Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Virus Infection and DNA Transfection
2.2. SDS-PAGE, Immunoblotting and Antibodies
2.3. Immunoprecipitation and Peptide Pull-Down Assays
2.4. Immunofluorescence Microscopy
2.5. Structural Predictions
3. Results
3.1. Identification of Novel HAdV-C5 E1B-55K Binding Proteins
3.2. Interaction of Adenovirus E1B-55K Proteins with the xWxxxPx Sequence
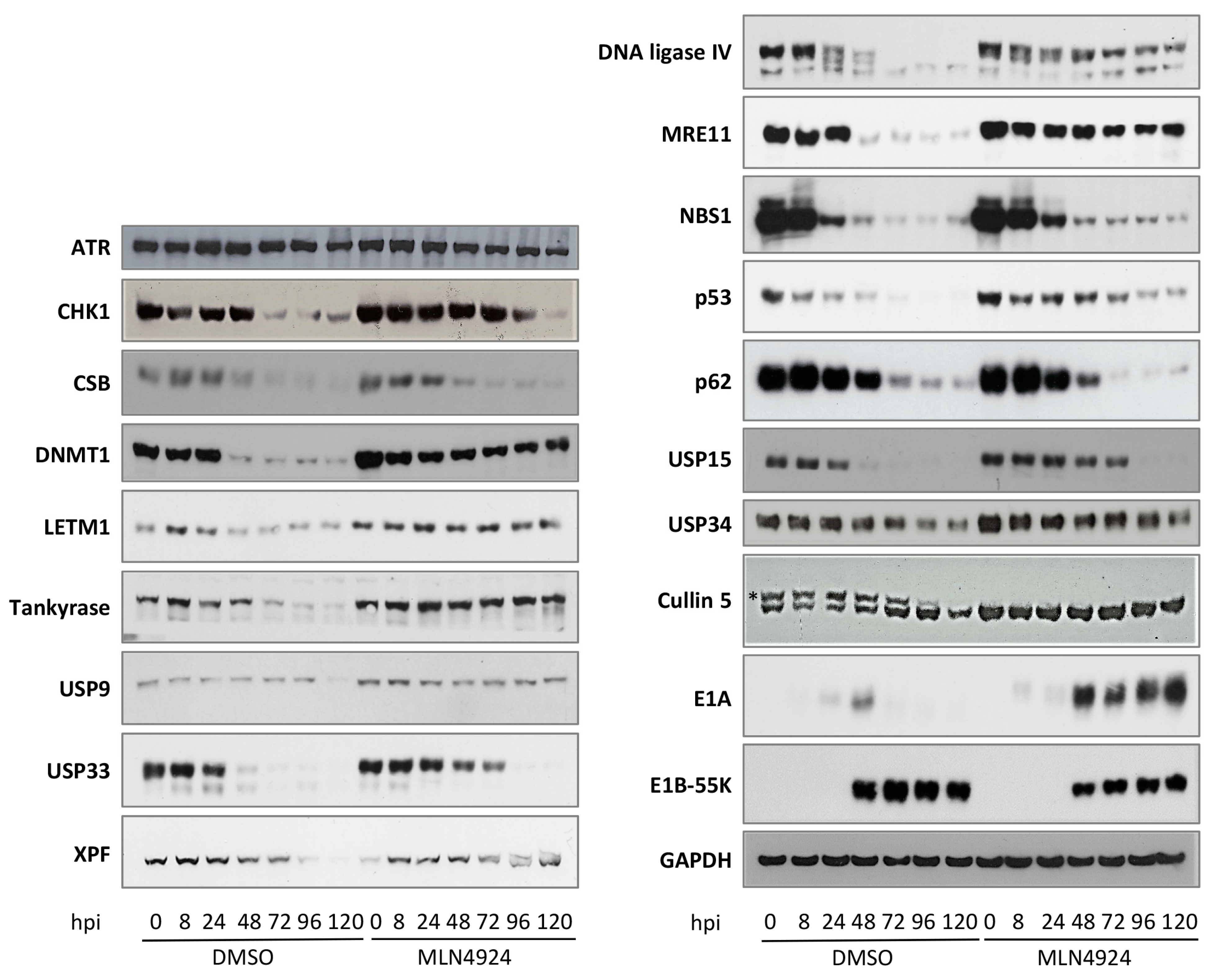
3.3. Adenovirus-Mediated Degradation of Novel Cellular Targets
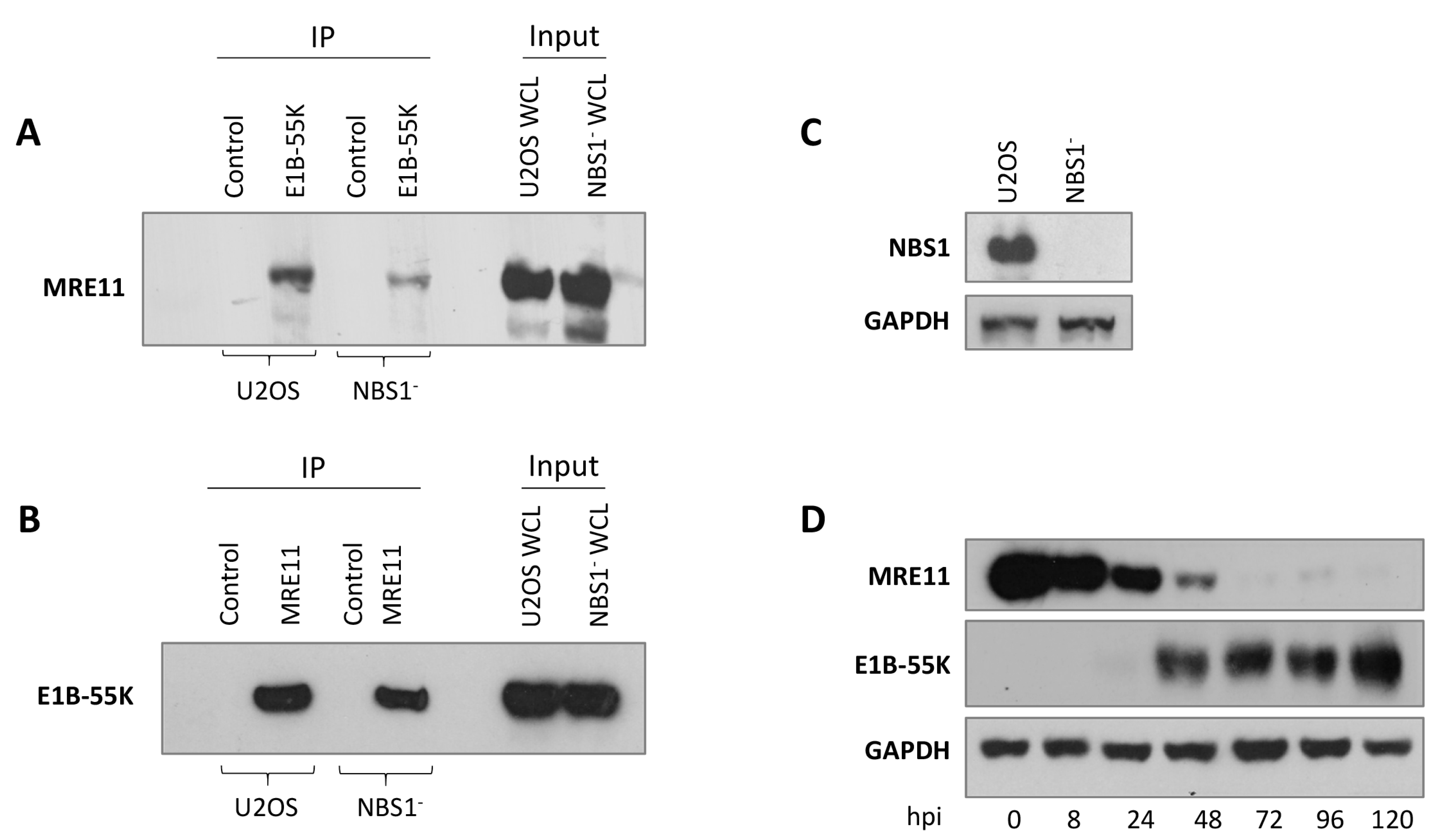
3.4. Interaction of MRE11 with HAdV-C5 E1B-55K
3.5. Sub-Cellular Localization of Novel Adenovirus E1B-55K Binding Proteins during Infection
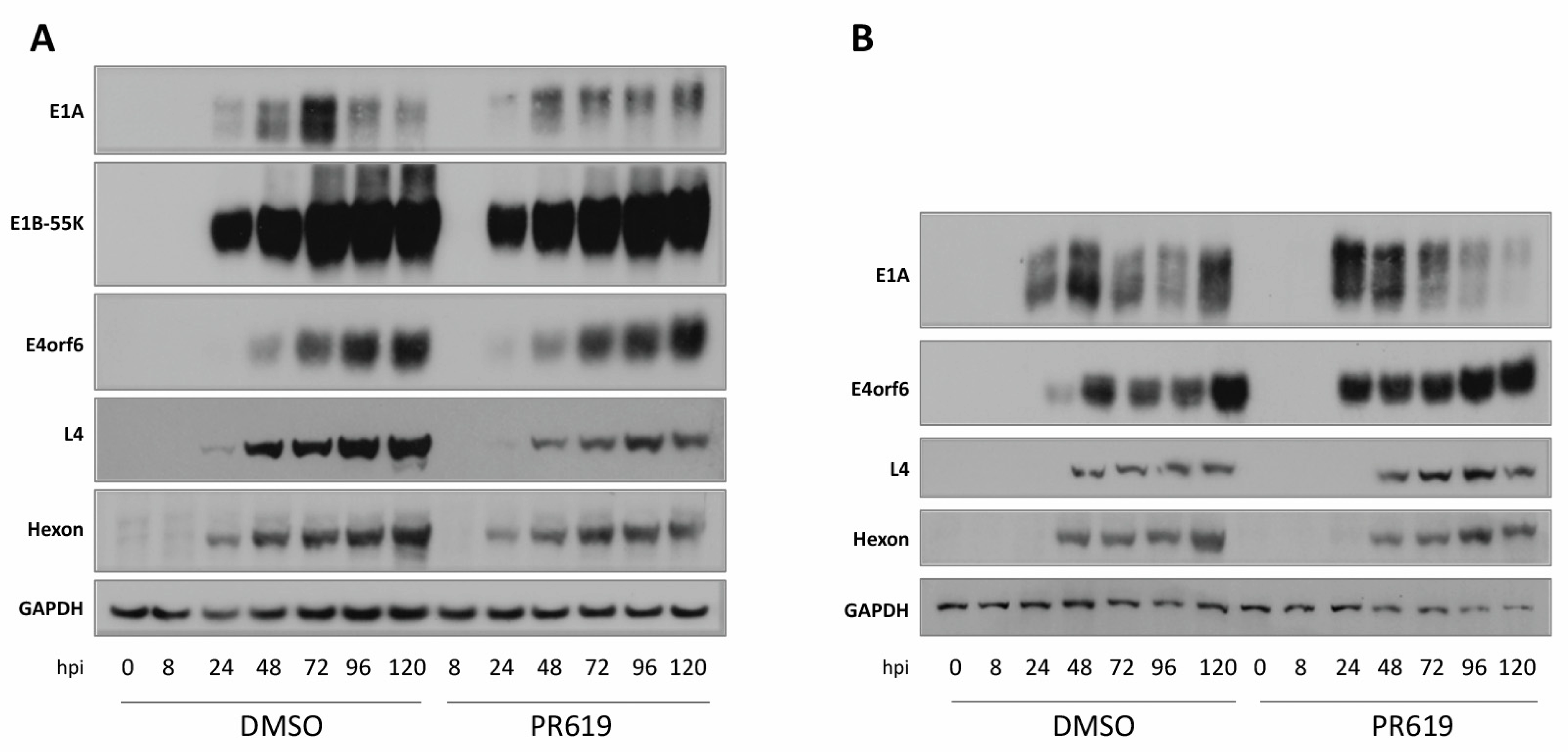
3.6. Effect of Deubiquitinase (DUB) Inhibition on HAdV-C5 Infection
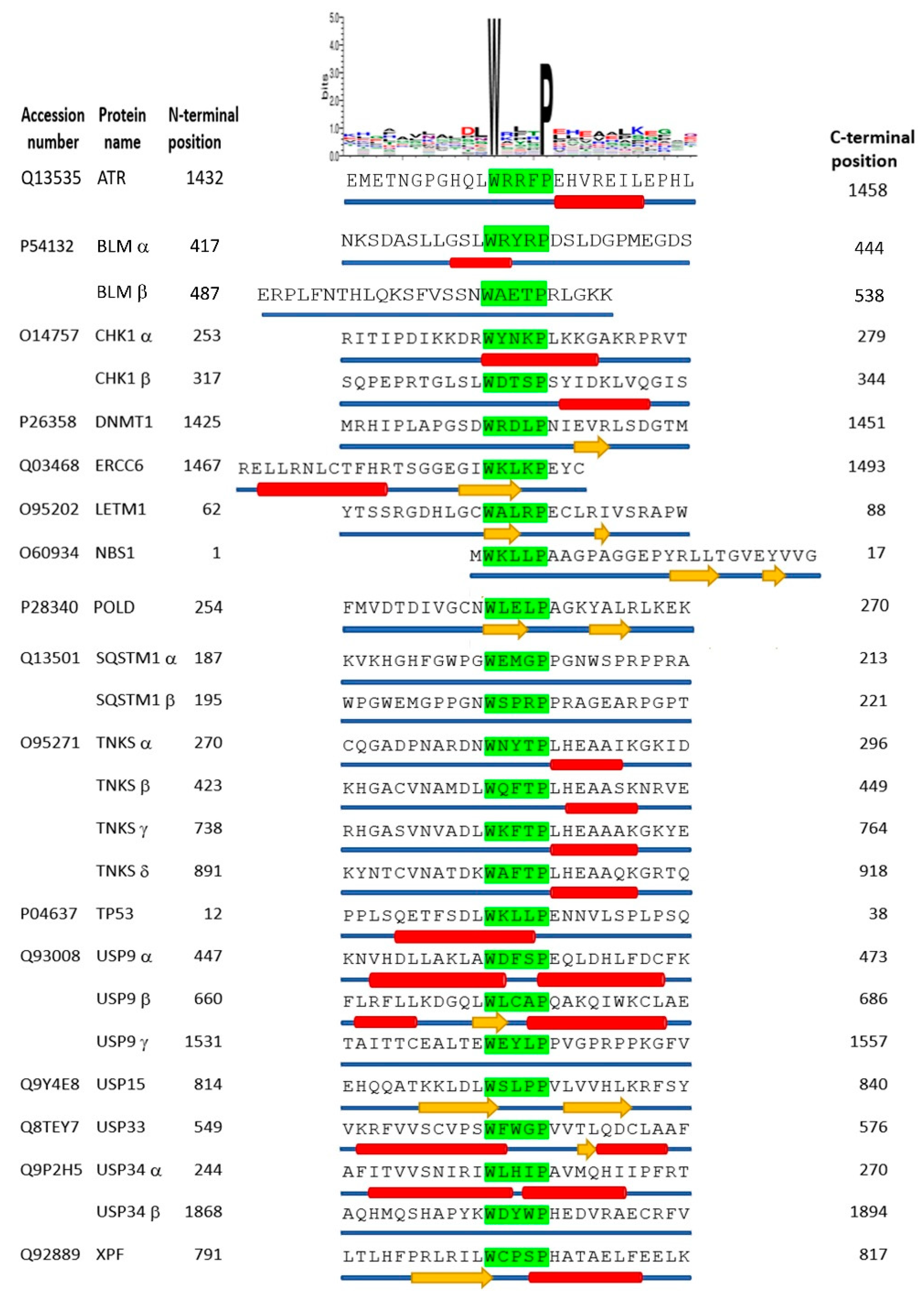
3.7. Structural Prediction for the xWxxxPx Motif
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Benko, M.; Aoki, K.; Arnberg, N.; Davison, A.J.; Echavarria, M.; Hess, M.; Jones, M.S.; Kajan, G.L.; Kajon, A.E.; Mittal, S.K.; et al. ICTV Virus Taxonomy Profile: Adenoviridae 2022. J. Gen. Virol. 2022, 103, 001721. [Google Scholar] [CrossRef]
- Khanal, S.; Ghimire, P.; Dhamoon, A.S. The Repertoire of Adenovirus in Human Disease: The Innocuous to the Deadly. Biomedicines 2018, 6, 30. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P., 3rd; Kajon, A.E. Adenovirus: Epidemiology, Global Spread of Novel Serotypes, and Advances in Treatment and Prevention. Semin. Respir. Crit. Care Med. 2016, 37, 586–602. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.J. Recent lessons in gene expression, cell cycle control, and cell biology from adenovirus. Oncogene 2005, 24, 7673–7685. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.H.; Dobner, T. Cell transformation by the adenovirus oncogenes E1 and E4. FEBS Lett. 2020, 594, 1848–1860. [Google Scholar] [CrossRef] [PubMed]
- King, C.R.; Zhang, A.; Tessier, T.M.; Gameiro, S.F.; Mymryk, J.S. Hacking the Cell: Network Intrusion and Exploitation by Adenovirus E1A. mBio 2018, 9, e00390-18. [Google Scholar] [CrossRef] [PubMed]
- Cuconati, A.; White, E. Viral homologs of BCL-2: Role of apoptosis in the regulation of virus infection. Genes Dev. 2002, 16, 2465–2478. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Gilson, T.; Dallaire, F.; Ketner, G.; Branton, P.E.; Blanchette, P. The E4orf6/E1B55K E3 ubiquitin ligase complexes of human adenoviruses exhibit heterogeneity in composition and substrate specificity. J. Virol. 2011, 85, 765–775. [Google Scholar] [CrossRef]
- Querido, E.; Blanchette, P.; Yan, Q.; Kamura, T.; Morrison, M.; Boivin, D.; Kaelin, W.G.; Conaway, R.C.; Conaway, J.W.; Branton, P.E. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 2001, 15, 3104–3117. [Google Scholar] [CrossRef]
- Querido, E.; Marcellus, R.C.; Lai, A.; Charbonneau, R.; Teodoro, J.G.; Ketner, G.; Branton, P.E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 1997, 71, 3788–3798. [Google Scholar] [CrossRef]
