Viral Entry Inhibitors Protect against SARS-CoV-2-Induced Neurite Shortening in Differentiated SH-SY5Y Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Amplification of Belgian SARS-CoV-2 Strain and Infection Paradigms
2.3. RNA Extraction and RT-qPCR
2.4. Protein Extraction and Resolution
2.5. Cell Viability Assays
2.6. Neuropeptide Y Assay
2.7. Fluorescent In Situ Hybridization/(Immuno)Staining
2.8. Immunofluorescence
2.9. Electron Microscopy
2.10. Image Analysis
2.11. Statistics
3. Results
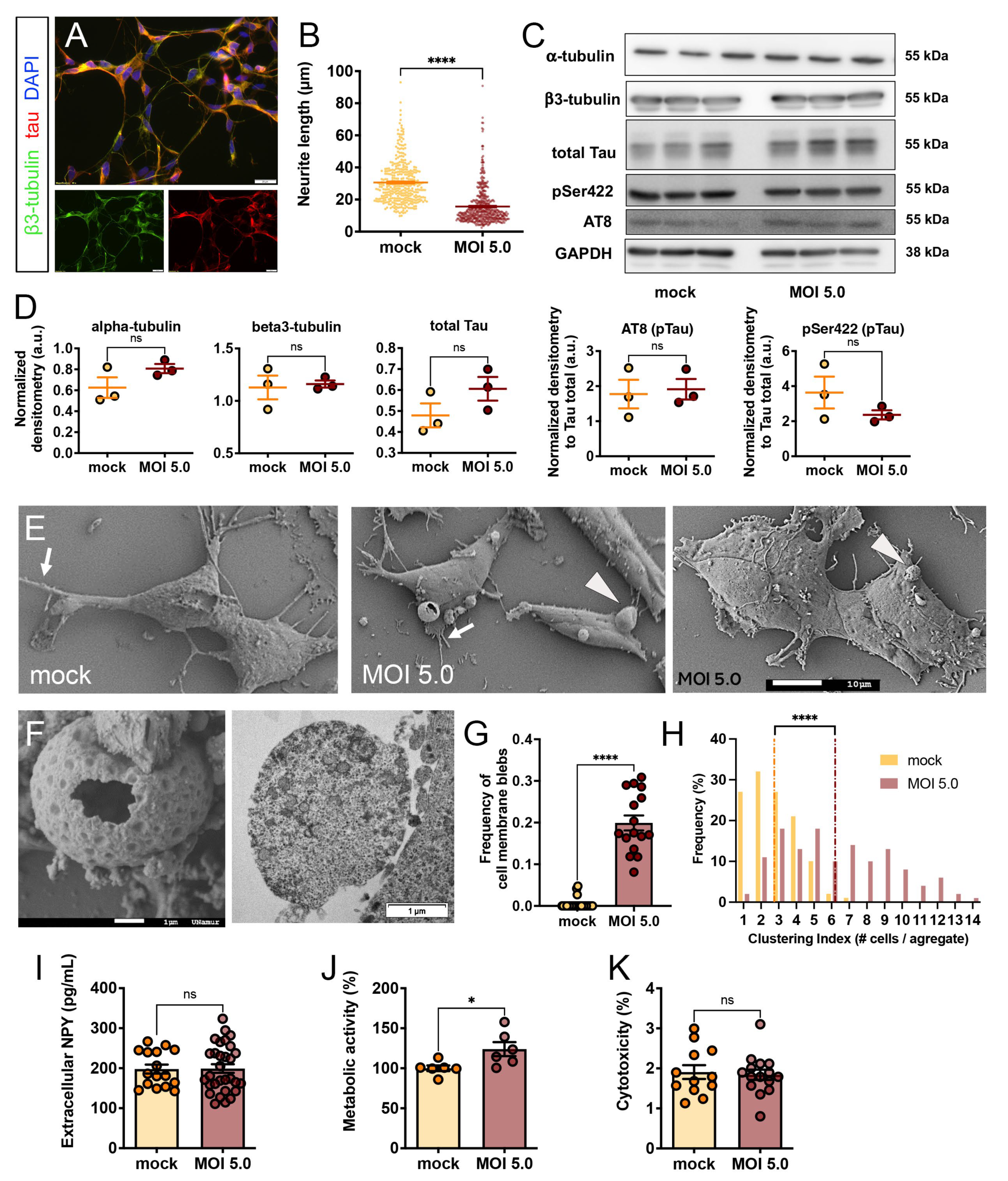
3.1. Differentiated SH-SY5Y Were Susceptible, but Not Permissive, to SARS-CoV-2
3.2. SARS-CoV-2 Induced Morphological Cytopathic Effects on SH-SY5Y Cell Body and Neurites
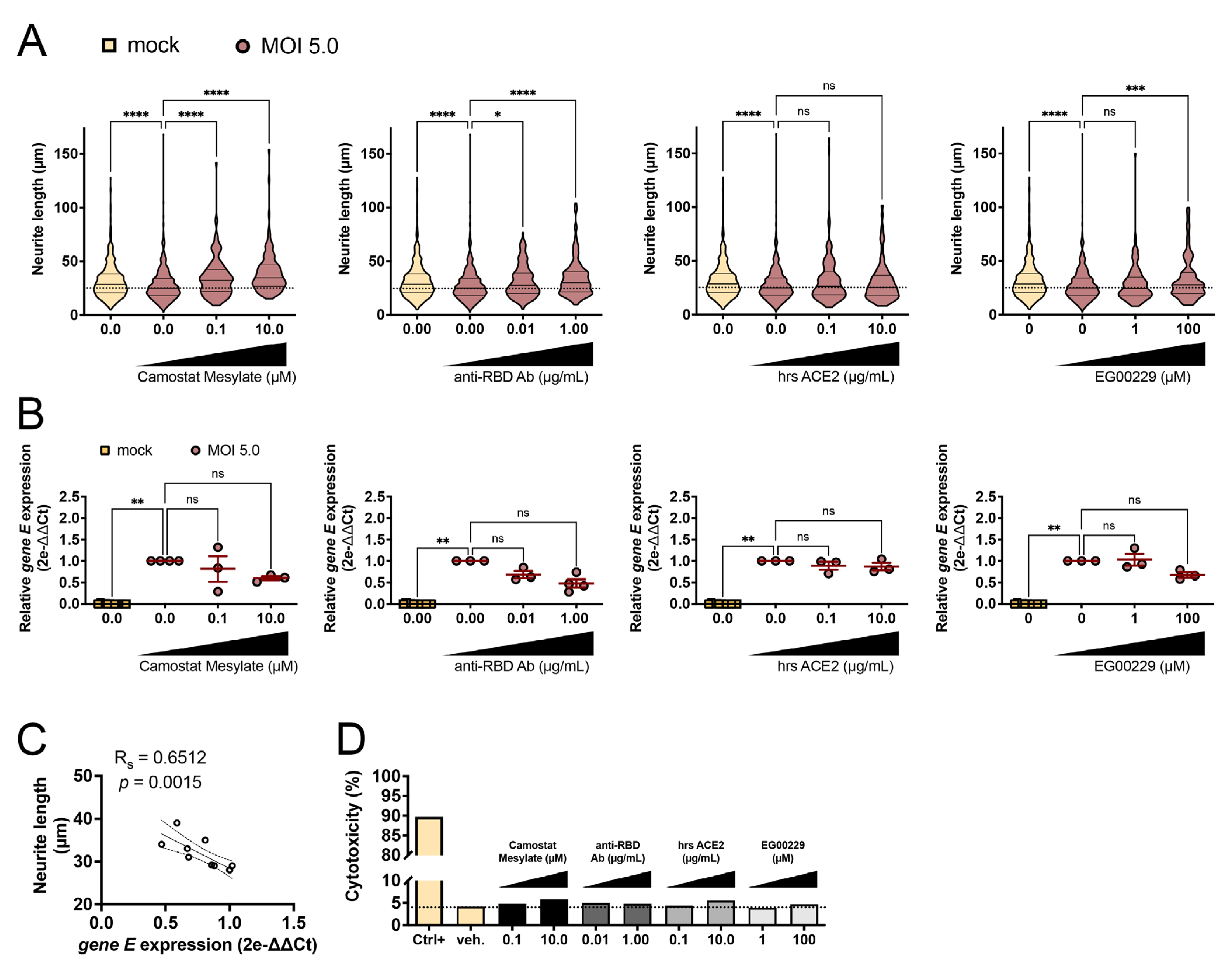
3.3. SARS-CoV-2-Induced Neurite Shortening Was Prevented by TMPRSS2, RBD, and NRP1 Inhibitors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, T.J.; Qasem, A.; Abdelli, L.S.; Naser, S.A. Extra-Pulmonary Complications in SARS-CoV-2 Infection: A Comprehensive Multi Organ-System Review. Microorganisms 2022, 10, 153. [Google Scholar] [CrossRef]
- Spudich, S.; Nath, A. Nervous System Consequences of COVID-19. Science 2022, 375, 267–269. [Google Scholar] [CrossRef] [PubMed]
- Douaud, G.; Lee, S.; Alfaro-Almagro, F.; Arthofer, C.; Wang, C.; McCarthy, P.; Lange, F.; Andersson, J.L.R.; Griffanti, L.; Duff, E.; et al. SARS-CoV-2 Is Associated with Changes in Brain Structure in UK Biobank. Nature 2022, 604, 697–707. [Google Scholar] [CrossRef] [PubMed]
- Hampshire, A.; Trender, W.; Chamberlain, S.R.; Jolly, A.E.; Grant, J.E.; Patrick, F.; Mazibuko, N.; Williams, S.C.; Barnby, J.M.; Hellyer, P.; et al. Cognitive Deficits in People Who Have Recovered from COVID-19. EClinicalMedicine 2021, 39, 101044. [Google Scholar] [CrossRef] [PubMed]
- Hosp, J.A.; Dressing, A.; Blazhenets, G.; Bormann, T.; Rau, A.; Schwabenland, M.; Thurow, J.; Wagner, D.; Waller, C.; Niesen, W.D.; et al. Cognitive impairment and Altered Cerebral Glucose Metabolism in the Subacute Stage of COVID-19. Brain 2021, 144, 1263–1276. [Google Scholar] [CrossRef] [PubMed]
- Castanares-Zapatero, D.; Chalon, P.; Kohn, L.; Dauvrin, M.; Detollenaere, J.; Maertens de Noordhout, C.; Primus-de Jong, C.; Cleemput, I.; Van den Heede, K. Pathophysiology and Mechanism of long COVID: A Comprehensive Review. Ann. Med. 2022, 54, 1473–1487. [Google Scholar] [CrossRef] [PubMed]
- Puelles, V.G.; Lütgehetmann, M.; Lindenmeyer, M.T.; Sperhake, J.P.; Wong, M.N.; Allweiss, L.; Chilla, S.; Heinemann, A.; Wanner, N.; Liu, S.; et al. Multiorgan and Renal Tropism of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 590–592. [Google Scholar] [CrossRef] [PubMed]
