Generation and Characterization of a SARS-CoV-2-Susceptible Mouse Model Using Adeno-Associated Virus (AAV6.2FF)-Mediated Respiratory Delivery of the Human ACE2 Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Cells and Viruses
2.3. Mouse Experiments
2.4. Infectious Virus Quantification
2.5. Reverse Transcriptase-Quantitative Polymerase Chain Reaction
2.6. Serum ELISA
2.7. Cytokine Analyses
2.8. Western Blot
2.9. Immunofluorescence Analysis of hACE2 Expression in Murine Lungs
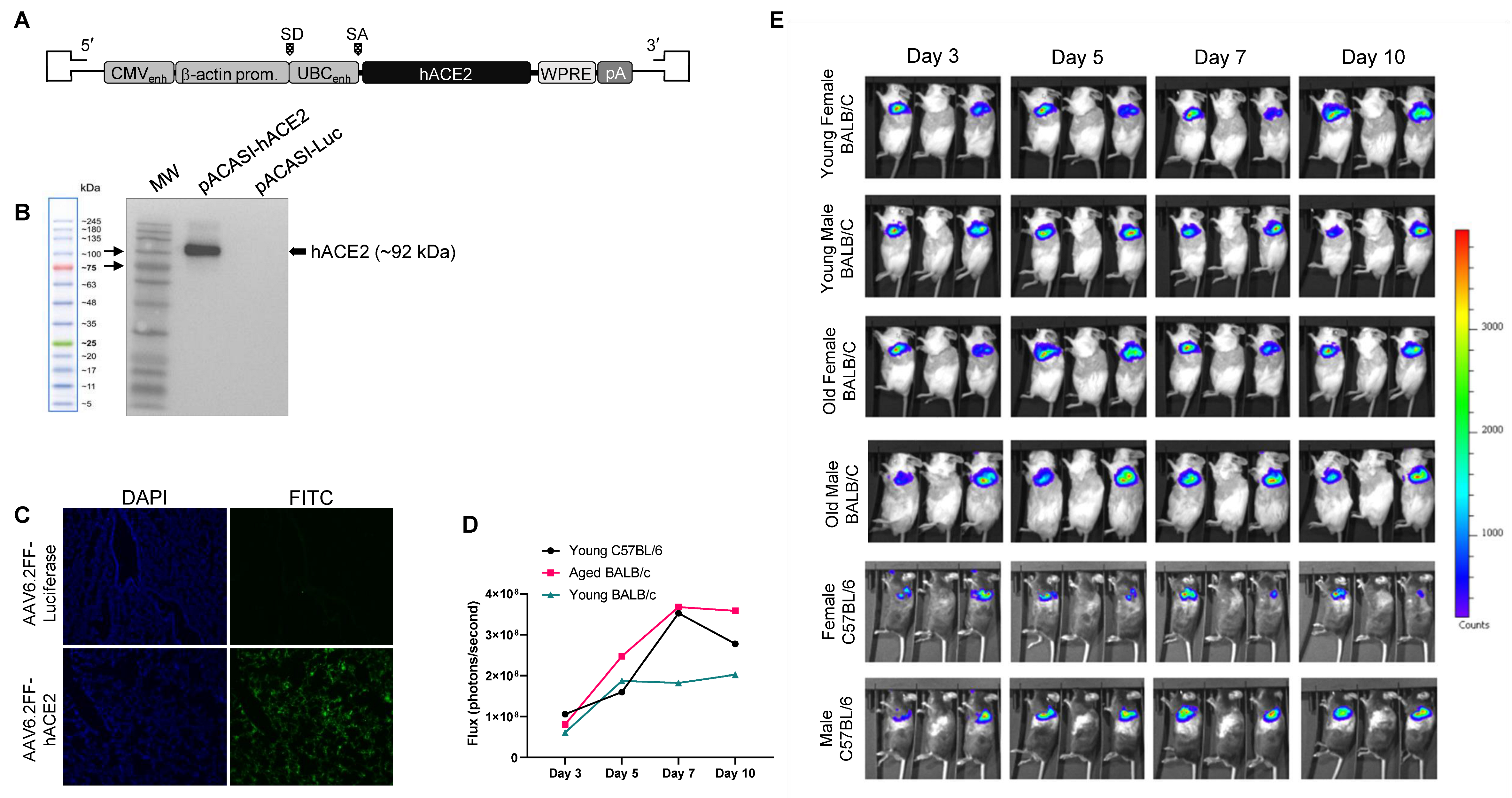
2.10. In Vivo Luciferase Imaging
2.11. Histology and Immunohistochemistry
2.12. Data Analysis
3. Results
3.1. Generation of an AAV6.2FF-hACE2 and AAV6.2FF-Luciferase
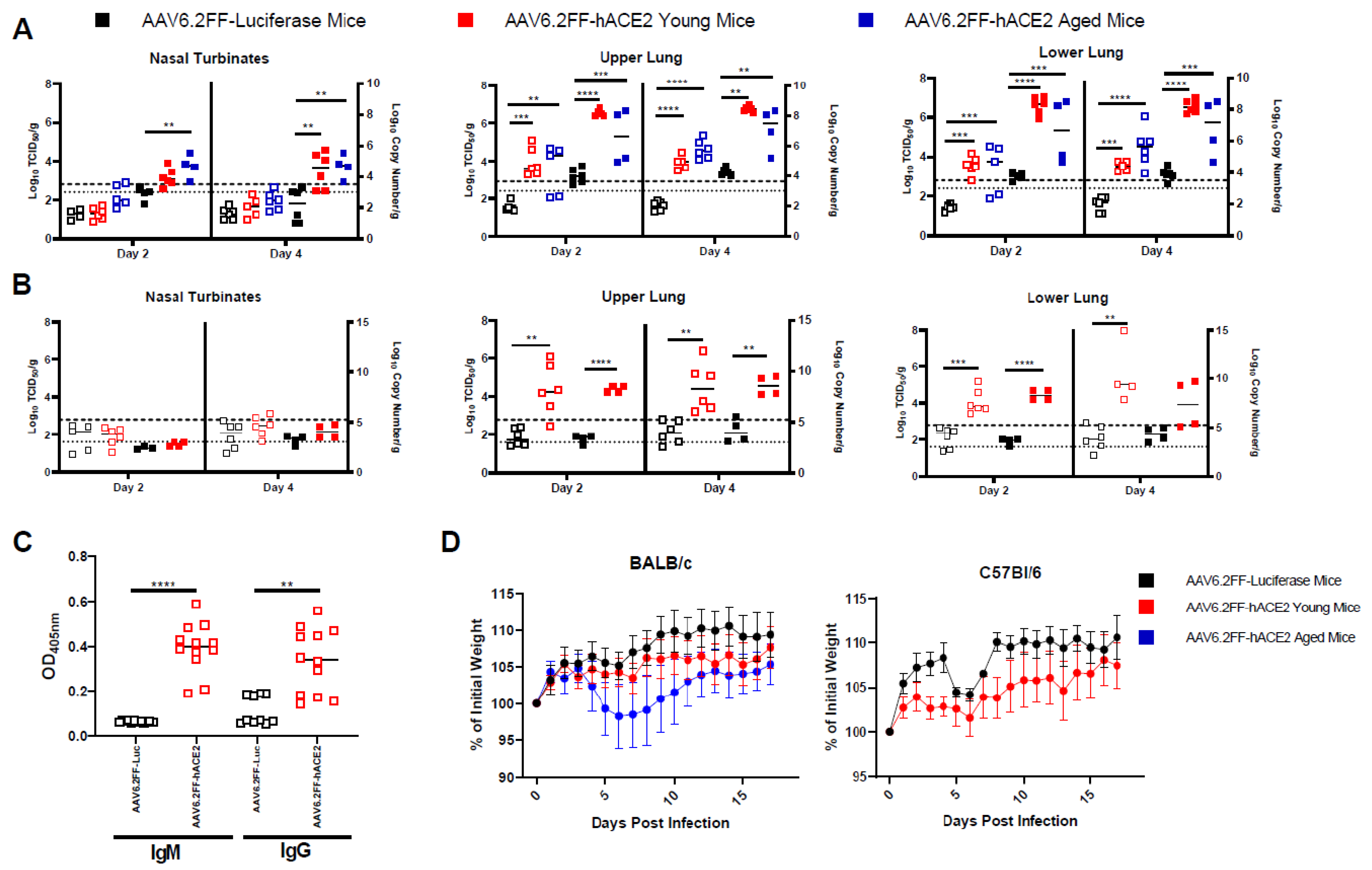
3.2. AAV6.2FF-hACE2-Transduced Mice Are Permissive to SARS-CoV-2 Infection
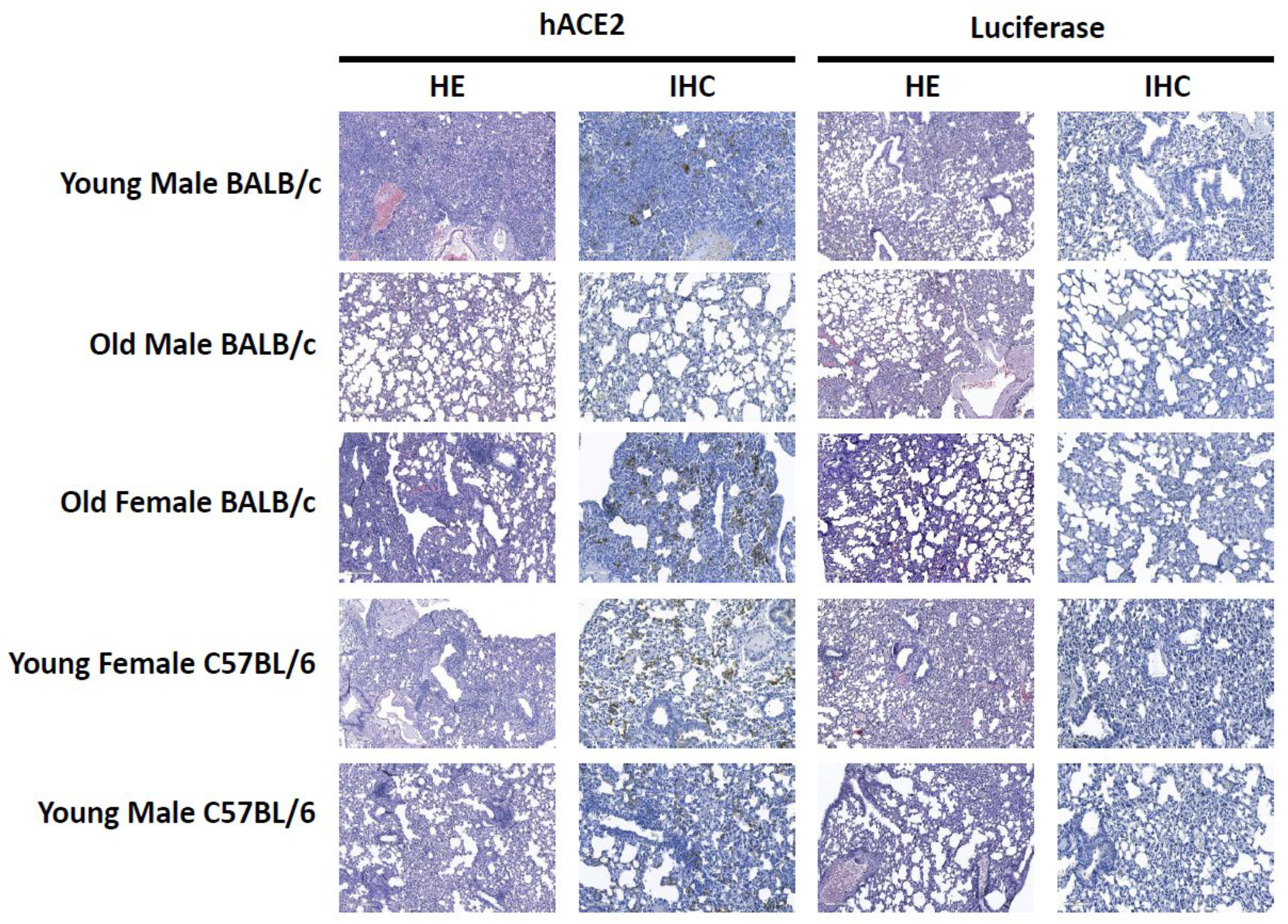
3.3. Histopathology and Immunohistochemistry
3.4. SARS-2-CoV Infection Induces a Modulation of Immune Responses in hACE2-Transduced Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Latinne, A.; Hu, B.; Olival, K.J.; Zhu, G.; Zhang, L.; Li, H.; Chmura, A.A.; Field, H.E.; Zambrana-Torrelio, C.; Epstein, J.H.; et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 2020, 11, 4235. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. mBio 2017, 8, e00373-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petersen, E.; Koopmans, M.; Go, U.; Hamer, D.H.; Petrosillo, N.; Castelli, F.; Storgaard, M.; Al Khalili, S.; Simonsen, L. Comparing SARS-CoV-2 with SARS-CoV and influenza pandemics. Lancet Infect. Dis. 2020, 20, e238–e244. [Google Scholar] [CrossRef]
- Pfefferbaum, B.; North, C.S. Mental Health and the COVID-19 Pandemic. N. Engl. J. Med. 2020, 383, 510–512. [Google Scholar] [CrossRef]
- Imai, M.; Iwatsuki-Horimoto, K.; Hatta, M.; Loeber, S.; Halfmann, P.J.; Nakajima, N.; Watanabe, T.; Ujie, M.; Takahashi, K.; Ito, M.; et al. Syrian hamsters as a small animal model for SARS-CoV-2 infection and countermeasure development. Proc. Natl. Acad. Sci. USA 2020, 117, 16587–16595. [Google Scholar] [CrossRef]
