A NitroPure Nitrocellulose Membrane-Based Grapevine Virus Sampling Kit: Development and Deployment to Survey Japanese Vineyards and Nurseries
Abstract
:1. Introduction
2. Materials and Methods
2.1. Development of the NPN Membrane-Based Grapevine Virus Sampling Kit
2.1.1. Validation of the NPN Membrane
2.1.2. Maceration Buffer
2.1.3. Membrane Preparation for PCR Reactions
2.1.4. PCR Templates
2.1.5. Validation Testing
2.1.6. Stability of Samples after 18 Months of Storage
2.2. The NPN Membrane Kit in the Real World
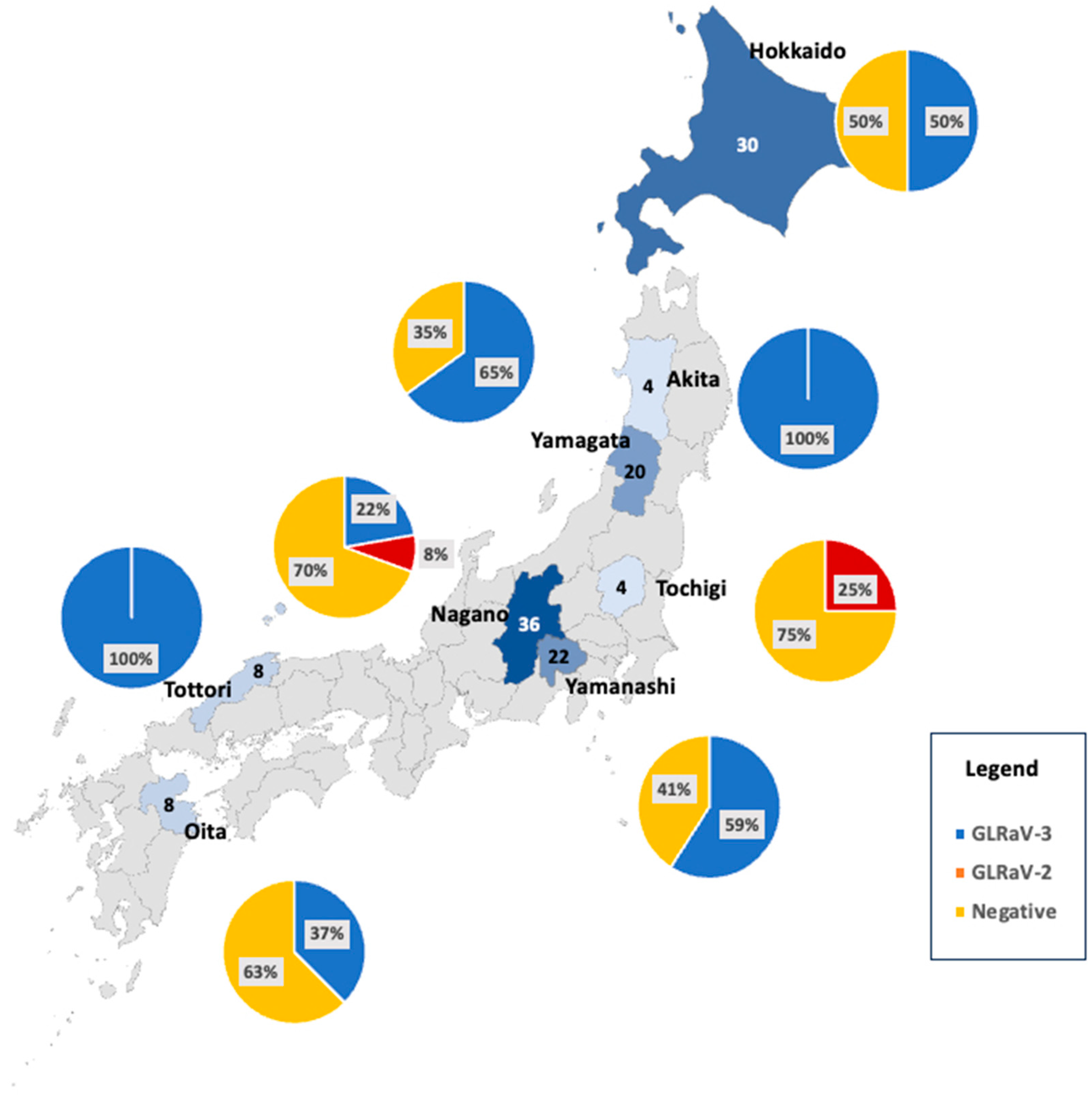
2.2.1. Grapevine Virus Surveys in Japan
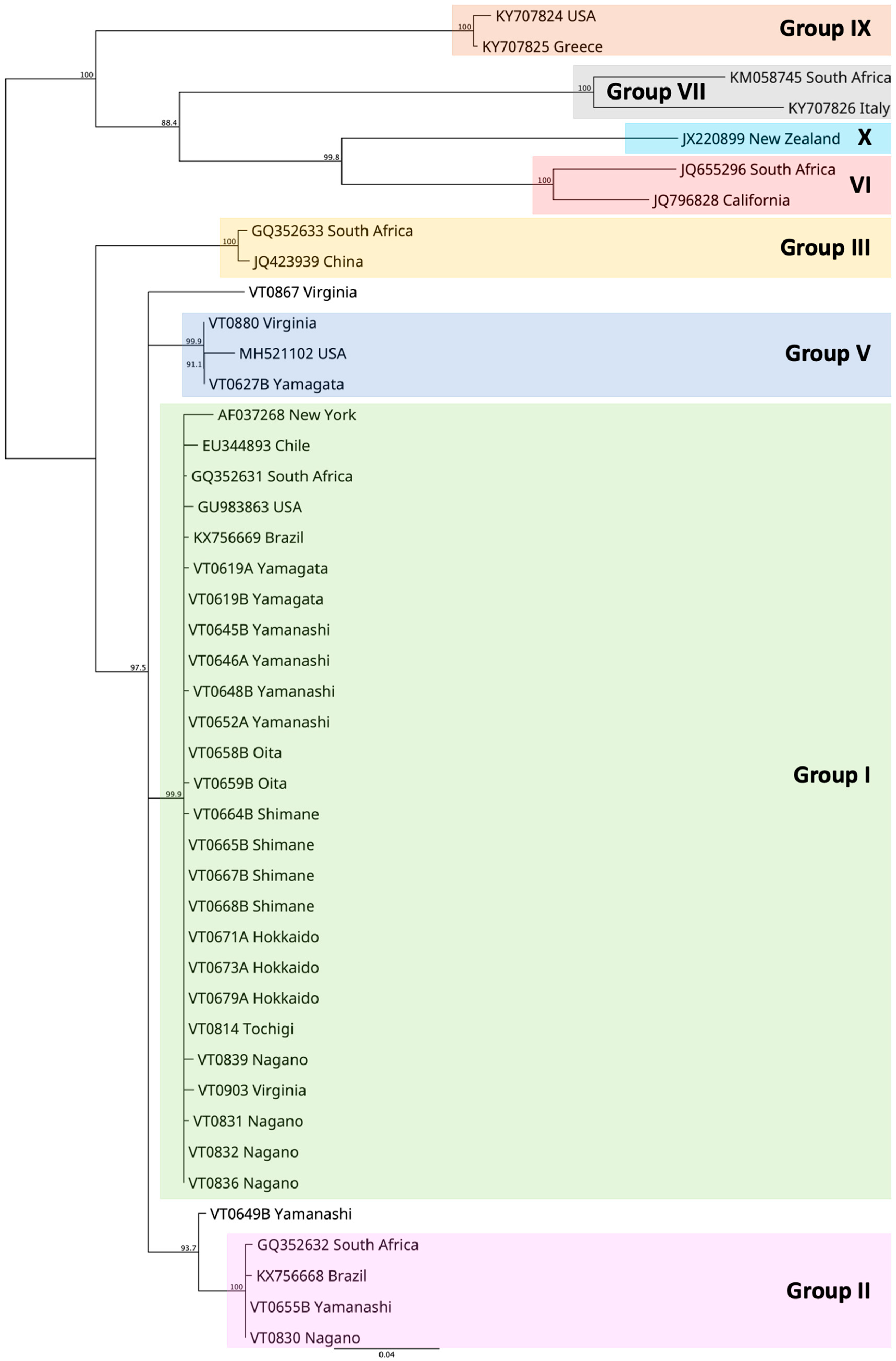
2.2.2. Genetic Variability of GLRaV3 Samples in Japan
3. Results
3.1. Development of the NPN Membrane-Based Grapevine Virus Sampling Kit
3.1.1. Screening for the Type of Membranes, Maceration, and Extraction Methods
3.1.2. Virus Detection after an 18-Month Storage
3.2. The NPN Membrane Kit in the Real World
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Martelli, G.P. Directory of virus and virus-like diseases of the grapevine and their agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar] [CrossRef]
- Meng, B.; Martelli, G.P.; Golino, D.A.; Fuchs, M. Grapevine Viruses: Molecular Biology, Diagnostics and Management. In Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer: Berlin/Heidelberg, Germany, 2017; pp. 1–698. [Google Scholar] [CrossRef]
- EFSA Panel on Plant Health (PLH); Bragard, C.; Dehnen-Schmutz, K.; Gonthier, P.; Jacques, M.; Miret, J.A.J.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. List of non-EU viruses and viroids of Cydonia Mill., Fragaria L., Malus Mill., Prunus L., Pyrus L., Ribes L., Rubus L. and Vitis L. EFSA J. 2019, 17, e05501. [Google Scholar] [CrossRef] [PubMed]
- Naidu, R.A.; Rowhani, A.; Fuchs, M.F.; Golino, D.; Martelli, G.P. Grapevine Leafroll: A Complex Viral Disease Affecting a High-Value Fruit Crop. Plant Dis. 2014, 98, 1172–1185. [Google Scholar] [CrossRef]
- Wallis, C.M.; Sudarshana, M.R. Effects of Grapevine red blotch-associated virus (GRBaV) infection on foliar metabolism of grapevines. Can. J. Plant Pathol. 2016, 38, 358–366. [Google Scholar] [CrossRef]
- Jones, T.; Nita, M. A Survey of Virginia Vineyards Revealed High Incidences of Grapevine Rupestris Stem Pitting-Associated Virus, Grapevine Red Blotch Virus, and Two Mealybug Species. Plant Heal. Prog. 2019, 20, 207–214. [Google Scholar] [CrossRef]
- Wang, C. Joining the Global Wine World: Japan’s Winemaking Industry. In Feeding Japan; Palgrave Macmillan: Cham, Switzerland, 2017; pp. 225–250. [Google Scholar] [CrossRef]
- Wang, C. Creating a wine heritage in Japan. Asian Anthr. 2020, 20, 61–76. [Google Scholar] [CrossRef]