- Song, E.; Zhang, C.; Israelow, B.; Lu-Culligan, A.; Prado, A.V.; Skriabine, S.; Lu, P.; Weizman, O.E.; Liu, F.; Dai, Y.; et al. Neuroinvasion of SARS-CoV-2 in Human and Mouse Brain. J. Exp. Med. 2021, 218, e20202135. [Google Scholar] [CrossRef]
- Rutkai, I.; Mayer, M.G.; Hellmers, L.M.; Ning, B.; Huang, Z.; Monjure, C.J.; Coyne, C.; Silvestri, R.; Golden, N.; Hensley, K.; et al. Neuropathology and Virus in Brain of SARS-CoV-2 Infected Non-Human Primates. Nat. Commun. 2022, 13, 1745. [Google Scholar] [CrossRef]
- Seehusen, F.; Clark, J.J.; Sharma, P.; Bentley, E.G.; Kirby, A.; Subramaniam, K.; Wunderlin-Giuliani, S.; Hughes, G.L.; Patterson, E.I.; Michael, B.D.; et al. Neuroinvasion and Neurotropism by SARS-CoV-2 Variants in the K18-Hace2 Mouse. Viruses 2022, 14, 1020. [Google Scholar] [CrossRef]
- Andrews, M.G.; Mukhtar, T.; Eze, U.C.; Simoneau, C.R.; Ross, J.; Parikshak, N.; Wang, S.; Zhou, L.; Koontz, M.; Velmeshev, D.; et al. Tropism of SARS-CoV-2 for Human Cortical Astrocytes. Proc. Natl. Acad. Sci. USA 2022, 119, e2122236119. [Google Scholar] [CrossRef] [PubMed]
- Jacob, F.; Pather, S.R.; Huang, W.K.; Zhang, F.; Wong, S.Z.H.; Zhou, H.; Cubitt, B.; Fan, W.; Chen, C.Z.; Xu, M.; et al. Human Pluripotent Stem Cell-Derived Neural Cells and Brain Organoids Reveal SARS-CoV-2 Neurotropism Predominates in Choroid Plexus Epithelium. Cell Stem Cell 2020, 27, 937–950.e9. [Google Scholar] [CrossRef] [PubMed]
- Bullen, C.K.; Hogberg, H.T.; Bahadirli-Talbott, A.; Bishai, W.R.; Hartung, T.; Keuthan, C.; Looney, M.M.; Pekosz, A.; Romero, J.C.; Sillé, F.C.M.; et al. Infectability of Human BrainSphere Neurons Suggests Neurotropism of SARS-CoV-2. Altex 2020, 37, 665–671. [Google Scholar] [CrossRef] [PubMed]
- Ramani, A.; Müller, L.; Ostermann, P.N.; Gabriel, E.; Abida-Islam, P.; Müller-Schiffmann, A.; Mariappan, A.; Goureau, O.; Gruell, H.; Walker, A.; et al. SARS-CoV-2 Targets Neurons of 3D Human Brain Organoids. EMBO J. 2020, 39, e106230. [Google Scholar] [CrossRef]
- Yang, L.; Han, Y.; Nilsson-Payant, B.E.; Gupta, V.; Wang, P.; Duan, X.; Tang, X.; Zhu, J.; Zhao, Z.; Jaffre, F.; et al. A Human Pluripotent Stem Cell-Based Platform to Study SARS-CoV-2 Tropism and Model Virus Infection in Human Cells and Organoids. Cell Stem Cell 2020, 27, 125–136.e7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.Z.; Chu, H.; Han, S.; Shuai, H.; Deng, J.; Hu, Y.F.; Gong, H.R.; Lee, A.C.; Zou, Z.; Yau, T.; et al. SARS-CoV-2 Infects Human Neural Progenitor Cells and Brain Organoids. Cell Res. 2020, 30, 928–931. [Google Scholar] [CrossRef]