- Steegenga, W.T.; Riteco, N.; Jochemsen, A.G.; Fallaux, F.J.; Bos, J.L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 1998, 16, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Forrester, N.A.; Sedgwick, G.G.; Thomas, A.; Blackford, A.N.; Speiseder, T.; Dobner, T.; Byrd, P.J.; Stewart, G.S.; Turnell, A.S.; Grand, R.J. Serotype-specific inactivation of the cellular DNA damage response during adenovirus infection. J. Virol. 2011, 85, 2201–2211. [Google Scholar] [CrossRef] [PubMed]
- von Stromberg, K.; Seddar, L.; Ip, W.H.; Günther, T.; Gornott, B.; Weinert, S.C.; Hüppner, M.; Bertzbach, L.D.; Dobner, T. The human adenovirus E1B-55K oncoprotein coordinates cell transformation through regulation of DNA-bound host transcription factors. Proc. Natl. Acad. Sci. USA 2023, 120, e2310770120. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Patel, R.N.; Forrester, N.A.; Theil, K.; Groitl, P.; Stewart, G.S.; Taylor, A.M.; Morgan, I.M.; Dobner, T.; Grand, R.J.; et al. Adenovirus 12 E4orf6 inhibits ATR activation by promoting TOPBP1 degradation. Proc. Natl. Acad. Sci. USA 2010, 107, 12251–12256. [Google Scholar] [CrossRef] [PubMed]
- Blanchette, P.; Cheng, C.Y.; Yan, Q.; Ketner, G.; Ornelles, D.A.; Dobner, T.; Conaway, R.C.; Conaway, J.W.; Branton, P.E. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 2004, 24, 9619–9629. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Wimmer, P.; Dobner, T. Adenovirus degradation of cellular proteins. Future Microbiol. 2012, 7, 211–225. [Google Scholar] [CrossRef] [PubMed]
- Ip, W.H.; Tatham, M.H.; Krohne, S.; Gruhne, J.; Melling, M.; Meyer, T.; Gornott, B.; Bertzbach, L.D.; Hay, R.T.; Rodriguez, E.; et al. Adenovirus E1B-55K controls SUMO-dependent degradation of antiviral cellular restriction factors. J. Virol. 2023, e0079123. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; Rohleder, K.J.; Hanakahi, L.A.; Ketner, G. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 2007, 81, 7034–7040. [Google Scholar] [CrossRef]
- Chalabi Hagkarim, N.; Ryan, E.L.; Byrd, P.J.; Hollingworth, R.; Shimwell, N.J.; Agathanggelou, A.; Vavasseur, M.; Kolbe, V.; Speiseder, T.; Dobner, T.; et al. Degradation of a Novel DNA Damage Response Protein, Tankyrase 1 Binding Protein 1, following Adenovirus Infection. J. Virol. 2018, 92, e02034-17. [Google Scholar] [CrossRef]
- Ching, W.; Koyuncu, E.; Singh, S.; Arbelo-Roman, C.; Härtl, B.; Kremmer, E.; Speiseder, T.; Meier, C.; Dobner, T. A ubiquitin-specific protease possesses a decisive role for adenovirus replication and oncogene-mediated transformation. PLoS Pathog. 2013, 9, e1003273. [Google Scholar] [CrossRef]
- Orazio, N.I.; Naeger, C.M.; Karlseder, J.; Weitzman, M.D. The adenovirus E1b55K/E4orf6 complex induces degradation of the Bloom helicase during infection. J. Virol. 2011, 85, 1887–1892. [Google Scholar] [CrossRef]
