baculovirus-Mediated Gene Transfer into Mammalian Cells. Proc. Natl. Acad. Sci. USA 1996, 93, 2348–2352. [Google Scholar] [CrossRef] [Green Version]
- Possee, R.D.; Chambers, A.C.; Graves, L.P.; Aksular, M.; King, L.A. Recent Developments in the Use of Baculovirus Expression Vectors. Curr. Issues Mol. Biol. 2020, 34, 215–230. [Google Scholar] [CrossRef]
- Mäkelä, A.R.; Matilainen, H.; White, D.J.; Ruoslahti, E.; Oker-Blom, C. Enhanced Baculovirus-Mediated Transduction of Human Cancer Cells by Tumor-Homing Peptides. J. Virol. 2006, 80, 6603–6611. [Google Scholar] [CrossRef] [Green Version]
- Ge, J.; Huang, Y.; Hu, X.; Zhong, J. A Surface-Modified Baculovirus Vector with Improved Gene Delivery to B-Lymphocytic Cells. J. Biotechnol. 2007, 129, 367–372. [Google Scholar] [CrossRef]
- Martyn, J.C.; Cardin, A.J.; Wines, B.D.; Cendron, A.; Li, S.; Mackenzie, J.; Powell, M.; Gowans, E.J. Surface Display of IgG Fc on Baculovirus Vectors Enhances Binding to Antigen-Presenting Cells and Cell Lines Expressing Fc Receptors. Arch. Virol. 2009, 154, 1129–1138. [Google Scholar] [CrossRef] [PubMed]
- Nobiron, I.; O’Reilly, D.R.; Olszewski, J.A. Autographa Californica Nucleopolyhedrovirus Infection of Spodoptera frugiperda Cells: A Global Analysis of Host Gene Regulation during Infection, Using a Differential Display Approach. J. Gen. Virol. 2003, 84, 3029–3039. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-H.; Wei, S.-C.; Lo, H.-R.; Chao, Y.-C. Baculovirus as Versatile Vectors for Protein Display and Biotechnological Applications. Curr. Issues Mol. Biol. 2020, 34, 231–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pelosse, M.; Crocker, H.; Gorda, B.; Lemaire, P.; Rauch, J.; Berger, I. MultiBac: From Protein Complex Structures to Synthetic Viral Nanosystems. BMC Biol. 2017, 15, 99. [Google Scholar] [CrossRef] [Green Version]
- Chambers, A.C.; Aksular, M.; Graves, L.P.; Irons, S.L.; Possee, R.D.; King, L.A. Overview of the Baculovirus Expression System. Curr. Protoc. Protein Sci. 2018, 91, 5.4.1–5.4.6. [Google Scholar] [CrossRef]
- Von Seggern, D.J.; Nemerow, G.R. Adenoviral vectors for protein expression. In Gene Expression Systems; Elsevier: Amsterdam, The Netherlands, 1999; pp. 111–156. [Google Scholar] [CrossRef]
- Warnock, J.N.; Daigre, C.; Al-Rubeai, M. Introduction to Viral Vectors. Methods Mol. Biol. 2011, 737, 1–25. [Google Scholar] [CrossRef]
- Vannucci, L.; Lai, M.; Chiuppesi, F.; Ceccherini-Nelli, L.; Pistello, M. Viral Vectors: A Look Back and Ahead on Gene Transfer Technology. New Microbiol. 2013, 36, 1–22. [Google Scholar]
- dos Coura, R.S.; Nardi, N.B. The State of the Art of Adeno-Associated Virus-Based Vectors in Gene Therapy. Virol. J. 2007, 4, 99. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.-X.; Wang, S.-M.; Bai, Y.-H.; Luo, T.-T.; Wang, J.-Q.; Dai, C.-Q.; Guo, B.-L.; Luo, S.-C.; Wang, D.-H.; Yang, Y.-L.; et al. Lentiviral Vectors and Adeno-Associated Virus Vectors: Useful Tools for Gene Transfer in Pain Research. Anat. Rec. 2018, 301, 825–836. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-Associated Virus Vector as a Platform for Gene Therapy Delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Anson, D.S. The Use of Retroviral Vectors for Gene Therapy-What Are the Risks? A Review of Retroviral Pathogenesis and Its Relevance to Retroviral Vector-Mediated Gene Delivery. Genet. Vaccines Ther. 2004, 2, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.H.; Robbins, P.D. Retroviral vectors for gene therapy. In Lysosomal Storage Disorders; Springer: Boston, MA, USA, 2007; pp. 53–67. [Google Scholar] [CrossRef]