- Bauer, L.; Lendemeijer, B.; Leijten, L.; Embregts, C.W.E.; Rockx, B.; Kushner, S.A.; de Vrij, F.M.S.; van Riel, D. Replication Kinetics, Cell Tropism, and Associated Immune Responses in SARS-CoV-2- and H5N1 Virus-Infected Human Induced Pluripotent Stem Cell-Derived Neural Models. mSphere 2021, 6, e0027021. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.M.; Tan, B.H.; Wu, S.; Gui, Y.; Suo, J.L.; Li, Y.C. Evidence of Central Nervous System Infection and Neuroinvasive Routes, as well as Neurological Involvement, in the Lethality of SARS-CoV-2 Infection. J. Med. Virol. 2021, 93, 1304–1313. [Google Scholar] [CrossRef]
- Mesci, P.; de Souza, J.S.; Martin-Sancho, L.; Macia, A.; Saleh, A.; Yin, X.; Snethlage, C.; Adams, J.W.; Avansini, S.H.; Herai, R.H.; et al. SARS-CoV-2 Infects Human Brain Organoids Causing Cell Death and Loss of Synapses That Can Be Rescued by Treatment with Sofosbuvir. PLoS Biol. 2022, 20, e3001845. [Google Scholar] [CrossRef]
- Tiwari, S.K.; Wang, S.; Smith, D.; Carlin, A.F.; Rana, T.M. Revealing Tissue-Specific SARS-CoV-2 Infection and Host Responses using Human Stem Cell-Derived Lung and Cerebral Organoids. Stem Cell Rep. 2021, 16, 437–445. [Google Scholar] [CrossRef]
- Bauer, L.; Rissmann, M.; Benavides, F.F.W.; Leijten, L.; van Run, P.; Begeman, L.; Veldhuis Kroeze, E.J.B.; Lendemeijer, B.; Smeenk, H.; de Vrij, F.M.S.; et al. In Vitro and In Vivo Differences in Neurovirulence between D614G, Delta And Omicron BA.1 SARS-CoV-2 Variants. Acta Neuropathol. Commun. 2022, 10, 124. [Google Scholar] [CrossRef] [PubMed]
- Ostermann, P.N.; Schaal, H. Human Brain Organoids to Explore SARS-CoV-2-Induced Effects on the Central Nervous System. Rev. Med. Virol. 2023, 33, e2430. [Google Scholar] [CrossRef] [PubMed]
- Sanclemente-Alaman, I.; Moreno-Jimenez, L.; Benito-Martin, M.S.; Canales-Aguirre, A.; Matias-Guiu, J.A.; Matias-Guiu, J.; Gomez-Pinedo, U. Experimental Models for the Study of Central Nervous System Infection by SARS-CoV-2. Front. Immunol. 2020, 11, 2163. [Google Scholar] [CrossRef] [PubMed]
- Bartolomeo, C.S.; Lemes, R.M.R.; Morais, R.L.; Pereria, G.C.; Nunes, T.A.; Costa, A.J.; de Barros Maciel, R.M.; Braconi, C.T.; Maricato, J.T.; Janini, L.M.R.; et al. SARS-CoV-2 Infection and Replication Kinetics in Different Human Cell Types: The role of autophagy, cellular metabolism and ACE2 expression. Life Sci. 2022, 308, 120930. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Li, W.; Bao, J.; Peng, Q.; Wen, D.; Wang, J.; Sun, B. The Expression of SARS-CoV-2 Receptor ACE2 and CD147, and Protease TMPRSS2 in Human and Mouse Brain Cells and Mouse Brain Tissues. Biochem. Biophys. Res. Commun. 2020, 533, 867–871. [Google Scholar] [CrossRef]