- Nakaune, R.; Toda, S.; Mochizuki, M.; Nakano, M. Identification and characterization of a new vitivirus from grapevine. Arch. Virol. 2008, 153, 1827–1832. [Google Scholar] [CrossRef]
- Nakaune, R.; Inoue, K.; Nasu, H.; Kakogawa, K.; Nitta, H.; Imada, J.; Nakano, M. Detection of viruses associated with rugose wood in Japanese grapevines and analysis of genomic variability of Rupestris stem pitting-associated virus. J. Gen. Plant Pathol. 2008, 74, 156–163. [Google Scholar] [CrossRef]
- Ito, T.; Nakaune, R. Molecular characterization of a novel putative ampelovirus tentatively named grapevine leafroll-associated virus 13. Arch. Virol. 2016, 161, 2555–2559. [Google Scholar] [CrossRef]
- Ito, T.; Nakaune, R.; Nakano, M.; Suzaki, K. Novel variants of grapevine leafroll-associated virus 4 and 7 detected from a grapevine showing leafroll symptoms. Arch. Virol. 2012, 158, 273–275. [Google Scholar] [CrossRef]
- Martinson, T.; Fuchs, M.; Loeb, G.; Hoch, H. Grapevine Leafroll—An Increasing Problem in the Finger Lakes, the US and the World. Finger Lakes Vineyard Notes 2008, 6, 1–6. [Google Scholar]
- Atallah, S.S.; Gomez, M.I.; Fuchs, M.F.; Martinson, T.E. Economic Impact of Grapevine Leafroll Disease on Vitis vinifera cv. Cabernet franc in Finger Lakes Vineyards of New York. Am. J. Enol. Vitic. 2011, 63, 73–79. [Google Scholar] [CrossRef]
- Fuchs, M.; Martinson, T.E.; Loeb, G.M.; Hoch, H.C. Survey for the Three Major Leafroll Disease-Associated Viruses in Finger Lakes Vineyards in New York. Plant Dis. 2009, 93, 395–401. [Google Scholar] [CrossRef] [PubMed]
- Alley, C.J.; Goheen, A.C.; Olmo, H.P.; Koyama, A.T. The Effect of Virus Infections on Vines, Fruit and Wines of Ruby Cabernet. Am. J. Enol. Vitic. 1963, 14, 164–170. [Google Scholar] [CrossRef]
- Cabaleiro, C.; Segura, A.; García-Berrios, J.J. Effects of Grapevine Leafroll-Associated Virus 3 on the Physiology and Must of Vitis vinifera L. cv. Albariño Following Contamination in the Field. Am. J. Enol. Vitic. 1999, 50, 40–44. [Google Scholar] [CrossRef]
- Wolpert, J.A.; Vilas, E.P. Effect of Mild Leaf Roll Disease on Growth, Yield, and Fruit Maturity Indicies of Riesling and Zinfandel. Am. J. Enol. Vitic. 1992, 43, 367–369. [Google Scholar] [CrossRef]
- Naidu, R.A.; Maree, H.J.; Burger, J.T. Grapevine Leafroll Disease and Associated Viruses: A Unique Pathosystem. Annu. Rev. Phytopathol. 2015, 53, 613–634. [Google Scholar] [CrossRef]
- Jones, T.J.; Nita, M. Spatio-temporal association of GLRaV-3-infected grapevines, and effect of insecticidal treatments on mealybug populations in Virginia vineyards. Eur. J. Plant Pathol. 2016, 145, 885–900. [Google Scholar] [CrossRef]