- Torashima, T.; Okoyama, S.; Nishizaki, T.; Hirai, H. In Vivo Transduction of Murine Cerebellar Purkinje Cells by HIV-Derived Lentiviral Vectors. Brain Res. 2006, 1082, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Meunier, A.; Pohl, M. Lentiviral Vectors for Gene Transfer into the Spinal Cord Glial Cells. Gene Ther. 2009, 16, 476–482. [Google Scholar] [CrossRef]
- Lachmann, R. Herpes Simplex Virus-Based Vectors. Int. J. Exp. Pathol. 2004, 85, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.S.; Bartlett, D.L. Vaccinia as a Vector for Gene Delivery. Expert Opin. Biol. Ther. 2004, 4, 901–917. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Li, G.; Ren, X.; Herrler, G. Select What You Need: A Comparative Evaluation of the Advantages and Limitations of Frequently Used Expression Systems for Foreign Genes. J. Biotechnol. 2007, 127, 335–347. [Google Scholar] [CrossRef]
- Gorda, B.; Toelzer, C.; Aulicino, F.; Berger, I. The MultiBac BEVS: Basics, Applications, Performance and Recent Developments. Methods Enzymol. 2021, 660, 129–154. [Google Scholar] [CrossRef]
- Ma, Y.; Lee, C.-J.; Park, J.-S. Strategies for Optimizing the Production of Proteins and Peptides with Multiple Disulfide Bonds. Antibiotics 2020, 9, 541. [Google Scholar] [CrossRef]
- Biswas, K.; Pandey, P.; Kumar, R.; Maurya, M. Microbial recombinant protein: An epic from fundamental to future panorama of life science. In Research Trends in Molecular Biology; Research Singpost: Kerala, India, 2016; pp. 1–34. [Google Scholar]
- Jarvis, D.L. Baculovirus-Insect Cell Expression Systems. Methods Enzymol. 2009, 463, 191–222. [Google Scholar] [CrossRef]
- Robinson, C.V.; Sali, A.; Baumeister, W. The Molecular Sociology of the Cell. Nature 2007, 450, 973–982. [Google Scholar] [CrossRef]
- Bieniossek, C.; Berger, I. Towards Eukaryotic Structural Complexomics. J. Struct. Funct. Genom. 2009, 10, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Sari, D.; Gupta, K.; Thimiri Govinda Raj, D.B.; Aubert, A.; Drncová, P.; Garzoni, F.; Fitzgerald, D.; Berger, I. The MultiBac baculovirus/insect cell expression vector system for producing complex protein biologics. In Advanced Technologies for Protein Complex Production and Characterization; Springer: Cham, Switzerland, 2016; pp. 199–215. [Google Scholar] [CrossRef] [Green Version]
- Berger, I.; Fitzgerald, D.J.; Richmond, T.J. Baculovirus Expression System for Heterologous Multiprotein Complexes. Nat. Biotechnol. 2004, 22, 1583–1587. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, D.J.; Berger, P.; Schaffitzel, C.; Yamada, K.; Richmond, T.J.; Berger, I. Protein Complex Expression by Using Multigene Baculoviral Vectors. Nat. Methods 2006, 3, 1021–1032. [Google Scholar] [CrossRef] [PubMed]
- Bieniossek, C.; Richmond, T.J.; Berger, I. MultiBac: Multigene Baculovirus-Based Eukaryotic Protein Complex Production. Curr. Protoc. Protein Sci. 2008, 51, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Bieniossek, C.; Imasaki, T.; Takagi, Y.; Berger, I. MultiBac: Expanding the Research Toolbox for Multiprotein Complexes. Trends Biochem. Sci. 2012, 37, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Trowitzsch, S.; Palmberger, D.; Fitzgerald, D.; Takagi, Y.; Berger, I. MultiBac Complexomics. Expert Rev. Proteom. 2012, 9, 363–373. [Google Scholar] [CrossRef]
- Nie, Y.; Chaillet, M.; Becke, C.; Haffke, M.; Pelosse, M.; Fitzgerald, D.; Collinson, I.; Schaffitzel, C.; Berger, I. ACEMBL Tool-Kits for High-Throughput Multigene Delivery and Expression in Prokaryotic and Eukaryotic Hosts. Adv. Exp. Med. Biol. 2016, 896, 27–42. [Google Scholar] [CrossRef] [Green Version]
- Shang, Y.; Wang, M.; Xiao, G.; Wang, X.; Hou, D.; Pan, K.; Liu, S.; Li, J.; Wang, J.; Arif, B.M.; et al. Construction and Rescue of a Functional Synthetic Baculovirus. ACS Synth. Biol. 2017, 6, 1393–1402. [Google Scholar] [CrossRef]