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Cantuti-Castelvetri, L.; Ojha, R.; Pedro, L.D.; Djannatian, M.; Franz, J.; Kuivanen, S.; van der Meer, F.; Kallio, K.; Kaya, T.; Anastasina, M.; et al. Neuropilin-1 Facilitates SARS-CoV-2 Cell Entry and Infectivity. Science 2020, 370, 856–860. [Google Scholar] [CrossRef]
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Anton-Plagaro, C.; Shoemark, D.K.; Simon-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 Is a Host Factor for SARS-CoV-2 Infection. Science 2020, 370, 861–865. [Google Scholar] [CrossRef]
- Kettunen, P.; Lesnikova, A.; Rasanen, N.; Ojha, R.; Palmunen, L.; Laakso, M.; Lehtonen, S.; Kuusisto, J.; Pietilainen, O.; Saber, S.H.; et al. SARS-CoV-2 Infection of Human Neurons Is TMPRSS2 Independent, Requires Endosomal Cell Entry, and Can Be Blocked by Inhibitors of Host Phosphoinositol-5 Kinase. J. Virol. 2023, 97, e0014423. [Google Scholar] [CrossRef]
- Bielarz, V.; Willemart, K.; Avalosse, N.; De Swert, K.; Lotfi, R.; Lejeune, N.; Poulain, F.; Ninanne, N.; Gilloteaux, J.; Gillet, N.; et al. Susceptibility of Neuroblastoma and Glioblastoma Cell Lines to SARS-CoV-2 Infection. Brain Res. 2021, 1758, 147344. [Google Scholar] [CrossRef] [PubMed]
- Benedetti, F.; Silvestri, G.; Mavian, C.; Weichseldorfer, M.; Munawwar, A.; Cash, M.N.; Dulcey, M.; Vittor, A.Y.; Ciccozzi, M.; Salemi, M.; et al. Comparison of SARS-CoV-2 Receptors Expression in Primary Endothelial Cells and Retinoic Acid-Differentiated Human Neuronal Cells. Viruses 2021, 13, 2193. [Google Scholar] [CrossRef] [PubMed]
- Coupeau, D.; Burton, N.; Lejeune, N.; Loret, S.; Petit, A.; Pejakovic, S.; Poulain, F.; Bonil, L.; Trozzi, G.; Wiggers, L.; et al. SARS-CoV-2 Detection for Diagnosis Purposes in the Setting of a Molecular Biology Research Lab. Methods Protoc. 2020, 3, 59. [Google Scholar] [CrossRef] [PubMed]
- Sprimont, L.; Janssen, P.; De Swert, K.; Van Bulck, M.; Rooman, I.; Gilloteaux, J.; Massie, A.; Nicaise, C. Cystine-Glutamate Antiporter Deletion Accelerates Motor Recovery and Improves Histological Outcomes Following Spinal Cord Injury in Mice. Sci. Rep. 2021, 11, 12227. [Google Scholar] [CrossRef] [PubMed]
- Shayegan, A.; Zucchi, A.; De Swert, K.; Balau, B.; Truyens, C.; Nicaise, C. Lipoteichoic acid Stimulates the Proliferation, Migration and Cytokine Production of Adult Dental Pulp Stem Cells without Affecting Osteogenic Differentiation. Int. Endod. J. 2021, 54, 585–600. [Google Scholar] [CrossRef] [PubMed]
- Bouchat, J.; Gilloteaux, J.; Suain, V.; Van Vlaender, D.; Brion, J.P.; Nicaise, C. Ultrastructural Analysis of Thalamus Damages in a Mouse Model of Osmotic-Induced Demyelination. Neurotox. Res. 2019, 36, 144–162. [Google Scholar] [CrossRef] [PubMed]