- Stracker, T.H.; Carson, C.T.; Weitzman, M.D. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 2002, 418, 348–352. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Bürck, C.; Glass, M.; Groitl, P.; Wimmer, P.; Kinkley, S.; Mund, A.; Everett, R.D.; Dobner, T. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013, 41, 3532–3550. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Kinkley, S.; Bürck, C.; Mund, A.; Wimmer, P.; Schubert, T.; Groitl, P.; Will, H.; Dobner, T. SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog. 2013, 9, e1003775. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, S.; Wimmer, P.; Sirma, H.; Everett, R.D.; Blanchette, P.; Groitl, P.; Dobner, T. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 2010, 84, 7029–7038. [Google Scholar] [CrossRef] [PubMed]
- Dallaire, F.; Blanchette, P.; Groitl, P.; Dobner, T.; Branton, P.E. Identification of integrin alpha3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 2009, 83, 5329–5338. [Google Scholar] [CrossRef]
- Herrmann, C.; Dybas, J.M.; Liddle, J.C.; Price, A.M.; Hayer, K.E.; Lauman, R.; Purman, C.E.; Charman, M.; Kim, E.T.; Garcia, B.A.; et al. Adenovirus-mediated ubiquitination alters protein-RNA binding and aids viral RNA processing. Nat. Microbiol. 2020, 5, 1217–1231. [Google Scholar] [CrossRef] [PubMed]
- Flint, S.J.; Gonzalez, R.A. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 2003, 272, 287–330. [Google Scholar] [CrossRef]
- Gales, J.P.; Kubina, J.; Geldreich, A.; Dimitrova, M. Strength in Diversity: Nuclear Export of Viral RNAs. Viruses 2020, 12, 1014. [Google Scholar] [CrossRef]
- Valdés Alemán, M.; Bertzbach, L.D.; Speiseder, T.; Ip, W.H.; González, R.A.; Dobner, T. Global Transcriptome Analyses of Cellular and Viral mRNAs During HAdV-C5 Infection Highlight New Aspects of Viral mRNA Biogenesis and Cytoplasmic Viral mRNA Accumulations. Viruses 2022, 14, 2428. [Google Scholar] [CrossRef]
- Yatherajam, G.; Huang, W.; Flint, S.J. Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor. J. Virol. 2011, 85, 1429–1438. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, P.; Garces, Y.; Mundo, E.; Lopez, R.E.; Bertzbach, L.D.; Dobner, T.; Gonzalez, R.A. E1B-55K is a phosphorylation-dependent transcriptional and post-transcriptional regulator of viral gene expression in HAdV-C5 infection. J. Virol. 2022, 96, JVI0206221. [Google Scholar] [CrossRef] [PubMed]
- Gabler, S.; Schutt, H.; Groitl, P.; Wolf, H.; Shenk, T.; Dobner, T. E1B 55-kilodalton-associated protein: A cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J. Virol. 1998, 72, 7960–7971. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, P.; Ip, W.H.; Dobner, T.; Gonzalez, R.A. The biology of the adenovirus E1B 55K protein. FEBS Lett. 2019, 593, 3504–3517. [Google Scholar] [CrossRef] [PubMed]
- Menendez-Conejero, R.; Nguyen, T.H.; Singh, A.K.; Condezo, G.N.; Marschang, R.E.; van Raaij, M.J.; San Martin, C. Structure of a Reptilian Adenovirus Reveals a Phage Tailspike Fold Stabilizing a Vertebrate Virus Capsid. Structure 2017, 25, 1562–1573.e5. [Google Scholar] [CrossRef] [PubMed]
