Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
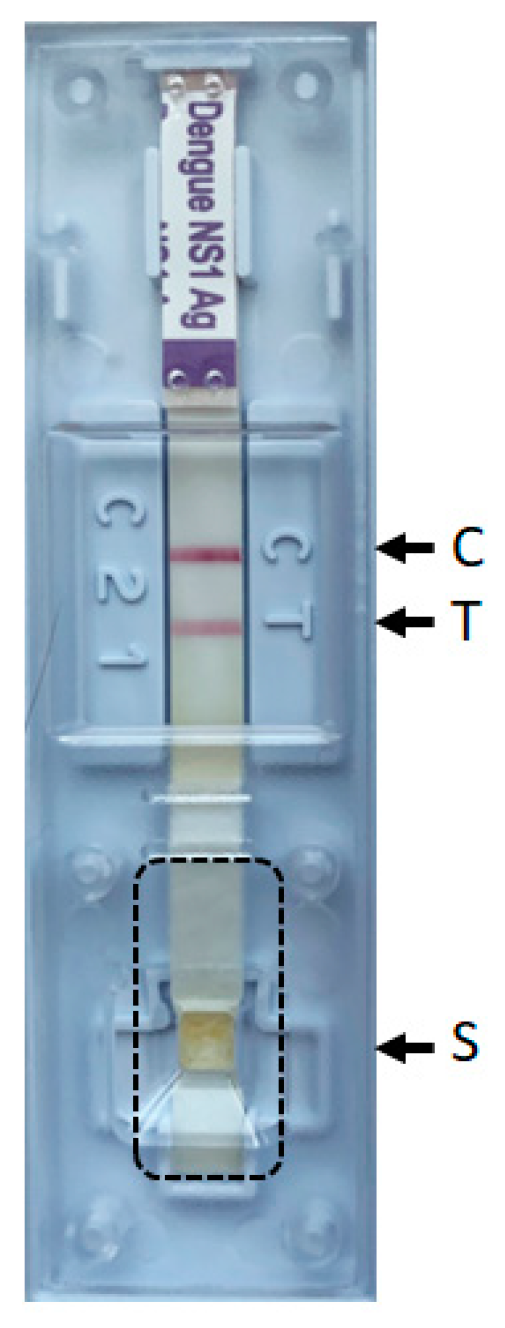
2.2. Dengue NS1 Antigen RDTs
2.3. Unbiased Metagenomic Next-Generation Sequencing (mNGS) from RDTs Strips and Sequence Analysis
2.4. Real-Time RT–PCR
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bhatt, S.; Gething, P.W.; Brady, O.J.; Messina, J.P.; Farlow, A.W.; Moyes, C.L.; Drake, J.M.; Brownstein, J.S.; Hoen, A.G.; Sankoh, O.; et al. The global distribution and burden of dengue. Nature 2013, 496, 504–507. [Google Scholar] [CrossRef] [PubMed]
- Brady, O.J.; Gething, P.W.; Bhatt, S.; Messina, J.P.; Brownstein, J.S.; Hoen, A.G.; Moyes, C.L.; Farlow, A.W.; Scott, T.W.; Hay, S.I. Refining the global spatial limits of dengue virus transmission by evidence-based consensus. PLoS Negl. Trop. Dis. 2012, 6, e1760. [Google Scholar] [CrossRef] [PubMed]
- Katzelnick, L.C.; Coello Escoto, A.; Huang, A.T.; Garcia-Carreras, B.; Chowdhury, N.; Maljkovic Berry, I.; Chavez, C.; Buchy, P.; Duong, V.; Dussart, P.; et al. Antigenic evolution of dengue viruses over 20 years. Science 2021, 374, 999–1004. [Google Scholar] [PubMed]
- Waggoner, J.J.; Balmaseda, A.; Gresh, L.; Sahoo, M.K.; Montoya, M.; Wang, C.; Abeynayake, J.; Kuan, G.; Pinsky, B.A.; Harris, E. Homotypic Dengue Virus Reinfections in Nicaraguan Children. J. Infect. Dis. 2016, 214, 986–993. [Google Scholar] [CrossRef] [PubMed]
- Yow, K.S.; Aik, J.; Tan, E.Y.; Ng, L.C.; Lai, Y.L. Rapid diagnostic tests for the detection of recent dengue infections: An evaluation of six kits on clinical specimens. PLoS ONE 2021, 16, e0249602. [Google Scholar] [CrossRef] [PubMed]
- Macori, G.; Russell, T.; Barry, G.; McCarthy, S.C.; Koolman, L.; Wall, P.; Sammin, D.; Mulcahy, G.; Fanning, S. Inactivation and Recovery of High Quality RNA From Positive SARS-CoV-2 Rapid Antigen Tests Suitable for Whole Virus Genome Sequencing. Front. Public Health 2022, 10, 863862. [Google Scholar] [CrossRef] [PubMed]
- Rector, A.; Bloemen, M.; Schiettekatte, G.; Maes, P.; Van Ranst, M.; Wollants, E. Sequencing directly from antigen-detection rapid diagnostic tests in Belgium, 2022: A gamechanger in genomic surveillance? Euro Surveill. 2023, 28, 2200618. [Google Scholar] [CrossRef] [PubMed]
- Gleizes, A.; Laubscher, F.; Guex, N.; Iseli, C.; Junier, T.; Cordey, S.; Fellay, J.; Xenarios, I.; Kaiser, L.; Mercier, P.L. Virosaurus A Reference to Explore and Capture Virus Genetic Diversity. Viruses 2020, 12, 1248. [Google Scholar] [CrossRef] [PubMed]
- Cordey, S.; Laubscher, F.; Hartley, M.A.; Junier, T.; Keitel, K.; Docquier, M.; Guex, N.; Iseli, C.; Vieille, G.; Le Mercier, P.; et al. Blood virosphere in febrile Tanzanian children. Emerg. Microbes Infect. 2021, 10, 982–993. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Schibler, M.; Yerly, S.; Vieille, G.; Docquier, M.; Turin, L.; Kaiser, L.; Tapparel, C. Critical analysis of rhinovirus RNA load quantification by real-time reverse transcription-PCR. J. Clin. Microbiol. 2012, 50, 2868–2872. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Trioplex Real-Time RT-PCR Assay. Available online: https://www.cdc.gov/zika/pdfs/trioplex-real-time-rt-pcr-assay-instructions-for-use.pdf (accessed on 25 September 2023).