- Tsai, C.-W.; Rowhani, A.; Golino, D.A.; Daane, K.M.; Almeida, R.P.P.; Jones, T.; Nita, M.; Poojari, S.; Boulé, J.; DeLury, N.; et al. Mealybug Transmission of Grapevine Leafroll Viruses: An Analysis of Virus–Vector Specificity. Phytopathology® 2010, 100, 830–834. [Google Scholar] [CrossRef]
- Tsai, C.-W.; Chau, J.; Fernandez, L.; Bosco, D.; Daane, K.M.; Almeida, R.P.P. Transmission of Grapevine leafroll-associated virus 3 by the Vine Mealybug (Planococcus ficus). Phytopathology® 2008, 98, 1093–1098. [Google Scholar] [CrossRef]
- Geiger, C.A.; Daane, K.M. Seasonal Movement and Distribution of the Grape Mealybug (Homoptera: Pseudococcidae): Developing a Sampling Program for San Joaquin Valley Vineyards. J. Econ. Èntomol. 2001, 94, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.; Nita, M. Gill’s mealybug, Ferrisia gilli, can Transmit Grapevine Leafroll-associated Virus-3 after a 24-hour Acquisition Time. Int. J. Phytopathol. 2020, 9, 139–144. [Google Scholar] [CrossRef]
- Ghanem-Sabanadzovic, N.A.; Sabanadzovic, S.; Gugerli, P.; Rowhani, A. Genome organization, serology and phylogeny of Grapevine leafroll-associated viruses 4 and 6: Taxonomic implications. Virus Res. 2012, 163, 120–128. [Google Scholar] [CrossRef] [PubMed]
- Martelli, G.; Ghanem-Sabanadzovic, N.A.; Agranovsky, A.; Al Rwahnih, M.; Dolja, V.; Dovas, C.; Fuchs, M.; Gugerli, P.; Hu, J.; Jelkmann, W.; et al. TAXONOMIC REVISION OF THE FAMILY CLOSTEROVIRIDAE WITH SPECIAL REFERENCE TO THE GRAPEVINE LEAFROLL-ASSOCIATED MEMBERS OF THE GENUS AMPELOVIRUS AND THE PUTATIVE SPECIES UNASSIGNED TO THE FAMILY. J. Plant Pathol. 2012, 94, 7–19. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Rowhani, A.; Golino, D.A.; Islas, C.M.; Preece, J.E.; Sudarshana, M.R. Detection and genetic diversity of Grapevine red blotch-associated virus isolates in table grape accessions in the National Clonal Germplasm Repository in California. Can. J. Plant Pathol. 2015, 37, 130–135. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Rowhani, A.; Golino, D. First Report of Grapevine red blotch-associated virus in Archival Grapevine Material From Sonoma County, California. Plant Dis. 2015, 99, 895. [Google Scholar] [CrossRef]
- Al Rwahnih, M.; Dave, A.; Anderson, M.M.; Rowhani, A.; Uyemoto, J.K.; Sudarshana, M.R.; Wilson, H.; Hogg, B.N.; Blaisdell, K.G.; Andersen, J.C.; et al. Association of a DNA Virus with Grapevines Affected by Red Blotch Disease in California. Phytopathology® 2013, 103, 1069–1076. [Google Scholar] [CrossRef]
- Bowen, P.; Bogdanoff, C.; Poojari, S.; Usher, K.; Lowery, T.; Úrbez-Torres, J.R. Effects of Grapevine Red Blotch Disease on Cabernet franc Vine Physiology, Bud Hardiness, and Fruit and Wine Quality. Am. J. Enol. Vitic. 2020, 71, 308–318. [Google Scholar] [CrossRef]