- Rosenke, K.; Meade-White, K.; Letko, M.; Clancy, C.; Hansen, F.; Liu, Y.; Okumura, A.; Tang-Huau, T.L.; Li, R.; Saturday, G.; et al. Defining the Syrian hamster as a highly susceptible preclinical model for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2673–2684. [Google Scholar] [CrossRef]
- Chan, J.F.; Zhang, A.J.; Yuan, S.; Poon, V.K.; Chan, C.C.; Lee, A.C.; Chan, W.M.; Fan, Z.; Tsoi, H.W.; Wen, L.; et al. Simulation of the Clinical and Pathological Manifestations of Coronavirus Disease 2019 (COVID-19) in a Golden Syrian Hamster Model: Implications for Disease Pathogenesis and Transmissibility. Clin. Infect. Dis. 2020, 71, 2428–2446. [Google Scholar] [CrossRef]
- Griffin, B.D.; Chan, M.; Tailor, N.; Mendoza, E.J.; Leung, A.; Warner, B.M.; Duggan, A.T.; Moffat, E.; He, S.; Garnett, L.; et al. SARS-CoV-2 infection and transmission in the North American deer mouse. Nat. Commun. 2021, 12, 3612. [Google Scholar] [CrossRef]
- Kim, Y.I.; Kim, S.G.; Kim, S.M.; Kim, E.H.; Park, S.J.; Yu, K.M.; Chang, J.H.; Kim, E.J.; Lee, S.; Casel, M.A.B.; et al. Infection and Rapid Transmission of SARS-CoV-2 in Ferrets. Cell Host Microbe 2020, 27, 704–709.e2. [Google Scholar] [CrossRef]
- Ryan, K.A.; Bewley, K.R.; Fotheringham, S.A.; Slack, G.S.; Brown, P.; Hall, Y.; Wand, N.I.; Marriott, A.C.; Cavell, B.E.; Tree, J.A.; et al. Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nat. Commun. 2021, 12, 81. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Richard, M.; Kok, A.; de Meulder, D.; Bestebroer, T.M.; Lamers, M.M.; Okba, N.M.A.; Fentener van Vlissingen, M.; Rockx, B.; Haagmans, B.L.; Koopmans, M.P.G.; et al. SARS-CoV-2 is transmitted via contact and via the air between ferrets. Nat. Commun. 2020, 11, 3496. [Google Scholar] [CrossRef]