- Sieber, T.; Scholz, R.; Spoerner, M.; Schumann, F.; Kalbitzer, H.R.; Dobner, T. Intrinsic disorder in the common N-terminus of human adenovirus 5 E1B-55K and its related E1BN proteins indicated by studies on E1B-93R. Virology 2011, 418, 133–143. [Google Scholar] [CrossRef]
- Tejera, B.; Lopez, R.E.; Hidalgo, P.; Cardenas, R.; Ballesteros, G.; Rivillas, L.; French, L.; Amero, C.; Pastor, N.; Santiago, A.; et al. The human adenovirus type 5 E1B 55kDa protein interacts with RNA promoting timely DNA replication and viral late mRNA metabolism. PLoS ONE 2019, 14, e0214882. [Google Scholar] [CrossRef]
- Gonzalez, R.A.; Flint, S.J. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 2002, 76, 4507–4519. [Google Scholar] [CrossRef]
- Kao, C.C.; Yew, P.R.; Berk, A.J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55k proteins. Virology 1990, 179, 806–814. [Google Scholar] [CrossRef]
- Rubenwolf, S.; Schutt, H.; Nevels, M.; Wolf, H.; Dobner, T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J. Virol. 1997, 71, 1115–1123. [Google Scholar] [CrossRef]
- Shen, Y.; Kitzes, G.; Nye, J.A.; Fattaey, A.; Hermiston, T. Analyses of single-amino-acid substitution mutants of adenovirus type 5 E1B-55K protein. J. Virol. 2001, 75, 4297–4307. [Google Scholar] [CrossRef] [PubMed]
- Yew, P.R.; Kao, C.C.; Berk, A.J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology 1990, 179, 795–805. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, M.; Ip, W.H.; Hofmann-Sieber, H.; Wilkens, B.; Nkrumah, F.K.; Zhang, W.; Ehrhardt, A.; Bertzbach, L.D.; Dobner, T. Protein–Protein Interactions Facilitate E4orf6-Dependent Regulation of E1B-55K SUMOylation in HAdV-C5 Infection. Viruses 2022, 14, 463. [Google Scholar] [CrossRef] [PubMed]
- Grand, R.J.; Parkhill, J.; Szestak, T.; Rookes, S.M.; Roberts, S.; Gallimore, P.H. Definition of a major p53 binding site on Ad2E1B58K protein and a possible nuclear localization signal on the Ad12E1B54K protein. Oncogene 1999, 18, 955–965. [Google Scholar] [CrossRef] [PubMed]
- Yew, P.R.; Berk, A.J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature 1992, 357, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Wimmer, P.; Berscheminski, J.; Blanchette, P.; Groitl, P.; Branton, P.E.; Hay, R.T.; Dobner, T.; Schreiner, S. PML isoforms IV and V contribute to adenovirus-mediated oncogenic transformation by functionally inhibiting the tumor-suppressor p53. Oncogene 2016, 35, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Bürck, C.; Mund, A.; Berscheminski, J.; Kieweg, L.; Müncheberg, S.; Dobner, T.; Schreiner, S. KAP1 Is a Host Restriction Factor That Promotes Human Adenovirus E1B-55K SUMO Modification. J. Virol. 2016, 90, 930–946. [Google Scholar] [CrossRef]
- Lin, J.; Chen, J.; Elenbaas, B.; Levine, A.J. Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev. 1994, 8, 1235–1246. [Google Scholar] [CrossRef]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef]