- Girardello, R.C.; Rich, V.; Smith, R.J.; Brenneman, C.; Heymann, H.; Oberholster, A. The impact of grapevine red blotch disease on Vitis vinifera L. Chardonnay grape and wine composition and sensory attributes over three seasons. J. Sci. Food Agric. 2019, 100, 1436–1447. [Google Scholar] [CrossRef]
- Krenz, B.; Thompson, J.R.; McLane, H.L.; Fuchs, M.; Perry, K.L. Grapevine red blotch-associated virus Is Widespread in the United States. Phytopathology® 2014, 104, 1232–1240. [Google Scholar] [CrossRef]
- Jones, T.J.; Rayapati, N.A.; Nita, M. Occurrence of Grapevine leafroll associated virus-2, −3 and Grapevine fleck virus in Virginia, U.S.A., and factors affecting virus infected vines. Eur. J. Plant Pathol. 2015, 142, 209–222. [Google Scholar] [CrossRef]
- Jones, T.J.; Westover, F.; Nita, M. First Report of Grapevine leafroll-associated virus-2 and -3 in Texas Vineyards. Plant Dis. 2014, 98, 1592. [Google Scholar] [CrossRef]
- Wallingford, A.K.; Fuchs, M.F.; Martinson, T.; Hesler, S.; Loeb, G.M. Slowing the Spread of Grapevine Leafroll-Associated Viruses in Commercial Vineyards with Insecticide Control of the Vector, Pseudococcus maritimus (Hemiptera: Pseudococcidae). J. Insect Sci. 2015, 15, 112. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.R.; Eastwell, K.C.; Wagner, A.; Lamprecht, S.; Tzanetakis, I.E. Survey for Viruses of Grapevine in Oregon and Washington. Plant Dis. 2005, 89, 763–766. [Google Scholar] [CrossRef]
- Naidu, R.A.; Mekuria, T.A. First Report of Grapevine fleck virus from Washington Vineyards. Plant Dis. 2010, 94, 784. [Google Scholar] [CrossRef] [PubMed]
- Adiputra, J.; Kesoju, S.R.; Naidu, R.A. The Relative Occurrence of Grapevine leafroll-associated virus 3 and Grapevine red blotch virus in Washington State Vineyards. Plant Dis. 2018, 102, 2129–2135. [Google Scholar] [CrossRef]
- Bertazzon, N.; Angelini, E. Advances in the Detection of Grapevine Leafroll-Associated Virus 2 Variants. J. Plant Pathol. 2004, 86, 283–290. [Google Scholar]
- Al Rwahnih, M.; Osman, F.; Sudarshana, M.; Uyemoto, J.; Minafra, A.; Saldarelli, P.; Martelli, G.; Rowhani, A. Detection of Grapevine leafroll-associated virus 7 using real time qRT-PCR and conventional RT-PCR. J. Virol. Methods 2012, 179, 383–389. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Real-time RT-PCR (TaqMan®) assays for the detection of Grapevine Leafroll associated viruses 1–5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Comparison of low-density arrays, RT-PCR and real-time TaqMan® RT-PCR in detection of grapevine viruses. J. Virol. Methods 2008, 149, 292–299. [Google Scholar] [CrossRef]