- Munster, V.J.; Feldmann, F.; Williamson, B.N.; van Doremalen, N.; Pérez-Pérez, L.; Schulz, J.; Meade-White, K.; Okumura, A.; Callison, J.; Brumbaugh, B.; et al. Respiratory disease in rhesus macaques inoculated with SARS-CoV-2. Nature 2020, 585, 268–272. [Google Scholar] [CrossRef]
- Rockx, B.; Kuiken, T.; Herfst, S.; Bestebroer, T.; Lamers, M.M.; Oude Munnink, B.B.; de Meulder, D.; van Amerongen, G.; van den Brand, J.; Okba, N.M.A.; et al. Comparative pathogenesis of COVID-19, MERS, and SARS in a nonhuman primate model. Science 2020, 368, 1012–1015. [Google Scholar] [CrossRef] [Green Version]
- Chandrashekar, A.; Liu, J.; Martinot, A.J.; McMahan, K.; Mercado, N.B.; Peter, L.; Tostanoski, L.H.; Yu, J.; Maliga, Z.; Nekorchuk, M.; et al. SARS-CoV-2 infection protects against rechallenge in rhesus macaques. Science 2020, 369, 812–817. [Google Scholar] [CrossRef]
- Deng, W.; Bao, L.; Liu, J.; Xiao, C.; Xue, J.; Lv, Q.; Qi, F.; Gao, H.; Yu, P.; Xu, Y.; et al. Primary exposure to SARS-CoV-2 protects against reinfection in rhesus macaques. Science 2020, 369, 818–823. [Google Scholar] [CrossRef]
- McMahan, K.; Yu, J.; Mercado, N.B.; Loos, C.; Tostanoski, L.H.; Chandrashekar, A.; Liu, J.; Peter, L.; Atyeo, C.; Zhu, A.; et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature 2020, 590, 630–634. [Google Scholar] [CrossRef]
- Muñoz-Fontela, C.; Dowling, W.E.; Funnell, S.G.P.; Gsell, P.S.; Riveros-Balta, A.X.; Albrecht, R.A.; Andersen, H.; Baric, R.S.; Carroll, M.W.; Cavaleri, M.; et al. Animal models for COVID-19. Nature 2020, 586, 509–515. [Google Scholar] [CrossRef]
- Tare, D.S.; Kode, S.S.; Hurt, A.C.; Pawar, S.D. Assessing the susceptibility of highly pathogenic avian influenza H5N1 viruses to oseltamivir using embryonated chicken eggs. Indian J. Med. Res. 2019, 150, 486–491. [Google Scholar] [PubMed]