- Mooser, C.; Symeonidou, I.E.; Leimbacher, P.A.; Ribeiro, A.; Shorrocks, A.K.; Jungmichel, S.; Larsen, S.C.; Knechtle, K.; Jasrotia, A.; Zurbriggen, D.; et al. Treacle controls the nucleolar response to rDNA breaks via TOPBP1 recruitment and ATR activation. Nat. Commun. 2020, 11, 123. [Google Scholar] [CrossRef]
- Byrd, P.; Brown, K.W.; Gallimore, P.H. Malignant transformation of human embryo retinoblasts by cloned adenovirus 12 DNA. Nature 1982, 298, 69–71. [Google Scholar] [CrossRef] [PubMed]
- Barker, D.D.; Berk, A.J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 1987, 156, 107–121. [Google Scholar] [CrossRef] [PubMed]
- Byrd, P.J.; Grand, R.J.; Breiding, D.; Williams, J.F.; Gallimore, P.H. Host range mutants of adenovirus type 12 E1 defective for lytic infection, transformation, and oncogenicity. Virology 1988, 163, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Harlow, E.; Franza, B.R., Jr.; Schley, C. Monoclonal antibodies specific for adenovirus early region 1A proteins: Extensive heterogeneity in early region 1A products. J. Virol. 1985, 55, 533–546. [Google Scholar] [CrossRef]
- Sarnow, P.; Ho, Y.S.; Williams, J.; Levine, A.J. Adenovirus E1b-58kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell 1982, 28, 387–394. [Google Scholar] [CrossRef]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Jones, D.T. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999, 292, 195–202. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef]
- Frishman, D.; Argos, P. Incorporation of non-local interactions in protein secondary structure prediction from the amino acid sequence. Protein Eng. 1996, 9, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Hung, G.; Flint, S.J. Normal human cell proteins that interact with the adenovirus type 5 E1B 55kDa protein. Virology 2017, 504, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Punga, T.; Akusjärvi, G. The adenovirus-2 E1B-55K protein interacts with a mSin3A/histone deacetylase 1 complex. FEBS Lett. 2000, 476, 248–252. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Jha, S.; Engel, D.A.; Ornelles, D.A.; Dutta, A. Tip60 degradation by adenovirus relieves transcriptional repression of viral transcriptional activator EIA. Oncogene 2013, 32, 5017–5025. [Google Scholar] [CrossRef] [PubMed]
- Nayak, R.; Farris, K.D.; Pintel, D.J. E4Orf6-E1B-55k-dependent degradation of de novo-generated adeno-associated virus type 5 Rep52 and capsid proteins employs a cullin 5-containing E3 ligase complex. J. Virol. 2008, 82, 3803–3808. [Google Scholar] [CrossRef] [PubMed]
- Sukhu, L.; Pintel, D. The large Rep protein of adeno-associated virus type 2 is polyubiquitinated. J. Gen. Virol. 2011, 92 Pt 12, 2792–2796. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.Y.; Gilson, T.; Wimmer, P.; Schreiner, S.; Ketner, G.; Dobner, T.; Branton, P.E.; Blanchette, P. Role of E1B55K in E4orf6/E1B55K E3 ligase complexes formed by different human adenovirus serotypes. J. Virol. 2013, 87, 6232–6245. [Google Scholar] [CrossRef]