- Osman, F.; Hodzic, E.; Omanska-Klusek, A.; Olineka, T.; Rowhani, A. Development and validation of a multiplex quantitative PCR assay for the rapid detection of Grapevine virus A, B and D. J. Virol. Methods 2013, 194, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Bendel, N.; Kicherer, A.; Backhaus, A.; Köckerling, J.; Maixner, M.; Bleser, E.; Klück, H.-C.; Seiffert, U.; Voegele, R.T.; Töpfer, R.J.R.S. Detection of grapevine leafroll-associated virus 1 and 3 in white and red grapevine cultivars using hyperspectral imaging. Remote Sens. 2020, 12, 1693. [Google Scholar] [CrossRef]
- Tarquini, G.; Dall’ara, M.; Ermacora, P.; Ratti, C. Traditional Approaches and Emerging Biotechnologies in Grapevine Virology. Viruses 2023, 15, 826. [Google Scholar] [CrossRef] [PubMed]
- Poojari, S.; Alabi, O.J.; Okubara, P.A.; Naidu, R.A. SYBR® Green-based real-time quantitative reverse-transcription PCR for detection and discrimination of grapevine viruses. J. Virol. Methods 2016, 235, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Buchs, N.; Braga-Lagache, S.; Uldry, A.-C.; Brodard, J.; Debonneville, C.; Reynard, J.-S.; Heller, M. Absolute Quantification of Grapevine Red Blotch Virus in Grapevine Leaf and Petiole Tissues by Proteomics. Front. Plant Sci. 2018, 9, 1735. [Google Scholar] [CrossRef]
- Chang, P.-G.S.; McLaughlin, W.A.; Tolin, S.A. Tissue blot immunoassay and direct RT-PCR of cucumoviruses and potyviruses from the same NitroPure nitrocellulose membrane. J. Virol. Methods 2011, 171, 345–351. [Google Scholar] [CrossRef]
- Osman, F.; Rowhani, A. Application of a spotting sample preparation technique for the detection of pathogens in woody plants by RT-PCR and real-time PCR (TaqMan). J. Virol. Methods 2006, 133, 130–136. [Google Scholar] [CrossRef]
- La Notte, P.; Minafra, A.; Saldarelli, P. A spot-PCR technique for the detection of phloem-limited grapevine viruses. J. Virol. Methods 1997, 66, 103–108. [Google Scholar] [CrossRef]
- Charles, J.G.; Cohen, D.; Walker, J.T.S.; Forgie, S.A.; Bell, V.A.; Breen, K.C. A Review of the Ecology of Grapevine Leafroll Asso-ciated Virus Type 3(GLRaV-3). N. Z. Plant Prot. 2006, 59, 330–337. [Google Scholar]
- Rowhani, A.; Biardi, L.; Johnson, R.; Saldarelli, P.; Zhang, Y.P.; Chin, J.; Green, M. Simplified Sample Preparation Method and One-Tube RT-PCR for Grapevine Viruses. In Proceedings of the XIII International Council for the Study of Viruses and Virus-Like Diseases of the Grapevine, Adelaide, Australia, 12–17 March 2000; Volume 148. [Google Scholar]
- Poojari, S.; Alabi, O.J.; A Naidu, R. Molecular characterization and impacts of a strain of Grapevine leafroll-associated virus 2 causing asymptomatic infection in a wine grape cultivar. Virol. J. 2013, 10, 324. [Google Scholar] [CrossRef]