- Pool, M.; Thiemann, J.; Bar-Or, A.; Fournier, A.E. NeuriteTracer: A Novel ImageJ Plugin for Automated Quantification of Neurite Outgrowth. J. Neurosci. Methods 2008, 168, 134–139. [Google Scholar] [CrossRef]
- Pepe, A.; Pietropaoli, S.; Vos, M.; Barba-Spaeth, G.; Zurzolo, C. Tunneling Nanotubes Provide a Route for SARS-CoV-2 Spreading. Sci. Adv. 2022, 8, eabo0171. [Google Scholar] [CrossRef]
- Devlin, L.; Gombolay, G.Y. Cerebrospinal Fluid Cytokines in COVID-19: A Review and Meta-Analysis. J. Neurol. 2023. [Google Scholar] [CrossRef]
- Hopfer, H.; Herzig, M.C.; Gosert, R.; Menter, T.; Hench, J.; Tzankov, A.; Hirsch, H.H.; Miller, S.E. Hunting Coronavirus by Transmission Electron Microscopy—A Guide to SARS-CoV-2-Associated Ultrastructural Pathology in COVID-19 Tissues. Histopathology 2021, 78, 358–370. [Google Scholar] [CrossRef]
- Williams, C.G.; Jureka, A.S.; Silvas, J.A.; Nicolini, A.M.; Chvatal, S.A.; Carlson-Stevermer, J.; Oki, J.; Holden, K.; Basler, C.F. Inhibitors of VPS34 and Fatty-Acid Metabolism Suppress SARS-CoV-2 Replication. Cell Rep. 2021, 36, 109479. [Google Scholar] [CrossRef] [PubMed]
- Nardacci, R.; Colavita, F.; Castilletti, C.; Lapa, D.; Matusali, G.; Meschi, S.; Del Nonno, F.; Colombo, D.; Capobianchi, M.R.; Zumla, A.; et al. Evidences for Lipid Involvement in SARS-CoV-2 Cytopathogenesis. Cell Death Dis. 2021, 12, 263. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Soares, V.C.; Ferreira, A.C.; Sacramento, C.Q.; Fintelman-Rodrigues, N.; Temerozo, J.R.; Teixeira, L.; Nunes da Silva, M.A.; Barreto, E.; Mattos, M.; et al. Lipid Droplets Fuel SARS-CoV-2 Replication and Production of Inflammatory Mediators. PLoS Pathog. 2020, 16, e1009127. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, N.; Fabiano, M.; Muller, U.C.; Walter, J. Carboxy-Terminal Fragment of Amyloid Precursor Protein Mediates Lipid Droplet Accumulation upon Gamma-Secretase Inhibition. Biochem. Biophys. Res. Commun. 2021, 570, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary Manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Bojkova, D.; Reus, P.; Panosch, L.; Bechtel, M.; Rothenburger, T.; Kandler, J.D.; Pfeiffer, A.; Wagner, J.U.G.; Shumliakivska, M.; Dimmeler, S.; et al. Identification of Novel Antiviral Drug Candidates Using an Optimized SARS-CoV-2 Phenotypic Screening Platform. iScience 2023, 26, 105944. [Google Scholar] [CrossRef] [PubMed]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Silva, L.S.; Nogueira, M.H.; Antunes, A.S.L.M.; Vendramini, P.H.; Valenca, A.G.F.; Brandao-Teles, C.; Zuccoli, G.D.S.; et al. Morphological, Cellular, and Molecular Basis of Brain Infection in COVID-19 Patients. Proc. Natl. Acad. Sci. USA 2022, 119, e2200960119. [Google Scholar] [CrossRef] [PubMed]