- Harding, J.D. Nonhuman Primates and Translational Research: Progress, Opportunities, and Challenges. ILAR J. 2017, 58, 141–150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, E.S.; Bailey, A.L.; Kafai, N.M.; Nair, S.; McCune, B.T.; Yu, J.; Fox, J.M.; Chen, R.E.; Earnest, J.T.; Keeler, S.P.; et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020, 21, 1327–1335. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.H.; Chen, Q.; Gu, H.J.; Yang, G.; Wang, Y.X.; Huang, X.Y.; Liu, S.S.; Zhang, N.N.; Li, X.F.; Xiong, R.; et al. A Mouse Model of SARS-CoV-2 Infection and Pathogenesis. Cell Host Microbe 2020, 28, 124–133.e4. [Google Scholar] [CrossRef]
- Bao, L.; Deng, W.; Huang, B.; Gao, H.; Liu, J.; Ren, L.; Wei, Q.; Yu, P.; Xu, Y.; Qi, F.; et al. The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 2020, 583, 830–833. [Google Scholar] [CrossRef]
- Jiang, R.D.; Liu, M.Q.; Chen, Y.; Shan, C.; Zhou, Y.W.; Shen, X.R.; Li, Q.; Zhang, L.; Zhu, Y.; Si, H.R.; et al. Pathogenesis of SARS-CoV-2 in Transgenic Mice Expressing Human Angiotensin-Converting Enzyme 2. Cell 2020, 182, 50–58.e8. [Google Scholar] [CrossRef]
- Netland, J.; Meyerholz, D.K.; Moore, S.; Cassell, M.; Perlman, S. Severe acute respiratory syndrome coronavirus infection causes neuronal death in the absence of encephalitis in mice transgenic for human ACE2. J. Virol. 2008, 82, 7264–7275. [Google Scholar] [CrossRef] [Green Version]
- Kumari, P.; Rothan, H.A.; Natekar, J.P.; Stone, S.; Pathak, H.; Strate, P.G.; Arora, K.; Brinton, M.A.; Kumar, M. Neuroinvasion and Encephalitis Following Intranasal Inoculation of SARS-CoV-2 in K18-hACE2 Mice. Viruses 2021, 13, 132. [Google Scholar] [CrossRef]
- Carossino, M.; Montanaro, P.; O’Connell, A.; Kenney, D.; Gertje, H.; Grosz, K.A.; Kurnick, S.A.; Bosmann, M.; Saeed, M.; Balasuriya, U.B.R.; et al. Fatal neuroinvasion of SARS-CoV-2 in K18-hACE2 mice is partially dependent on hACE2 expression. bioRxiv 2021. [Google Scholar] [CrossRef]