- Blackford, A.N.; Bruton, R.K.; Dirlik, O.; Stewart, G.S.; Taylor, A.M.; Dobner, T.; Grand, R.J.; Turnell, A.S. A role for E1B-AP5 in ATR signaling pathways during adenovirus infection. J. Virol. 2008, 82, 7640–7652. [Google Scholar] [CrossRef]
- Ornelles, D.A.; Shenk, T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: Association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 1991, 65, 424–429. [Google Scholar] [CrossRef]
- Kleinberger, T. En Guard! The Interactions between Adenoviruses and the DNA Damage Response. Viruses 2020, 12, 996. [Google Scholar] [CrossRef]
- Cowell, I.G.; Ling, E.M.; Swan, R.L.; Brooks, M.L.W.; Austin, C.A. The Deubiquitinating Enzyme Inhibitor PR-619 is a Potent DNA Topoisomerase II Poison. Mol. Pharmacol. 2019, 96, 562–572. [Google Scholar] [CrossRef] [PubMed]
- Seiberlich, V.; Goldbaum, O.; Zhukareva, V.; Richter-Landsberg, C. The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: Implications for neurodegenerative diseases. Biochim. Biophys. Acta 2012, 1823, 2057–2068. [Google Scholar] [CrossRef] [PubMed]
- Blackford, A.N.; Grand, R.J. Adenovirus E1B 55-kilodalton protein: Multiple roles in viral infection and cell transformation. J. Virol. 2009, 83, 4000–4012. [Google Scholar] [CrossRef] [PubMed]
- Pelka, P.; Ablack, J.N.; Fonseca, G.J.; Yousef, A.F.; Mymryk, J.S. Intrinsic structural disorder in adenovirus E1A: A viral molecular hub linking multiple diverse processes. J. Virol. 2008, 82, 7252–7263. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Tan, X. Viral Manipulations of the Cullin-RING Ubiquitin Ligases. Adv. Exp. Med. Biol. 2020, 1217, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Charman, M.; Herrmann, C.; Weitzman, M.D. Viral and cellular interactions during adenovirus DNA replication. FEBS Lett. 2019, 593, 3531–3550. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, P.; Anzures, L.; Hernandez-Mendoza, A.; Guerrero, A.; Wood, C.D.; Valdes, M.; Dobner, T.; Gonzalez, R.A. Morphological, Biochemical, and Functional Study of Viral Replication Compartments Isolated from Adenovirus-Infected Cells. J. Virol. 2016, 90, 3411–3427. [Google Scholar] [CrossRef]
- Charman, M.; Weitzman, M.D. Replication Compartments of DNA Viruses in the Nucleus: Location, Location, Location. Viruses 2020, 12, 151. [Google Scholar] [CrossRef]
- Sy, S.M.; Jiang, J.; O, W.S.; Deng, Y.; Huen, M.S. The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 2013, 41, 8572–8580. [Google Scholar] [CrossRef]
- Kusakabe, S.; Suzuki, T.; Sugiyama, Y.; Haga, S.; Horike, K.; Tokunaga, M.; Hirano, J.; Zhang, H.; Chen, D.V.; Ishiga, H.; et al. USP15 Participates in Hepatitis C Virus Propagation through Regulation of Viral RNA Translation and Lipid Droplet Formation. J. Virol. 2019, 93, e01708-18. [Google Scholar] [CrossRef]