- Crunfli, F.; Carregari, V.C.; Veras, F.P.; Vendramini, P.H.; Valenca, A.G.F.; Antunes, A.S.L.M.; Brandao-Teles, C.; Zuccoli, G.D.S.; Reis-de-Oliveira, G.; Silva-Costa, L.C.; et al. SARS-CoV-2 Infects Brain Astrocytes of COVID-19 Patients and Impairs Neuronal Viability. medRxiv 2020. [Google Scholar] [CrossRef]
- Yuan, S.; Peng, L.; Park, J.J.; Hu, Y.; Devarkar, S.C.; Dong, M.B.; Shen, Q.; Wu, S.; Chen, S.; Lomakin, I.B.; et al. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Mol. Cell 2020, 80, 1055–1066.e6. [Google Scholar] [CrossRef]
- Hou, Y.; Li, C.; Yoon, C.; Leung, O.W.; You, S.; Cui, X.; Chan, J.F.; Pei, D.; Cheung, H.H.; Chu, H. Enhanced Replication of SARS-CoV-2 Omicron BA.2 in Human Forebrain and Midbrain Organoids. Signal Transduct. Target. Ther. 2022, 7, 381. [Google Scholar] [CrossRef]
- Eden, A.; Grahn, A.; Bremell, D.; Aghvanyan, A.; Bathala, P.; Fuchs, D.; Gostner, J.; Hagberg, L.; Kanberg, N.; Kanjananimmanont, S.; et al. Viral Antigen and Inflammatory Biomarkers in Cerebrospinal Fluid in Patients With COVID-19 Infection and Neurologic Symptoms Compared With Control Participants Without Infection or Neurologic Symptoms. JAMA Netw. Open 2022, 5, e2213253. [Google Scholar] [CrossRef] [PubMed]
- Prajapati, P.; Sripada, L.; Singh, K.; Bhatelia, K.; Singh, R.; Singh, R. TNF-alpha Regulates miRNA Targeting Mitochondrial Complex-I and Induces Cell Death in Dopaminergic Cells. Biochim. Biophys. Acta 2015, 1852, 451–461. [Google Scholar] [CrossRef]
- Seitz, T.; Setz, C.; Rauch, P.; Schubert, U.; Hellerbrand, C. Lipid Accumulation in Host Cells Promotes SARS-CoV-2 Replication. Viruses 2023, 15, 1026. [Google Scholar] [CrossRef] [PubMed]
- Dias, S.S.G.; Cunha-Fernandes, T.; Souza-Moreira, L.; Soares, V.C.; Lima, G.B.; Azevedo-Quintanilha, I.G.; Santos, J.; Pereira-Dutra, F.; Freitas, C.; Reis, P.A.; et al. Metabolic Reprogramming and Lipid Droplets Are Involved in Zika Virus Replication in Neural Cells. J. Neuroinflammation 2023, 20, 61. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yuan, H.; Li, X.; Wang, H. Spike Protein Mediated Membrane Fusion during SARS-CoV-2 Infection. J. Med. Virol. 2023, 95, e28212. [Google Scholar] [CrossRef] [PubMed]
- Reuter, N.; Chen, X.; Kropff, B.; Peter, A.S.; Britt, W.J.; Mach, M.; Uberla, K.; Thomas, M. SARS-CoV-2 Spike Protein Is Capable of Inducing Cell-Cell Fusions Independent from Its Receptor ACE2 and This Activity Can Be Impaired by Furin Inhibitors or a Subset of Monoclonal Antibodies. Viruses 2023, 15, 1500. [Google Scholar] [CrossRef] [PubMed]