- Von Weyhern, C.H.; Kaufmann, I.; Neff, F.; Kremer, M. Early evidence of pronounced brain involvement in fatal COVID-19 outcomes. Lancet 2020, 395, e109. [Google Scholar] [CrossRef]
- Dinnon, K.H.; Leist, S.R.; Schäfer, A.; Edwards, C.E.; Martinez, D.R.; Montgomery, S.A.; West, A.; Yount, B.L.; Hou, Y.J.; Adams, L.E.; et al. A mouse-adapted model of SARS-CoV-2 to test COVID-19 countermeasures. Nature 2020, 586, 560–566. [Google Scholar] [CrossRef]
- Zhao, J.; Li, K.; Wohlford-Lenane, C.; Agnihothram, S.S.; Fett, C.; Gale, M.J.; Baric, R.S.; Enjuanes, L.; Gallagher, T.; McCray, P.B.; et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc. Natl. Acad. Sci. USA 2014, 111, 4970–4975. [Google Scholar] [CrossRef] [Green Version]
- Israelow, B.; Song, E.; Mao, T.; Lu, P.; Meir, A.; Liu, F.; Alfajaro, M.M.; Wei, J.; Dong, H.; Homer, R.J.; et al. Mouse model of SARS-CoV-2 reveals inflammatory role of type I interferon signaling. J. Exp. Med. 2020, 217, e20201241. [Google Scholar] [CrossRef]
- Rathnasinghe, R.; Strohmeier, S.; Amanat, F.; Gillespie, V.L.; Krammer, F.; García-Sastre, A.; Coughlan, L.; Schotsaert, M.; Uccellini, M.B. Comparison of transgenic and adenovirus hACE2 mouse models for SARS-CoV-2 infection. Emerg. Microbes Infect. 2020, 9, 2433–2445. [Google Scholar] [CrossRef]
- Hassan, A.O.; Case, J.B.; Winkler, E.S.; Thackray, L.B.; Kafai, N.M.; Bailey, A.L.; McCune, B.T.; Fox, J.M.; Chen, R.E.; Alsoussi, W.B.; et al. A SARS-CoV-2 Infection Model in Mice Demonstrates Protection by Neutralizing Antibodies. Cell 2020, 182, 744–753.e4. [Google Scholar] [CrossRef]
- Han, K.; Blair, R.V.; Iwanaga, N.; Liu, F.; Russell-Lodrigue, K.E.; Qin, Z.; Midkiff, C.C.; Golden, N.A.; Doyle-Meyers, L.A.; Kabir, M.E.; et al. Lung Expression of Human Angiotensin-Converting Enzyme 2 Sensitizes the Mouse to SARS-CoV-2 Infection. Am. J. Respir. Cell Mol. Biol. 2021, 64, 79–88. [Google Scholar] [CrossRef]