- Ming, S.L.; Zhang, S.; Wang, Q.; Zeng, L.; Zhou, L.Y.; Wang, M.D.; Ma, Y.X.; Han, L.Q.; Zhong, K.; Zhu, H.S.; et al. Inhibition of USP14 influences alphaherpesvirus proliferation by degrading viral VP16 protein via ER stress-triggered selective autophagy. Autophagy 2022, 18, 1801–1821. [Google Scholar] [CrossRef] [PubMed]
- Carson, C.T.; Schwartz, R.A.; Stracker, T.H.; Lilley, C.E.; Lee, D.V.; Weitzman, M.D. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J. 2003, 22, 6610–6620. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, R.A.; Lakdawala, S.S.; Eshleman, H.D.; Russell, M.R.; Carson, C.T.; Weitzman, M.D. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 2008, 82, 9043–9055. [Google Scholar] [CrossRef] [PubMed]
- Lakdawala, S.S.; Schwartz, R.A.; Ferenchak, K.; Carson, C.T.; McSharry, B.P.; Wilkinson, G.W.; Weitzman, M.D. Differential requirements of the C terminus of Nbs1 in suppressing adenovirus DNA replication and promoting concatemer formation. J. Virol. 2008, 82, 8362–8372. [Google Scholar] [CrossRef]


| Group 1 | Protein | Amino Acid Sequence | Residues (WxxxP) | Reference |
|---|---|---|---|---|
| I | CEP170 | NSRWRRFPTDYA | 1230–1234 | [62] |
| I | CNOT3 | EAAWHHMPHPSD | 622–626 | [19] |
| I | Cullin 5 | LFAWNQRPREKI | 506–510 | [62] |
| I | hnRNPR | QQNWGSQPIAQQ | 597–601 | [62] |
| I | Integrin α3 (ITGA3) | NGKWLLYPTEIT NGSWPCRPPGDL HCVWLECPIPDA | 829–833 843–847 913–917 | [26] |
| I | mSin3a | NDTWVSFPSWSE | 585–589 | [63] |
| I | MYCB2 | AGKWVELPITKS VPYWNAKPAPMP DVIWRFRPNTRE | 571–575 1034–1038 1138–1142 | [62] |
| I | SQSTM1 (p62) | WPGWEMGPPGNW PGNWSPRPPRAG | 198–202 206–210 | [62] |
| I | SRSF3 | PPSWGRRPRDDY | 96–100 | [62] |
| I | UBR5 | LYWWGVVPFSQR DPDWLDLPPISS PPSWVPDPPAMD | 494–498 835–839 1027–1031 | [62] |
| I | USP8 | IEIWKLPPVLLV | 1001–1005 | [62] |
| I | USP15 | LDLWSLPPVLVV | 825–829 | [62] |
| I | USP34 | IRIWLHIPAVMQ PYKWDYWPHEDV | 255–259 1879–1883 | [62] |
| I | ZNF638 | GSRWDDEPHISA | 104–108 | [62] |
| II | BLM | GSLWRYRPDSLD | 428–432 | [21] |
| II | DNA ligase IV | RYSWDCSPLSMF | 805–809 | [18] |
| II | NBS1 (NBN) | MWKLLPAAGP | 2–6 | [22] |
| II | p53 | SDLWKLLPENNV | 23–27 | [55] |
| II | Tab182 (TNKS1BP1) | PPSWRPQPDGEA | 1507–1511 | [19] |
| II | TIP60 | LKPWYFSPYPQE | 245–249 | [64] |
| II | TOPBP1 | NLQWPSCPTQYS | 163–1167 | [14] |
| III | AAV Rep52 | NTIWLFGPATTG | 108–112 | [65] |
| III | AAV Rep68 | EKEWELPPDSDM NTIWLFGPATTG | 35–39 331–335 | [66] |
| III | AAV Capsid | YKNWFPGPMGRT GPIWAKIPETGA | 464–468 608–612 | [65] |
| IV | TNFRSF10A | TQQWEHSPLGEL | 123–127 | [27] |
| IV | RP2 | ELNWSLLPEDAV | 186–190 | [27] |
| IV | CLPTM1 | YLSWILFPLLGC | 481–485 | [27] |
| IV | PDGKRB | IMLWQKKPRYEI | 556–560 | [27] |
| IV | FAS | LGIWTLLPLVLT | 4–8 | [27] |
| IV | CXADR | DIEWLISPADNQ | 57–61 | [27] |
| IV | EPHA7 | DIEWLISPADNQ ELEWISSPPNGW | 57–61 397–401 | [27] |
| IV | STK11IP | HGSWSLSPPPER | 774–778 | [27] |
| IV | TRPC4AP | WGGWGGRPRPGN | 25–29 | [27] |
