Oncolytic Adenovirus for the Targeting of Paclitaxel-Resistant Breast Cancer Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Adenoviral Vectors
2.3. Binding Assay
2.4. Crystal Violet Assay
2.5. Immunofluorescent Analyses
- (1)
- Cell line preparation: Cells were grown in 96-well plates (10,000–20,000 cells per well), treated with 0.001–0.5 pfu/cell in 200 μL of growth medium, and incubated for up to 5 days. Cells were washed in PBS (Corning, New York, NY, USA), fixed (4% paraformaldehyde (VWR, Radnor, PA, USA); 20 min; ice), and permeabilized (0.2% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA); 1 h; room temperature).
- (2)
- Immune staining: Fixed and permeabilized cells were blocked for 2 h at room temperature in a 5% nonfat dry milk (Bio-Rad, Hercules, CA, USA) and 0.2% Triton X-100 solution (Sigma-Aldrich, St. Louis, MO, USA). Cells were washed with PBS (Corning, New York, NY, USA) and incubated at 4 °C overnight with a 5% bovine serum albumin (Roche, Basel, Switzerland), 1% glycine (Sigma-Aldrich, St. Louis, MO, USA), 2% goat serum (MP Biomedicals, Santa Ana, CA, USA), and 0.1% Triton X-100 solution (Sigma-Aldrich, St. Louis, MO, USA) containing the primary