- Van Lieshout, L.P.; Domm, J.M.; Rindler, T.N.; Frost, K.L.; Sorensen, D.L.; Medina, S.J.; Booth, S.A.; Bridges, J.P.; Wootton, S.K. A Novel Triple-Mutant AAV6 Capsid Induces Rapid and Potent Transgene Expression in the Muscle and Respiratory Tract of Mice. Mol. Ther. Methods Clin. Dev. 2018, 9, 323–329. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.H.; van Lieshout, L.P.; Xu, L.; Domm, J.M.; Vadivel, A.; Renesme, L.; Mühlfeld, C.; Hurskainen, M.; Mižíková, I.; Pei, Y.; et al. A lung tropic AAV vector improves survival in a mouse model of surfactant B deficiency. Nat. Commun. 2020, 11, 3929. [Google Scholar] [CrossRef]
- DeMatteo, R.P.; Chu, G.; Ahn, M.; Chang, E.; Barker, C.F.; Markmann, J.F. Long-lasting adenovirus transgene expression in mice through neonatal intrathymic tolerance induction without the use of immunosuppression. J. Virol. 1997, 71, 5330–5335. [Google Scholar] [CrossRef]
- Balazs, A.B.; Chen, J.; Hong, C.M.; Rao, D.S.; Yang, L.; Baltimore, D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature 2011, 481, 81–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gary, E.N.; Warner, B.M.; Parzych, E.M.; Griffin, B.D.; Zhu, X.; Tailor, N.; Tursi, N.J.; Chan, M.; Purwar, M.; Vendramelli, R.; et al. A novel mouse AAV6 hACE2 transduction model of wild-type SARS-CoV-2 infection studied using synDNA immunogens. iScience 2021, 24, 102699. [Google Scholar] [CrossRef] [PubMed]
- Van Lieshout, L.P.; Domm, J.M.; Wootton, S.K. AAV-Mediated Gene Delivery to the Lung. Methods Mol. Biol. 2019, 1950, 361–372. [Google Scholar] [PubMed]
- Aurnhammer, C.; Haase, M.; Muether, N.; Hausl, M.; Rauschhuber, C.; Huber, I.; Nitschko, H.; Busch, U.; Sing, A.; Ehrhardt, A.; et al. Universal real-time PCR for the detection and quantification of adeno-associated virus serotype 2-derived inverted terminal repeat sequences. Hum. Gene Ther. Methods 2012, 23, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Rghei, A.D.; Stevens, B.A.Y.; Thomas, S.P.; Yates, J.G.E.; McLeod, B.M.; Karimi, K.; Susta, L.; Bridle, B.W.; Wootton, S.K. Production of Adeno-Associated Virus Vectors in Cell Stacks for Preclinical Studies in Large Animal Models. JoVE (J. Vis. Exp.) 2021, 172, e62727. [Google Scholar] [CrossRef]