| IV | CLCC1 | NPIWLVPPTKAL | 242–246 | [27] |
| IV | BABAM1 | PKSWQVPPPAPE | 73–77 | [27] |
| V | HAX1 | DDVWPMDPHPRT | 173–177 | [27] |
| V | SCAMP3 | QNNWPPLPSFCP | 135–139 | [27] |
| V | SPTLC1 | IEEWQPEPLVPP | 64–68 | [27] |
| V | EGFR | EGCWGPEPRDCV | 471–475 | [27] |
| V | RPN2 | ASTWALTPTHYL | 21–25 | [27] |
| V | ITGB4 | GSFWWLIPLLLL | 712–716 | [27] |
| V | TARS | AESWKTTPYQIA PRSWRELPLRLA | 98–102 423–427 | [27] |
| V | CANX | PEDWDERPKIPD | 341–345 | [27] |
| V | EPHA2 | ELGWLTHPYGKG SVSWSIPPPQQS | 42–46 456–460 | [27] |
| V | EPHB4 | DLKWVTFPQVDG | 32–36 | [27] |
| V | RPL15 | DTQWITKPVHKH | 147–151 | [27] |
| V | GTF2I | SPSWYGIPRLEK | 518–522 | [27] |
| SPTWFGIPRLER | 623–627 | |||
| V | CDK6 | VTLWYRAPEVLL | 184–188 | [27] |
| V | hnRNPU | GQFWGQKPWSQH | 811–815 | [27] |
| V | SLC7A5 | GVWWKNKPKWLL | 478–482 | [27] |
| V | TAP1 | LLHWGSHPTAFV | 193–197 | [27] |
| V | DYNC1H1 | KMVWRINPAHRK TPSWLGLPNNAE | 450–454 4320–4324 | [27] |
| VI | ATR | HQLWRRFPEHVR | 1443–1447 | This work |
| VI | CHK1 | KDRWYNKPLKKG LSLWDTSPSYID | 264–268 328–332 | This work |
| VI | CSB (ERCC6) | EGIWKLKPEYC | 1486–1490 | This work |
| VI | DNMT1 | GSDWRDLPNIEV | 1436–1440 | This work |
| VI | LETM1 | LGCWALRPECLR | 73–77 | This work |
| VI | SQSTM1 (p62) | WPGWEMGPPGNW PGNWSPRPPRAG | 198–202 206–210 | [62] |
| VI | TNKS1 | RDNWNYTPLHEA MDLWQFTPLHEA ADLWKFTPLHEA TDKWAFTPLHEA | 281–285 434–438 748–752 902–906 | This work |
| VI | TNKS2 | RDNWNYTPLHEA MDLWQFTPLHEA ADLWKFTPLHEA TDKWAFTPLHEA | 123–127 276–280 591–595 744–748 | This work |
| VI | USP9 | KLAWDFSPEQLD GQLWLCAPQAKQ LTEWEYLPPVGP | 458–462 671–675 1542–1546 | This work |
| VI | USP15 | LDLWSLPPVLVV | 825–829 | [62] |
| VI | USP33 | VPSWFWGPVVTL | 561–565 | This work |
| VI | USP34 | IRIWLHIPAVMQ PYKWDYWPHEDV | 255–259 1879–1883 | [62] |
| VI | XPF | RILWCPSPHATA | 802–806 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chalabi Hagkarim, N.; Ip, W.-H.; Bertzbach, L.D.; Abualfaraj, T.; Dobner, T.; Molloy, D.P.; Stewart, G.S.; Grand, R.J. Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif. Viruses 2023, 15, 2356. https://doi.org/10.3390/v15122356
Chalabi Hagkarim N, Ip W-H, Bertzbach LD, Abualfaraj T, Dobner T, Molloy DP, Stewart GS, Grand RJ. Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif. Viruses. 2023; 15(12):2356. https://doi.org/10.3390/v15122356
Chicago/Turabian StyleChalabi Hagkarim, Nafiseh, Wing-Hang Ip, Luca D. Bertzbach, Tareq Abualfaraj, Thomas Dobner, David P. Molloy, Grant S. Stewart, and Roger J. Grand. 2023. "Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif" Viruses 15, no. 12: 2356. https://doi.org/10.3390/v15122356
APA StyleChalabi Hagkarim, N., Ip, W.-H., Bertzbach, L. D., Abualfaraj, T., Dobner, T., Molloy, D. P., Stewart, G. S., & Grand, R. J. (2023). Identification of Adenovirus E1B-55K Interaction Partners through a Common Binding Motif. Viruses, 15(12), 2356. https://doi.org/10.3390/v15122356