- Shang, Y.; Hu, H.; Wang, X.; Wang, H.; Deng, F.; Wang, M.; Hu, Z. Construction and Characterization of a Novel Bacmid AcBac-Syn Based on a Synthesized Baculovirus Genome. Virol. Sin. 2021, 36, 1566–1574. [Google Scholar] [CrossRef]
- Thimiri Govinda Raj, D.B.; Garzoni, F.; Gavin, A.C.; Gibson, T.; Berger, I. FEBS-EMBO Concurrent Session (Lecture): CS III-6-3 SynBac: Designer Minimal Baculovirus Genome for Drug Discovery. FEBS J. 2014, 281, 9–55. [Google Scholar] [CrossRef] [Green Version]
- Adli, M. The CRISPR Tool Kit for Genome Editing and Beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendriks, D.; Clevers, H.; Artegiani, B. CRISPR-Cas Tools and Their Application in Genetic Engineering of Human Stem Cells and Organoids. Cell Stem Cell 2020, 27, 705–731. [Google Scholar] [CrossRef] [PubMed]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide Sequence of the Iap Gene, Responsible for Alkaline Phosphatase Isozyme Conversion in Escherichia coli, and Identification of the Gene Product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolotin, A.; Quinquis, B.; Sorokin, A.; Ehrlich, S.D. Clustered Regularly Interspaced Short Palindrome Repeats (CRISPRs) Have Spacers of Extrachromosomal Origin. Microbiology 2005, 151 Pt 8, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR Provides Acquired Resistance against Viruses in Prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Wiedenheft, B.; Sternberg, S.H.; Doudna, J.A. RNA-Guided Genetic Silencing Systems in Bacteria and Archaea. Nature 2012, 482, 331–338. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Pickar-Oliver, A.; Gersbach, C.A. The next Generation of CRISPR-Cas Technologies and Applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Katti, A.; Diaz, B.J.; Caragine, C.M.; Sanjana, N.E.; Dow, L.E. CRISPR in Cancer Biology and Therapy. Nat. Rev. Cancer 2022, 22, 259–279. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koonin, E.V.; Makarova, K.S. Origins and Evolution of CRISPR-Cas Systems. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2019, 374, 20180087. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary Classification of CRISPR-Cas Systems: A Burst of Class 2 and Derived Variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, L.; Liu, H.; Cheng, K. Delivery Strategies of the CRISPR-Cas9 Gene-Editing System for Therapeutic Applications. J. Control. Release 2017, 266, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Sander, J.D.; Joung, J.K. CRISPR-Cas Systems for Editing, Regulating and Targeting Genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [CrossRef]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA Interrogation by the CRISPR RNA-Guided Endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar] [CrossRef] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-Programmed Genome Editing in Human Cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided Human Genome Engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [Green Version]
- Frock, R.L.; Hu, J.; Meyers, R.M.; Ho, Y.-J.; Kii, E.; Alt, F.W. Genome-Wide Detection of DNA Double-Stranded Breaks Induced by Engineered Nucleases. Nat. Biotechnol. 2015, 33, 179–186. [Google Scholar] [CrossRef]