- Schlaberg, R.; Chiu, C.Y.; Miller, S.; Procop, G.W.; Weinstock, G. Validation of Metagenomic Next-Generation Sequencing Tests for Universal Pathogen Detection. Arch. Pathol. Lab. Med. 2017, 141, 776–786. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, J.F.; Laubscher, F.; Held, J.; Eckerle, I.; Docquier, M.; Grobusch, M.P.; Mordmüller, B.; Kaiser, L.; Cordey, S. Unbiased metagenomic next-generation sequencing of blood from hospitalized febrile children in Gabon. Emerg. Microbes Infect. 2020, 9, 1242–1244. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.H.; Cordey, S.; Bhuva, N.; Laubscher, F.; Hartley, M.A.; Boillat-Blanco, N.; Mbarack, Z.; Samaka, J.; Mlaganile, T.; Jain, K.; et al. Investigation of the Plasma Virome from Cases of Unexplained Febrile Illness in Tanzania from 2013 to 2014: A Comparative Analysis between Unbiased and VirCapSeq-VERT High-Throughput Sequencing Approaches. mSphere 2018, 3, e00311-18. [Google Scholar] [PubMed]
- Zanella, M.C.; Cordey, S.; Laubscher, F.; Docquier, M.; Vieille, G.; Van Delden, C.; Braunersreuther, V.; Ta, M.K.; Lobrinus, J.A.; Masouridi-Levrat, S.; et al. Unmasking viral sequences by metagenomic next-generation sequencing in adult human blood samples during steroid-refractory/dependent graft-versus-host disease. Microbiome 2021, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Cabello, L.; Utrilla-Trigo, S.; Lorenzo, G.; Ortego, J.; Calvo-Pinilla, E. Epizootic Hemorrhagic Disease Virus: Current Knowledge and Emerging Perspectives. Microorganisms 2023, 11, 1339. [Google Scholar] [CrossRef]
| Sample | Traveler Returning From | RDT (Standard Q Duo) Results | Unbiased mNGS | Real-Time RT-PCR Ct Values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Antigen NS1 (Number of Days between Onset of Symptoms and Sample Collection) | IgM | IgG | Number of Days the Ag-RDT Remained at Room Temperature before Being Processed for Sequencing | Total Number of Reads | Human Reads (%) | Number of Reads Mapping to Dengue Virus Genome | Genome Coverage (%) | Depth | Serotype | Genotype | Best Matched GenBank Reference (Accession Number) | Ability to Perform Phylogenetic Analysis (yes/no) | |||
| 1 | Thailand | positive (3) | negative | negative | 31 | 17,565,488 | 11.112 | 1616 | 88.9 | 10 | dengue 3 | G III | MW788884 | yes | 11.3 |
| 2 | Singapore | positive (5) | positive | positive | 28 | 22,705,906 | 7.115 | 2860 | 99.4 | 19 | dengue 2 | Cosmopolitan | MW512481 | yes | 20.3 |
| 3 | Cuba | positive (5) | positive | negative | 23 | 25,572,466 | 8.082 | 8 | 5.1 | 1 | dengue 4 | undetermined | OR162320 | no | 21.9 |
| 4 | Mexico | positive (unknown) | equivocal | negative | 8 | 25,912,062 | 5.918 | 70 | 17.48 | 2 | dengue 1 | G V | GQ868510 | yes | 31.3 |
| 5 | Costa Rica | positive (6) | equivocal | negative | 15 | 7,569,600 | 7.086 | 409 | 54.87 | 4 | dengue 1 | G V | OQ603263 | yes | 21.5 |
| 6 | Cambodia/Indonesia | positive (8) | positive | negative | 14 | 7,179,842 | 6.517 | 0 | 0 | 0 | undetermined | undetermined | not applicable | no | undetected |
| 7 | Cambodia/Indonesia | positive (8) | equivocal | negative | 7 | 33,997,742 | 6.03 | 72 | 16.59 | 2 | dengue 1 | G I | MZ619038 | yes | 31.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Rodríguez, F.-J.; Laubscher, F.; Chudzinski, V.; Kaiser, L.; Cordey, S. Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay. Viruses 2023, 15, 2167. https://doi.org/10.3390/v15112167
Pérez-Rodríguez F-J, Laubscher F, Chudzinski V, Kaiser L, Cordey S. Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay. Viruses. 2023; 15(11):2167. https://doi.org/10.3390/v15112167
Chicago/Turabian StylePérez-Rodríguez, Francisco-Javier, Florian Laubscher, Valentin Chudzinski, Laurent Kaiser, and Samuel Cordey. 2023. "Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay" Viruses 15, no. 11: 2167. https://doi.org/10.3390/v15112167
APA StylePérez-Rodríguez, F.-J., Laubscher, F., Chudzinski, V., Kaiser, L., & Cordey, S. (2023). Direct Dengue Virus Genome Sequencing from Antigen NS1 Rapid Diagnostic Tests: A Proof-of-Concept with the Standard Q Dengue Duo Assay. Viruses, 15(11), 2167. https://doi.org/10.3390/v15112167