- Charras, G.; Paluch, E. Blebs Lead the Way: How to Migrate without Lamellipodia. Nat. Rev. Mol. Cell Biol. 2008, 9, 730–736. [Google Scholar] [CrossRef]
- Paluch, E.K.; Raz, E. The Role and Regulation of Blebs in Cell Migration. Curr. Opin. Cell Biol. 2013, 25, 582–590. [Google Scholar] [CrossRef]
- Wickman, G.R.; Julian, L.; Mardilovich, K.; Schumacher, S.; Munro, J.; Rath, N.; Zander, S.A.; Mleczak, A.; Sumpton, D.; Morrice, N.; et al. Blebs Produced by Actin-Myosin Contraction during Apoptosis Release Damage-Associated Molecular Pattern Proteins before Secondary Necrosis Occurs. Cell Death Differ. 2013, 20, 1293–1305. [Google Scholar] [CrossRef]
- Yang, R.C.; Huang, K.; Zhang, H.P.; Li, L.; Tan, C.; Chen, H.C.; Jin, M.L.; Wang, X.R. Transcriptional Landscape Of Human Neuroblastoma Cells in Response to SARS-CoV-2. BMC Neurosci. 2022, 23, 43. [Google Scholar] [CrossRef]
- Di Primio, P.; Quaranta, P.; Mignanelli, M.; Siano, G.; Bimbati, M.; Piazza, C.R.; Spezia, P.G.; Perrera, P.; Basolo, F.; Poma, A.M.; et al. SARS-CoV-2 Infection Leads to Tau Pathological Signature in Neurons. bioRxiv 2023. [Google Scholar] [CrossRef]
- Datta, G.; Miller, N.M.; Halcrow, P.W.; Khan, N.; Colwell, T.; Geiger, J.D.; Chen, X. SARS-CoV-2 S1 Protein Induces Endolysosome Dysfunction and Neuritic Dystrophy. Front. Cell. Neurosci. 2021, 15, 777738. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Marmol, R.; Giordano-Santini, R.; Kaulich, E.; Cho, A.N.; Przybyla, M.; Riyadh, M.A.; Robinson, E.; Chew, K.Y.; Amor, R.; Meunier, F.A.; et al. SARS-CoV-2 infection and Viral Fusogens Cause Neuronal and Glial Fusion That Compromises Neuronal Activity. Sci. Adv. 2023, 9, eadg2248. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Rubin, L.; Zhou, Z.; Zhang, H.; Su, Q.; Hou, S.T.; Lazarovici, P.; Zheng, W. Pharmacological Therapies and Drug Development Targeting SARS-CoV-2 infection. Cytokine Growth Factor Rev. 2022, 68, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkruys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Hurtado Del Pozo, C.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913.e7. [Google Scholar] [CrossRef] [PubMed]
- Zoufaly, A.; Poglitsch, M.; Aberle, J.H.; Hoepler, W.; Seitz, T.; Traugott, M.; Grieb, A.; Pawelka, E.; Laferl, H.; Wenisch, C.; et al. Human Recombinant Soluble ACE2 in Severe COVID-19. Lancet Respir. Med. 2020, 8, 1154–1158. [Google Scholar] [CrossRef]

| Gene Name | Primer Sense 5′-3′ | Primer Antisense 5′-3′ |
|---|---|---|
| SARS-CoV-2 gene E | 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′ | 5′-ATATTGCAGCAGTACGCACACA-3′ |
| Taqman probe: (FAM)-ACACTAGCCATCCTTACTGCGCTTCG-(BHQ1) | ||