- Su, T.; Liu, F.; Gu, P.; Jin, H.; Chang, Y.; Wang, Q.; Liang, Q.; Qi, Q. A CRISPR-Cas9 Assisted Non-Homologous End-Joining Strategy for One-Step Engineering of Bacterial Genome. Sci. Rep. 2016, 6, 37895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brinkman, E.K.; Chen, T.; de Haas, M.; Holland, H.A.; Akhtar, W.; van Steensel, B. Kinetics and Fidelity of the Repair of Cas9-Induced Double-Strand DNA Breaks. Mol. Cell 2018, 70, 801–813.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schep, R.; Brinkman, E.K.; Leemans, C.; Vergara, X.; van der Weide, R.H.; Morris, B.; van Schaik, T.; Manzo, S.G.; Peric-Hupkes, D.; van den Berg, J.; et al. Impact of Chromatin Context on Cas9-Induced DNA Double-Strand Break Repair Pathway Balance. Mol. Cell 2021, 81, 2216–2230.e10. [Google Scholar] [CrossRef] [PubMed]
- Nambiar, T.S.; Baudrier, L.; Billon, P.; Ciccia, A. CRISPR-Based Genome Editing through the Lens of DNA Repair. Mol. Cell 2022, 82, 348–388. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef] [Green Version]
- Shariati, S.A.; Dominguez, A.; Xie, S.; Wernig, M.; Qi, L.S.; Skotheim, J.M. Reversible Disruption of Specific Transcription Factor-DNA Interactions Using CRISPR/Cas9. Mol. Cell 2019, 74, 622–633.e4. [Google Scholar] [CrossRef]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/DCas9 Platforms in Plants: Strategies and Applications beyond Genome Editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef] [Green Version]
- Brezgin, S.; Kostyusheva, A.; Kostyushev, D.; Chulanov, V. Dead Cas Systems: Types, Principles, and Applications. Int. J. Mol. Sci. 2019, 20, 6041. [Google Scholar] [CrossRef]
- Saifaldeen, M.; Al-Ansari, D.E.; Ramotar, D.; Aouida, M. CRISPR FokI Dead Cas9 System: Principles and Applications in Genome Engineering. Cells 2020, 9, 2518. [Google Scholar] [CrossRef]
- Karlson, C.K.S.; Mohd-Noor, S.N.; Nolte, N.; Tan, B.C. CRISPR/DCas9-Based Systems: Mechanisms and Applications in Plant Sciences. Plants 2021, 10, 2055. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, X.; Qi, L.S. A CRISPR-DCas Toolbox for Genetic Engineering and Synthetic Biology. J. Mol. Biol. 2019, 431, 34–47. [Google Scholar] [CrossRef] [PubMed]
- Aulicino, F.; Pelosse, M.; Toelzer, C.; Capin, J.; Ilegems, E.; Meysami, P.; Rollarson, R.; Berggren, P.O.; Dillingham, M.S.; Schaffitzel, C.; et al. Highly Efficient CRISPR-Mediated Large DNA Docking and Multiplexed Prime Editing Using a Single Baculovirus. Nucleic Acids Res. 2022, 50, 7783–7799. [Google Scholar] [CrossRef] [PubMed]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic Genome Editing: Prospects and Challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR Technologies in Research and Beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef]
- Zhou, Q.; Krebs, J.F.; Snipas, S.J.; Price, A.; Alnemri, E.S.; Tomaselli, K.J.; Salvesen, G.S. Interaction of the Baculovirus Anti-Apoptotic Protein P35 with Caspases. Specificity, Kinetics, and Characterization of the Caspase/P35 Complex. Biochemistry 1998, 37, 10757–10765. [Google Scholar] [CrossRef]
- Lo, W.D.; Akhmametyeva, E.M.; Zhu, L.; Friesen, P.D.; Chang, L.-S. Induction of Apoptosis by the P53-Related P73 and Partial Inhibition by the Baculovirus-Encoded P35 in Neuronal Cells. Mol. Brain Res. 2003, 113, 1–12. [Google Scholar] [CrossRef]
- Rong, R.; Li, T.; Zhang, Y.; Gu, Y.; Xia, N.; Li, S. Progress in Vaccine Development Based on Baculovirus Expression Vector System. Sheng Wu Gong Cheng Xue Bao 2019, 35, 577–588. [Google Scholar] [CrossRef]
- Dong, Z.; Qin, Q.; Hu, Z.; Chen, P.; Huang, L.; Zhang, X.; Tian, T.; Lu, C.; Pan, M. Construction of a One-Vector Multiplex CRISPR/Cas9 Editing System to Inhibit Nucleopolyhedrovirus Replication in Silkworms. Virol. Sin. 2019, 34, 444–453. [Google Scholar] [CrossRef]
- Bruder, M.R.; Walji, S.-D.; Aucoin, M.G. Comparison of CRISPR–Cas9 Tools for Transcriptional Repression and Gene Disruption in the BEVS. Viruses 2021, 13, 1925. [Google Scholar] [CrossRef]