- Esteves, F.; Santos, M.T.; Eiras-Dias, J.E.; Fonseca, F. Molecular data mining to improve antibody-based detection of Grapevine leafroll-associated virus 1 (GLRaV-1). J. Virol. Methods 2013, 194, 258–270. [Google Scholar] [CrossRef] [PubMed]
- Beuve, M.; Sempé, L.; Lemaire, O. A sensitive one-step real-time RT-PCR method for detecting Grapevine leafroll-associated virus 2 variants in grapevine. J. Virol. Methods 2007, 141, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Lara, A.; Klaassen, V.; Stevens, K.; Sudarshana, M.R.; Rowhani, A.; Maree, H.J.; Chooi, K.M.; Blouin, A.G.; Habili, N.; Song, Y.; et al. Characterization of grapevine leafroll-associated virus 3 genetic variants and application towards RT-qPCR assay design. PLoS ONE 2018, 13, e0208862. [Google Scholar] [CrossRef] [PubMed]
- Meng, B.; Johnson, R.; Peressini, S.; Forsline, P.L.; Gonsalves, D. Rupestris Stem Pitting Associated Virus-1 is Consistently Detected in Grapevines that are Infected with Rupestris Stem Pitting. Eur. J. Plant Pathol. 1999, 105, 191–199. [Google Scholar] [CrossRef]
- Minafra, A.; Hadidi, A. Sensitive detection of grapevine virus A, B, or leafroll-associated III from viruliferous mealybugs and infected tissue by cDNA amplification. J. Virol. Methods 1994, 47, 175–187. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Mock, R.; Fuchs, M.; Halbrendt, J.; Howell, B.; Liu, Z. Characterization of the partial RNA1 and RNA2 3′ untranslated region of Tomato ringspot virus isolates from North America. Can. J. Plant Pathol. 2011, 33, 94–99. [Google Scholar] [CrossRef]
- Klaassen, V.A.; Sim, S.T.; Dangl, G.S.; Osman, F.; Al Rwahnih, M.; Rowhani, A.; Golino, D.A. Vitis californica and Vitis californica × Vitis vinifera Hybrids are Hosts for Grapevine leafroll-associated virus-2 and -3 and Grapevine virus A and B. Plant Dis. 2011, 95, 657–665. [Google Scholar] [CrossRef]
- Weller, S.A.; Elphinstone, J.G.; Smith, N.C.; Boonham, N.; Stead, D.E. Detection of Ralstonia solanacearum Strains with a Quantitative, Multiplex, Real-Time, Fluorogenic PCR (TaqMan) Assay. Appl. Environ. Microbiol. 2000, 66, 2853–2858. [Google Scholar] [CrossRef]
- Lemoine, F.; Correia, D.; Lefort, V.; Doppelt-Azeroual, O.; Mareuil, F.; Cohen-Boulakia, S.; Gascuel, O. NGPhylogeny.fr: New generation phylogenetic services for non-specialists. Nucleic Acids Res. 2019, 47, W260–W265. [Google Scholar] [CrossRef]
- Maliogka, V.I.; Martelli, G.P.; Fuchs, M. Control of Viruses Infecting Grapevine. Adv. Virus Res. 2015, 91, 175–227. [Google Scholar] [CrossRef]
- Lim, S.; Igori, D.; Zhao, F.; Moon, J.S.; Cho, I.-S.; Choi, G.-S. First Report of Grapevine red blotch-associated virus on Grapevine in Korea. Plant Dis. 2016, 100, 1957. [Google Scholar] [CrossRef]