- Santry, L.A.; Ingrao, J.C.; Darrick, L.Y.; de Jong, J.G.; van Lieshout, L.P.; Wood, G.A.; Wootton, S.K. AAV vector distribution in the mouse respiratory tract following four different methods of administration. BMC Biotechnol. 2017, 17, 43. [Google Scholar] [CrossRef] [Green Version]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Corman, V.M.; Landt, O.; Kaiser, M.; Molenkamp, R.; Meijer, A.; Chu, D.K.; Bleicker, T.; Brünink, S.; Schneider, J.; Schmidt, M.L.; et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020, 25, 2000045. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Li, S.; Li, X.; Pei, D.; Liu, Y.; Ding, Y.; Liu, L.; Bi, H.; Shi, X.; Guo, Y.; et al. Development of an Adeno-Associated Virus-Vectored SARS-CoV-2 Vaccine and Its Immunogenicity in Mice. Front. Cell. Infect. Microbiol. 2022, 192, 802147. [Google Scholar] [CrossRef]
- Yang, M.S.; Park, M.J.; Lee, J.; Oh, B.; Kang, K.W.; Kim, Y.; Lee, S.M.; Lim, J.O.; Jung, T.Y.; Park, J.H.; et al. Non-invasive administration of AAV to target lung parenchymal cells and develop SARS-CoV-2-susceptible mice. Mol. Ther. 2022, 30, 1994–2004. [Google Scholar] [CrossRef]
- Oladunni, F.S.; Park, J.G.; Pino, P.A.; Gonzalez, O.; Akhter, A.; Allué-Guardia, A.; Olmo-Fontánez, A.; Gautam, S.; Garcia-Vilanova, A.; Ye, C.; et al. Lethality of SARS-CoV-2 infection in K18 human angiotensin-converting enzyme 2 transgenic mice. Nat. Commun. 2020, 11, 6122. [Google Scholar] [CrossRef]
- Forero, C.; Ritter, J.M.; Seixas, J.N.; Coleman-McCray, J.D.; Brake, M.; Condrey, J.A.; Tansey, C.; Welch, S.R.; Genzer, S.C.; Spengler, J.R. Volume-Associated Clinical and Histopathological Effects of Intranasal Instillation in Syrian Hamsters: Considerations for Infection and Therapeutic Studies. Pathogens 2022, 11, 898. [Google Scholar] [CrossRef]