- Pazmiño-Ibarra, V.; Mengual-Martí, A.; Targovnik, A.M.; Herrero, S. Improvement of Baculovirus as Protein Expression Vector and as Biopesticide by CRISPR/Cas9 Editing. Biotechnol. Bioeng. 2019, 116, 2823–2833. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, X.; Hu, Z.; Li, M.; O’Reilly, D.R.; Zuidema, D.; Vlak, J.M. Genetic Engineering of Helicoverpa armigera Single-Nucleocapsid Nucleopolyhedrovirus as an Improved Pesticide. J. Invertebr. Pathol. 2000, 76, 140–146. [Google Scholar] [CrossRef] [PubMed]
- Cory, J.S.; Clarke, E.E.; Brown, M.L.; Hails, R.S.; O’Reilly, D.R. Microparasite Manipulation of an Insect: The Influence of the Egt Gene on the Interaction between a Baculovirus and Its Lepidopteran Host. Funct. Ecol. 2004, 18, 443–450. [Google Scholar] [CrossRef]
- Georgievska, L.; Hoover, K.; van der Werf, W.; Muñoz, D.; Caballero, P.; Cory, J.S.; Vlak, J.M. Dose Dependency of Time to Death in Single and Mixed Infections with a Wildtype and Egt Deletion Strain of Helicoverpa armigera Nucleopolyhedrovirus. J. Invertebr. Pathol. 2010, 104, 44–50. [Google Scholar] [CrossRef] [Green Version]
- Simón, O.; Williams, T.; López-Ferber, M.; Caballero, P. Deletion of Egt Is Responsible for the Fast-Killing Phenotype of Natural Deletion Genotypes in a Spodoptera frugiperda Multiple Nucleopolyhedrovirus Population. J. Invertebr. Pathol. 2012, 111, 260–263. [Google Scholar] [CrossRef] [Green Version]
- Antoniou, P.; Miccio, A.; Brusson, M. Base and Prime Editing Technologies for Blood Disorders. Front. Genome Ed. 2021, 3, 618406. [Google Scholar] [CrossRef]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-Replace Genome Editing without Double-Strand Breaks or Donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Schwinn, M.K.; Machleidt, T.; Zimmerman, K.; Eggers, C.T.; Dixon, A.S.; Hurst, R.; Hall, M.P.; Encell, L.P.; Binkowski, B.F.; Wood, K.V. CRISPR-Mediated Tagging of Endogenous Proteins with a Luminescent Peptide. ACS Chem. Biol. 2018, 13, 467–474. [Google Scholar] [CrossRef]
- García-Fernández, A.; Vivo-Llorca, G.; Sancho, M.; García-Jareño, A.B.; Ramírez-Jiménez, L.; Barber-Cano, E.; Murguía, J.R.; Orzáez, M.; Sancenón, F.; Martínez-Máñez, R. Nanodevices for the Efficient Codelivery of CRISPR-Cas9 Editing Machinery and an Entrapped Cargo: A Proposal for Dual Anti-Inflammatory Therapy. Pharmaceutics 2022, 14, 1495. [Google Scholar] [CrossRef]
- Aulicino, F.; Capin, J.; Berger, I. Synthetic Virus-Derived Nanosystems (Svns) for Delivery and Precision Docking of Large Multifunctional Dna Circuitry in Mammalian Cells. Pharmaceutics 2020, 12, 759. [Google Scholar] [CrossRef]
- Mansouri, M.; Berger, P. Baculovirus for Gene Delivery to Mammalian Cells: Past, Present and Future. Plasmid 2018, 98, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.H.; Bayat, H.; Rajabibazl, M.; Sabri, S.; Rahimpour, A. Humanizing Glycosylation Pathways in Eukaryotic Expression Systems. World J. Microbiol. Biotechnol. 2017, 33, 4. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, P.P.; Callewaert, N. N-Glycosylation Engineering of Biopharmaceutical Expression Systems. Curr. Mol. Med. 2009, 9, 774–800. [Google Scholar] [CrossRef]
- Dalziel, M.; Crispin, M.; Scanlan, C.N.; Zitzmann, N.; Dwek, R.A. Emerging Principles for the Therapeutic Exploitation of Glycosylation. Science 2014, 343, 1235681. [Google Scholar] [CrossRef]
- Wormald, M.R.; Dwek, R.A. Glycoproteins: Glycan Presentation and Protein-Fold Stability. Structure 1999, 7, R155–R160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, J.R.; Schuksz, M.; Esko, J.D. Heparan Sulphate Proteoglycans Fine-Tune Mammalian Physiology. Nature 2007, 446, 1030–1037. [Google Scholar] [CrossRef] [PubMed]