- Gasperin-Bulbarela, J.; Licea-Navarro, A.F.; Pino-Villar, C.; Hernandez-Martinez, R.; Carrillo-Tripp, J. First Report of Grapevine Red Blotch Virus in Mexico. Plant Dis. 2019, 103, 381. [Google Scholar] [CrossRef]
- Hoffmann, M.; Talton, W.; Nita, M.; Jones, T.; Al Rwahnih, M.; Sudarshana, M.R.; Almeyda, C. First Report of Grapevine red blotch virus, the Causal Agent of Grapevine Red Blotch Disease, in Vitis vinifera in North Carolina. Plant Dis. 2020, 104, 1266. [Google Scholar] [CrossRef]
- Marwal, A.; Kumar, R.; Khurana, S.M.P.; Gaur, R.K. Complete nucleotide sequence of a new geminivirus isolated from Vitis vinifera in India: A symptomless host of Grapevine red blotch virus. VirusDisease 2018, 30, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Reynard, J.-S.; Brodard, J.; Dubuis, N.; Kellenberger, I.; Spilmont, A.-S.; Roquis, D.; Maliogka, V.; Marchal, C.; Dedet, S.; Gning, O.; et al. Screening of grapevine red blotch virus in two European ampelographic collections. J. Plant Pathol. 2021, 104, 9–15. [Google Scholar] [CrossRef]
- Bertazzon, N.; Migliaro, D.; Rossa, A.; Filippin, L.; Casarin, S.; Giust, M.; Brancadoro, L.; Crespan, M.; Angelini, E. Grapevine red blotch virus is sporadically present in a germplasm collection in Northern Italy. J. Plant Dis. Prot. 2021, 128, 1115–1119. [Google Scholar] [CrossRef]
| Virus | Primer Name and Reference | PCR Parameters (Temp in °C) | Number of Samples Tested a |
|---|---|---|---|
| GLRaV1 | dCP1-1/dCP1-2 [54] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 9 |
| GLRaV2 | P19qtF4 and p24qtR [55] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 25 |
| GLRaV3 | GEN-11112F/GEN-11233R [56] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 25 |
| GLRaV4 | LRAmp-F/LRAmp-R [25] | 52°, 1 h; 94, 2 min; 40 x (94, 30 s; 50, 35 s; 72, 45 s); 72, 7 min | 6 |
| GRSPaV | RSP13/RSP14 [57] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 25 |
| GFkV | GFkV-585 F/GFkV-1117 R [37] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 6 |
| GVA | H587/C995 [58] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 25 |
| GVB | C410/H28 [58] | 52°, 1 h; 35 x (94, 30 s; 54, 45 s; 72, 1 min); 72, 2 min | 15 |
| ToRSV | ToRSV5/ToRSV6 [59] | 50°, 30 min; 94, 2 min; 30 x (94, 45 s; 60, 45 s; 68, 2 min); 68, 5 min | 8 |
| GRBV | GVGF1/GVGR1 [28] | 94°, 2 min; 35 x (94, 30 s; 60, 30 s; 72, 1 min); 72, 5 min | 25 |
| Maceration Buffer | Method | Washing Buffer a | Template for PCR b | GLRaV3 c | GRBV c |
|---|---|---|---|---|---|
| EB | A | No treatment | Disc | 0/48 | 0/48 |
| B | Triton X-100 | Disc | 16/48 | 23/48 | |
| C | FTA reagent | Disc | 0/48 | 0/48 | |
| D1 | GES | Disc | 33/48 | 41/48 | |
| D2 | GES | 2 μL solution | 37/48 | 44/48 | |