- O’Driscoll, M.; Ribeiro Dos Santos, G.; Wang, L.; Cummings, D.A.; Azman, A.S.; Paireau, J.; Fontanet, A.; Cauchemez, S.; Salje, H. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature 2020, 590, 140–145. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tailor, N.; Warner, B.M.; Griffin, B.D.; Tierney, K.; Moffat, E.; Frost, K.; Vendramelli, R.; Leung, A.; Willman, M.; Thomas, S.P.; et al. Generation and Characterization of a SARS-CoV-2-Susceptible Mouse Model Using Adeno-Associated Virus (AAV6.2FF)-Mediated Respiratory Delivery of the Human ACE2 Gene. Viruses 2023, 15, 85. https://doi.org/10.3390/v15010085
Tailor N, Warner BM, Griffin BD, Tierney K, Moffat E, Frost K, Vendramelli R, Leung A, Willman M, Thomas SP, et al. Generation and Characterization of a SARS-CoV-2-Susceptible Mouse Model Using Adeno-Associated Virus (AAV6.2FF)-Mediated Respiratory Delivery of the Human ACE2 Gene. Viruses. 2023; 15(1):85. https://doi.org/10.3390/v15010085
Chicago/Turabian StyleTailor, Nikesh, Bryce M. Warner, Bryan D. Griffin, Kevin Tierney, Estella Moffat, Kathy Frost, Robert Vendramelli, Anders Leung, Marnie Willman, Sylvia P. Thomas, and et al. 2023. "Generation and Characterization of a SARS-CoV-2-Susceptible Mouse Model Using Adeno-Associated Virus (AAV6.2FF)-Mediated Respiratory Delivery of the Human ACE2 Gene" Viruses 15, no. 1: 85. https://doi.org/10.3390/v15010085
APA StyleTailor, N., Warner, B. M., Griffin, B. D., Tierney, K., Moffat, E., Frost, K., Vendramelli, R., Leung, A., Willman, M., Thomas, S. P., Pei, Y., Booth, S. A., Embury-Hyatt, C., Wootton, S. K., & Kobasa, D. (2023). Generation and Characterization of a SARS-CoV-2-Susceptible Mouse Model Using Adeno-Associated Virus (AAV6.2FF)-Mediated Respiratory Delivery of the Human ACE2 Gene. Viruses, 15(1), 85. https://doi.org/10.3390/v15010085