- Varki, A. Sialic Acids in Human Health and Disease. Trends Mol. Med. 2008, 14, 351–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sperandio, M.; Gleissner, C.A.; Ley, K. Glycosylation in Immune Cell Trafficking. Immunol. Rev. 2009, 230, 97–113. [Google Scholar] [CrossRef]
- Marchal, I.; Jarvis, D.L.; Cacan, R.; Verbert, A. Glycoproteins from Insect Cells: Sialylated or Not? Biol. Chem. 2001, 382, 151–159. [Google Scholar] [CrossRef] [Green Version]
- Geisler, C.; Jarvis, D.L. Innovative Use of a Bacterial Enzyme Involved in Sialic Acid Degradation to Initiate Sialic Acid Biosynthesis in Glycoengineered Insect Cells. Metab. Eng. 2012, 14, 642–652. [Google Scholar] [CrossRef] [Green Version]
- Kim, A.; Lee, G.; Hwang, J.-H.; Park, J.-H.; Lee, M.J.; Kim, B.; Kim, S.-M. BacMam Expressing Highly Glycosylated Porcine Interferon Alpha Induces Robust Antiviral and Adjuvant Effects against Foot-and-Mouth Disease Virus in Pigs. J. Virol. 2022, 96, e00528-22. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Gómez, A.; Sánchez-Mirón, A.; García-Camacho, F.; Molina-Grima, E.; Chisti, Y. Protein Production Using the Baculovirus-Insect Cell Expression System. Biotechnol. Prog. 2014, 30, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Palomares, L.A.; Srivastava, I.K.; Ramírez, O.T.; Cox, M.M.J. Glycobiotechnology of the Insect Cell-Baculovirus Expression System Technology. Adv. Biochem. Eng. Biotechnol. 2021, 175, 71–92. [Google Scholar] [CrossRef] [PubMed]
- Legardinier, S.; Klett, D.; Poirier, J.-C.; Combarnous, Y.; Cahoreau, C. Mammalian-like Nonsialyl Complex-Type N-Glycosylation of Equine Gonadotropins in Mimic Insect Cells. Glycobiology 2005, 15, 776–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollister, J.; Grabenhorst, E.; Nimtz, M.; Conradt, H.; Jarvis, D.L. Engineering the Protein N-Glycosylation Pathway in Insect Cells for Production of Biantennary, Complex N-Glycans. Biochemistry 2002, 41, 15093–15104. [Google Scholar] [CrossRef] [PubMed]
- Hollister, J.R.; Jarvis, D.L. Engineering Lepidopteran Insect Cells for Sialoglycoprotein Production by Genetic Transformation with Mammalian Beta 1,4-Galactosyltransferase and Alpha 2,6-Sialyltransferase Genes. Glycobiology 2001, 11, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aumiller, J.J.; Hollister, J.R.; Jarvis, D.L. A Transgenic Insect Cell Line Engineered to Produce CMP-Sialic Acid and Sialylated Glycoproteins. Glycobiology 2003, 13, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Aumiller, J.J.; Mabashi-Asazuma, H.; Hillar, A.; Shi, X.; Jarvis, D.L. A New Glycoengineered Insect Cell Line with an Inducibly Mammalianized Protein N-Glycosylation Pathway. Glycobiology 2012, 22, 417–428. [Google Scholar] [CrossRef]
- Mabashi-Asazuma, H.; Shi, X.; Geisler, C.; Kuo, C.-W.; Khoo, K.-H.; Jarvis, D.L. Impact of a Human CMP-Sialic Acid Transporter on Recombinant Glycoprotein Sialylation in Glycoengineered Insect Cells. Glycobiology 2013, 23, 199–210. [Google Scholar] [CrossRef]
- Mabashi-Asazuma, H.; Jarvis, D.L. CRISPR-Cas9 Vectors for Genome Editing and Host Engineering in the Baculovirus–Insect Cell System. Proc. Natl. Acad. Sci. USA 2017, 114, 9068–9073. [Google Scholar] [CrossRef] [Green Version]
- Nweke, E.E.; Thimiri Govinda Raj, D.B. Development of Insect Cell Line Using CRISPR Technology. Prog. Mol. Biol. Transl. Sci. 2021, 180, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Geisler, C.; Aumiller, J.J.; Jarvis, D.L. A Fused Lobes Gene Encodes the Processing Beta-N-Acetylglucosaminidase in Sf9 Cells. J. Biol. Chem. 2008, 283, 11330–11339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, N.Y.; Baek, J.Y.; Choi, H.S.; Chung, I.S.; Shin, S.; Lee, J.I.; Choi, J.Y.; Yang, J.M. Short-Hairpin RNA-Mediated Gene Expression Interference in Trichoplusia Ni Cells. J. Microbiol. Biotechnol. 2012, 22, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mabashi-Asazuma, H.; Kuo, C.-W.; Khoo, K.-H.; Jarvis, D.L. Modifying an Insect Cell N-Glycan Processing Pathway Using CRISPR-Cas Technology. ACS Chem. Biol. 2015, 10, 2199–2208. [Google Scholar] [CrossRef] [PubMed]