| E1 | GES + beta-m | Disc | 40/48 | 43/48 | |
| E2 | GES + beta-m | 2 μL solution | 48/48 | 48/48 | |
| GSB | A | No treatment | Disc | 0/48 | 0/48 |
| B | Triton X-100 | Disc | 13/48 | 24/48 | |
| C | FTA reagent | Disc | 0/48 | 0/48 | |
| D1 | GES | Disc | 31/48 | 43/48 | |
| D2 | GES | 2 μL solution | 34/48 | 47/48 | |
| E1 | GES + beta-m | Disc | 42/48 | 45/48 | |
| E2 | GES + beta-m | 2 μL solution | 48/48 | 48/48 | |
| Water control | A | No treatment | Disc | 0/48 | 0/48 |
| B | Triton X-100 | Disc | 0/48 | 0/48 | |
| C | FTA reagent | Disc | 0/48 | 0/48 | |
| D1 | GES | Disc | 0/48 | 0/48 | |
| D2 | GES | 2 μL solution | 0/48 | 0/48 | |
| E1 | GES + beta-m | Disc | 0/48 | 0/48 | |
| E2 | GES + beta-m | 2 μL solution | 0/48 | 0/48 | |
| Positive control | Traditional nucleic acid extraction method without the membrane [6,18] | 48/48 | 48/48 | ||
| Petiole collection |
|
|
|
| Blotting |
|
|
|
|
|
|
| Nucleic acid extraction and PCR |
|
|
|
|
|
| Virus | Gene | Primers and Probes | Reference |
|---|---|---|---|
| RT-qPCR | |||
| GLRaV2 | heat shock protein (HSP70) | LR2-124f | [60] |
| LR2-284r1 | |||
| LR2-284r2 | |||
| LR2-284r3 | |||
| LR2-214p | |||
| GLRaV3 | 3′ untranslated terminal region (UTR) | LR3-FPST-F1 | [56] |
| LR3_FPST-F2 | |||
| LR3_FPST-F3 | |||
| LR3_FPST-F4 | |||
| LR3_FPST-R1 | |||
| LR3_FPST-R2 | |||
| LR3_FPST-P1 | |||
| LR3_FPST-P2 | |||
| qPCR | |||
| GRBV | V2 | RB3-F | [20] |
| RB3-R | |||
| RB3-P | |||
| Plant | COX | COX-F | [61] |
| COX-R | |||
| COX-P | |||
| RT-PCR for sequencing | |||
| GLRaV2 | heat shock protein (HSP70) | LR2-U2 | [39] |
| LR2-L2 | |||
| GLRaV3 | heat shock protein (HSP70) | LC1 | [49] |
| LC2 | |||
| PCR for sequencing | |||
| GRBV | V2 | GVGF1 | [29] |
| GVGR1 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nita, M.; Jones, T.; McHenry, D.; Bush, E.; Oliver, C.; Kawaguchi, A.; Nita, A.; Katori, M. A NitroPure Nitrocellulose Membrane-Based Grapevine Virus Sampling Kit: Development and Deployment to Survey Japanese Vineyards and Nurseries. Viruses 2023, 15, 2102. https://doi.org/10.3390/v15102102
Nita M, Jones T, McHenry D, Bush E, Oliver C, Kawaguchi A, Nita A, Katori M. A NitroPure Nitrocellulose Membrane-Based Grapevine Virus Sampling Kit: Development and Deployment to Survey Japanese Vineyards and Nurseries. Viruses. 2023; 15(10):2102. https://doi.org/10.3390/v15102102
Chicago/Turabian StyleNita, Mizuho, Taylor Jones, Diana McHenry, Elizabeth Bush, Charlotte Oliver, Akira Kawaguchi, Akiko Nita, and Miyuki Katori. 2023. "A NitroPure Nitrocellulose Membrane-Based Grapevine Virus Sampling Kit: Development and Deployment to Survey Japanese Vineyards and Nurseries" Viruses 15, no. 10: 2102. https://doi.org/10.3390/v15102102