| SARS-CoV-2 gene RdRp | 5′-AGAATAGAGCTCGCACCGTA-3′ | 5′-CTCCTCTAGTGGCGGCTATT-3′ |
| Human hprt | 5′-TGACACTGGCAAAACAATGCA-3′ | 5′-GGTCCTTTTCACCAGCAAGCT-3′ |
| Human ace2 | 5′-GTGCACAAAGGTGACAATGG-3′ | 5′-GGCTGCAGAAAGTGACATGA-3′ |
| Human tmprss2 | 5′-CACTGTGCA TCACCTTGACC-3′ | 5′-ACACGCCATCACACCAGTTA-3′ |
| Human cd147 | 5′-ATGCTGGTCTGCAAGTCAGA-3′ | 5′-GCGAGGAACTCACGAAGAAC-3′ |
| Human nrp1 | 5′-GAAGCACCGAGAGAACAAGG-3′ | 5′-AGTCCGCAGCTCAGGTGTAT-3′ |
| Antibody Name | Species | Reference | Application | Dilution |
|---|---|---|---|---|
| Anti-βIII-tubulin | Mouse | Ab78078, Abcam | Western Blotting Immunofluorescence | 1:5000 1:500 |
| Anti-α-tubulin | Mouse | T5168, Sigma-Aldrich | Western Blotting Immunofluorescence | 1:1000 1:500 |
| Anti-Tau total | Rabbit | A0024, Dako | Western Blotting Immunofluorescence | 1:5000 1:100 |
| Anti phospho-Tau (Ser202, Thr205) AT8 | Mouse | MN1020, Invitrogen | Western Blotting | 1:1000 |
| Anti-phospho-Tau (Ser422) | Mouse | 4BDX-1501, 4BioDX | Western Blotting | 1:5000 |
| Anti-GAPDH | Mouse | G8795-100 UL, Sigma-Aldrich | Western Blotting | 1:10,000 |
| SARS Coronavirus Nucleocapsid | Rabbit | PA1-41098, Invitrogen | Immunofluorescence | 1:100 |
| Antibody Name | Reference | Application | Dilution |
|---|---|---|---|
| Anti-Mouse IgG, HRP-Linked antibody | 7076S, Cell Signaling | Western Blotting | 1:1000 |
| Anti-Rabbit IgG, HRP-Linked antibody | 7074S, Cell signaling | Western Blotting | 1:1000 |
| Alexa fluor 488 Goat anti-mouse | A11001, Invitrogen | Immunofluorescence | 1:200 |
| Alexa fluor 568 Goat anti-rabbit | A11011, Life Technologies | Immunofluorescence | 1:200 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mignolet, M.; Gilloteaux, J.; Halloin, N.; Gueibe, M.; Willemart, K.; De Swert, K.; Bielarz, V.; Suain, V.; Pastushenko, I.; Gillet, N.A.; et al. Viral Entry Inhibitors Protect against SARS-CoV-2-Induced Neurite Shortening in Differentiated SH-SY5Y Cells. Viruses 2023, 15, 2020. https://doi.org/10.3390/v15102020
Mignolet M, Gilloteaux J, Halloin N, Gueibe M, Willemart K, De Swert K, Bielarz V, Suain V, Pastushenko I, Gillet NA, et al. Viral Entry Inhibitors Protect against SARS-CoV-2-Induced Neurite Shortening in Differentiated SH-SY5Y Cells. Viruses. 2023; 15(10):2020. https://doi.org/10.3390/v15102020
Chicago/Turabian StyleMignolet, Margaux, Jacques Gilloteaux, Nicolas Halloin, Matthieu Gueibe, Kévin Willemart, Kathleen De Swert, Valéry Bielarz, Valérie Suain, Ievgenia Pastushenko, Nicolas Albert Gillet, and et al. 2023. "Viral Entry Inhibitors Protect against SARS-CoV-2-Induced Neurite Shortening in Differentiated SH-SY5Y Cells" Viruses 15, no. 10: 2020. https://doi.org/10.3390/v15102020