- Chavez-Pena, C.; Kamen, A.A. RNA Interference Technology to Improve the Baculovirus-Insect Cell Expression System. Biotechnol. Adv. 2018, 36, 443–451. [Google Scholar] [CrossRef]
| Vector System | Advantages | Disadvantages |
|---|---|---|
| Adenovirus | Large quantities of high-titer viral stocks can be produced easily (1010 pfu/mL) | No integration into host cell genome can also be an advantage since errors related to random insertion are avoided |
| Ability to infect both dividing and non-dividing cells | High levels of pre-existing immunity in humans | |
| Non-oncogenic | Highly immunogenic | |
| Good insert capacity (carry up to 8 kbp) | Transient gene expression | |
| Adeno-Associated Virus | Highly safe since they have never been shown to cause any human disease | Small packaging size (~5.0 kb, including inverted terminal repeats; ITRs) |
| Some serotypes have the capacity to bypass the blood–brain barrier (BBB), allowing for the transduction of the central nervous system (CNS) via systemic administration to be used as vector for gene therapy in neurodegenerative diseases. | Slow onset of gene expression, due to the requirement of conversion of the single-stranded AAV DNA into double- stranded DNA | |
| Broad host and cell type tropism range | They persist as non-replicating episomes and are therefore gradually lost in mitotic cells | |
| Have the ability to transduce both dividing and non-dividing cells | ||
| High levels of gene expression for long-term (over years) | ||
| Heat stability and resistance to solvents and changes in pH and temperature | ||
| Low immunogenicity and cytotoxicity | ||
| Retrovirus | Non-immunogenic | They have relatively small carrying capacity |
| Wide range of target species and cells | Unable to infect non-dividing cells | |
| Become a permanent part of the host cell genome allowing for stable expression | Random integration into host chromosome, resulting in possible insertional mutagenesis or oncogene activation | |
| Lentivirus | The vector genome integrates into the host cell genome stably leading to long term expression of the transgene | Non-specific integration in the host genome may lead to insertional mutations |
| Relatively large carrying capacity (~12–15 kbp) | Uncertainty of biosafety | |
| Capable of infecting a wide variety of dividing and non-dividing cells | ||
| The normal function of infected cells is not affected both in vitro and in vivo | ||
| Enhanced proneness to transduce terminally differentiated tissues from neuronal origin | ||
| Herpes-Simplex Virus | Have natural tropism for neuronal cells | Possible cytotoxicity (low safety) |
| Vector particles are easily obtained in high titers from tissue culture (1012 pfu/mL) | High level of pre-existing immunity in humans | |
| Can accommodate large amounts of foreign DNA (~50 kbp) | Transient expression of the transgene | |
| Establish a latent infection during which the viral genome persists indefinitely without any discernible adverse effects on the host cell | The vector genome does not integrate into the host cell genome (can also be an advantage since errors related to random insertion are avoided) | |
| Baculovirus | Baculovirus arrests most host gene transcription, thus prioritizing viral gene expression. | Transient expression of heterologous gene |
| Inherent biosafety since the virus does not infect human cells | The bioactivity and immunogenicity of insect expression products are somewhat different from those of the natural product because insect and mammalian cells differ in their glycosylation patterns | |
| They have flexible capsid and envelope which simply increase in size proportional to the genomic DNA they harbor | ||
| Because it infects insect cells, BEVS affords eukaryotic post-translational modifications and folding of heterologous proteins. | ||
| High levels of recombinant protein production | ||
| BEVS manufacturing is cost-efficient | ||
| Yeast | Cost-effective | Hypermannosylation |
| Rapid growth in culture leading to high yield production of proteins | Cannot perform N- and O-linked glycosylation the same way as mammalian cells | |
| Share many features with higher eukaryotes allowing for protein processing similar to mammalian cells | ||
| Some intracellularly synthesized proteins to be secreted into the extracellular environment due to the enriched endomembrane system | ||
| Can produce correctly folded recombinant proteins that have undergone all the post-translational modifications that are essential for their functions | ||
| Easy to culture and manipulate | ||
| Safe systems | ||
| Escherichia coli | Rapid expression | Proteins with disulfide bonds difficult to express. |
| High yields | Produce unglycosylated proteins. | |
| Ease of culture and genome modifications | Acetate formation resulting in cell toxicity. | |
| Inexpensive | Proteins produced with endotoxins. | |
| Mass production is fast and cost effective | Proteins produced as inclusion bodies, are inactive; require refolding. | |
| Drosophila melanogaster | Quick turn round time | Regulatory records are less than other expression systems |
| The protein expressed in its native form | ||
| No endotoxin release from host cell organism | Possible mammalian virus infection | |
| Less expensive than mammalian culture | ||
| Integration of DNA of interest is very stable | Expensive compared to E. coli and yeast expression | |
| Safer than working with mammalian cell lines | ||
| Usually expresses and secretes even complex post-transitional modified proteins | Proteases present in the cells degrade the protein of interest | |
| Minor secretion of host cell proteins | ||
| The vectors are not pathogenic to human | Characteristic N-linked glycan structures of proteins are different when compared to typical mammalian proteins | |
| Extra-cellular expression to low viscosity medium of correctly folded protein | ||
| Lactobacillus zeae | Adapted to temperature sensitive products | No post-translational modifications |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sari-Ak, D.; Alomari, O.; Shomali, R.A.; Lim, J.; Thimiri Govinda Raj, D.B. Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review. Viruses 2023, 15, 54. https://doi.org/10.3390/v15010054
Sari-Ak D, Alomari O, Shomali RA, Lim J, Thimiri Govinda Raj DB. Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review. Viruses. 2023; 15(1):54. https://doi.org/10.3390/v15010054
Chicago/Turabian StyleSari-Ak, Duygu, Omar Alomari, Raghad Al Shomali, Jackwee Lim, and Deepak B. Thimiri Govinda Raj. 2023. "Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review" Viruses 15, no. 1: 54. https://doi.org/10.3390/v15010054
APA StyleSari-Ak, D., Alomari, O., Shomali, R. A., Lim, J., & Thimiri Govinda Raj, D. B. (2023). Advances in CRISPR-Cas9 for the Baculovirus Vector System: A Systematic Review. Viruses, 15(1), 54. https://doi.org/10.3390/v15010054