antibody anti-NIS (mouse anti-FP5A, MA5-12308, 1:500, Invitrogen, Waltham, MA, USA) and/or anti-CD44 (rabbit anti-CD44, ab189524, 1:200, Abcam, Cambridge, United Kingdom). Cells were then washed with PBS (Corning, New York, NY, USA) and incubated at room temperature, protected from light, for 2 h in the same primary antibody solution, containing either goat anti-mouse AF-555-conjugated (A21424, 1:1000, Invitrogen, Waltham, MA, USA; red; NIS-only staining), or goat anti-rabbit AF-488-conjugated (A11008, 1:1000, Invitrogen, Waltham, MA, USA; green; CD44-only staining), or AF-555- and Ad hexon FITC-conjugated (AB1056F, 1:1000, Millipore-Sigma, Burlington, MA, USA; green; costaining of NIS and Ad hexon proteins) secondary antibodies. For CD44 costaining with NIS, a combination of goat anti-rabbit AF-488 secondary antibody (for CD44, A11008, 1:1000, Invitrogen, Waltham, MA, USA; green) and goat anti-mouse AF-568 secondary antibody (for NIS, A11004, 1:1000, Invitrogen, Waltham, MA, USA; red) was used. For CD44 costaining with Ad hexon, a combination of goat anti-rabbit AF-568 secondary antibody (for CD44, A11011, 1:1000, Invitrogen, Waltham, MA, USA; red) and Ad hexon FITC-conjugated (AB1056F, 1:1000, Millipore-Sigma, Burlington, MA; green) was used. Cells were washed with PBS (Corning, New York, NY, USA) again and counterstained with a nuclear stain (DAPI, Sigma-Aldrich, St. Louis, MO, USA, 0.1 µg/mL, 20 min incubation, room temperature, in the dark) just prior to image capture using a fluorescent microscope (EVOS FL Auto, Life Technologies, Carlsbad, CA, USA). Plug-in functions of ImageJ software (version 1.53t, NIH, Madison, WI, USA) were used to quantify NIS and Ad hexon expression in cells using % area measurement and normalized to DAPI area.
2.6. Gene Expression Analysis
2.7. Tumorsphere Formation Assay
2.8. Statistical Analysis
3. Results
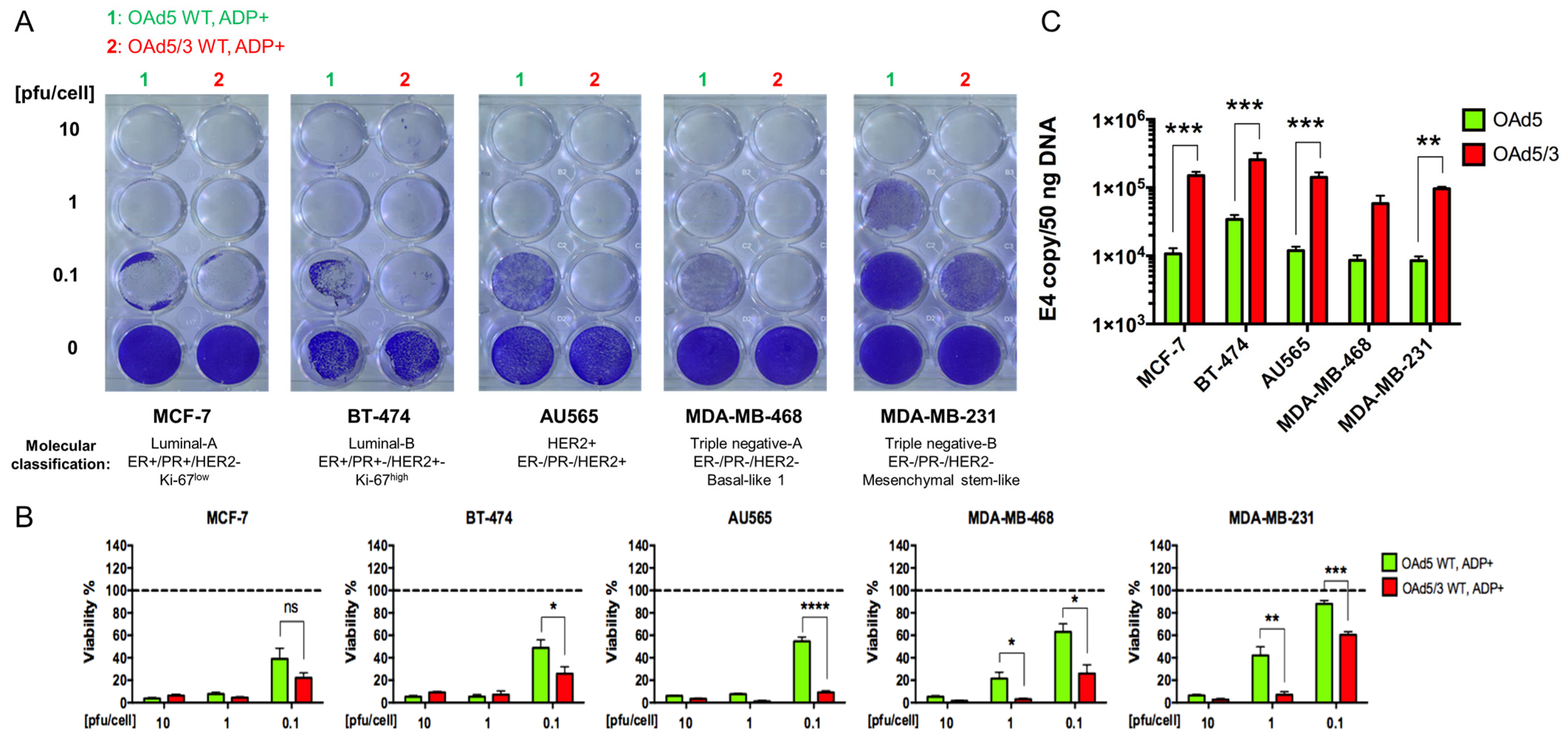
- Superiority of genetically modified Ad5/3 fiber for human breast cancer cells.
- 2.
- The effect of Cox-2 promoter and ADP deletion on OAd-hNIS replication and killing ability.
- 3.
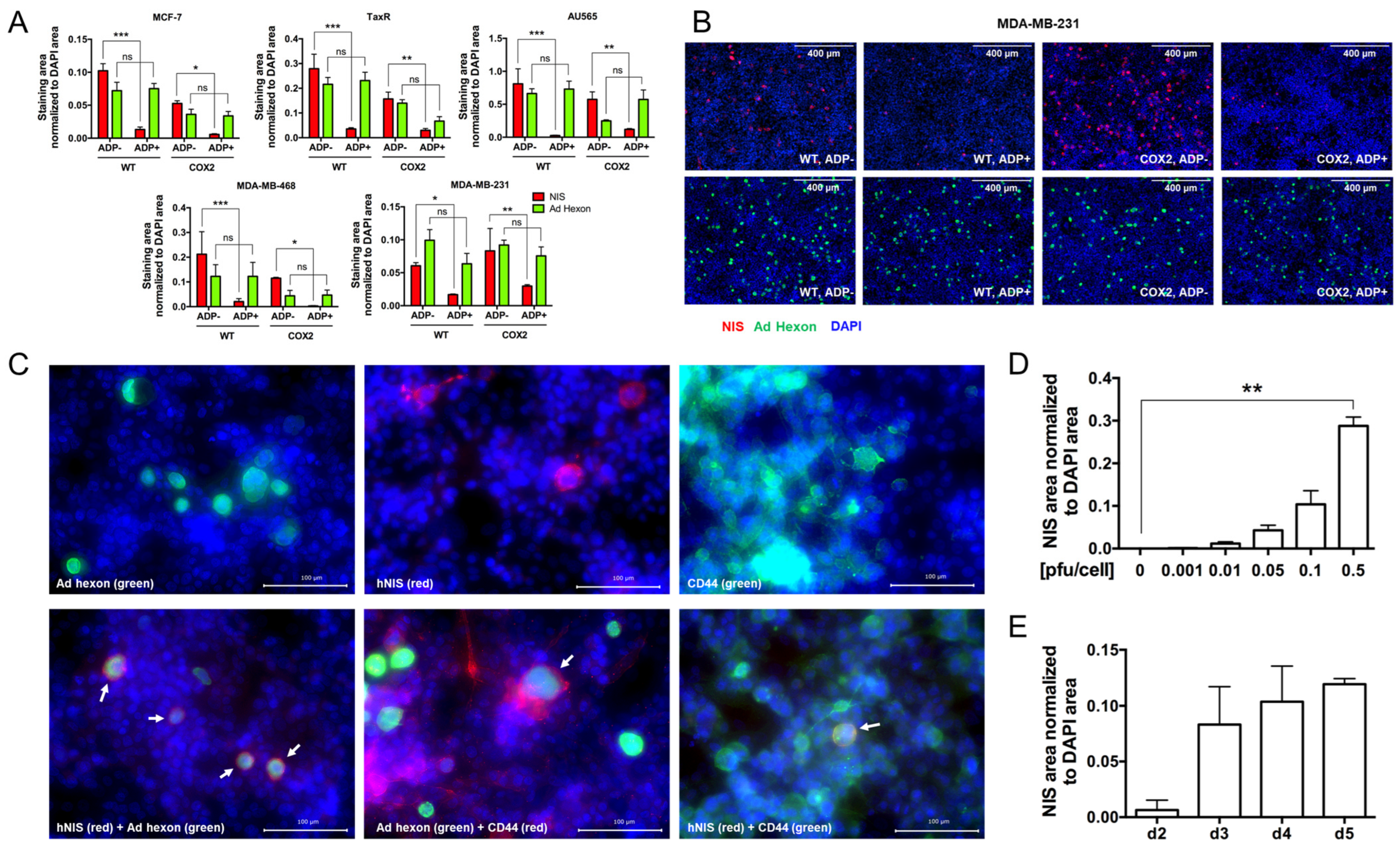
- The effect of ADP deletion on hNIS expression in breast cancer cells.
- 4.
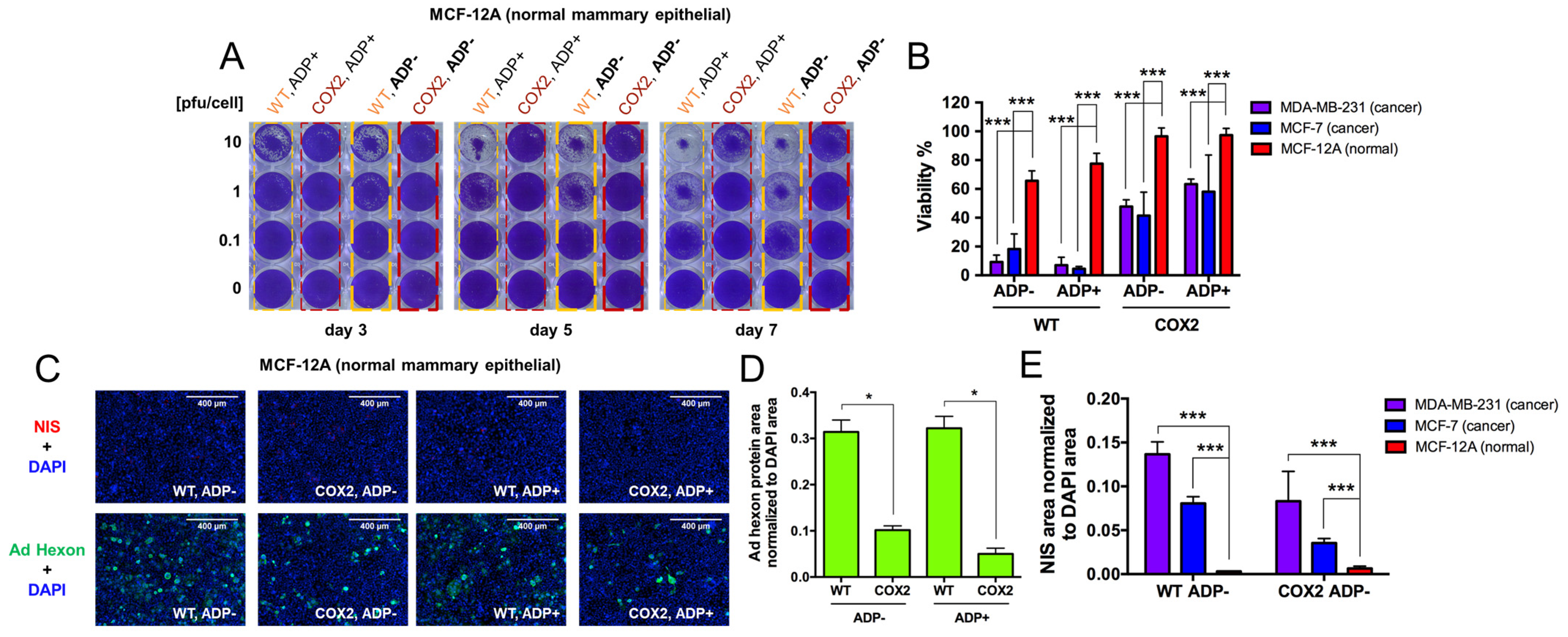
- Evaluation of OAd5/3-hNIS vectors’ selectivity.
- 5.
- Evaluation of OAd-hNIS vectors’ replication potential in chemoresistant TaxR cells.
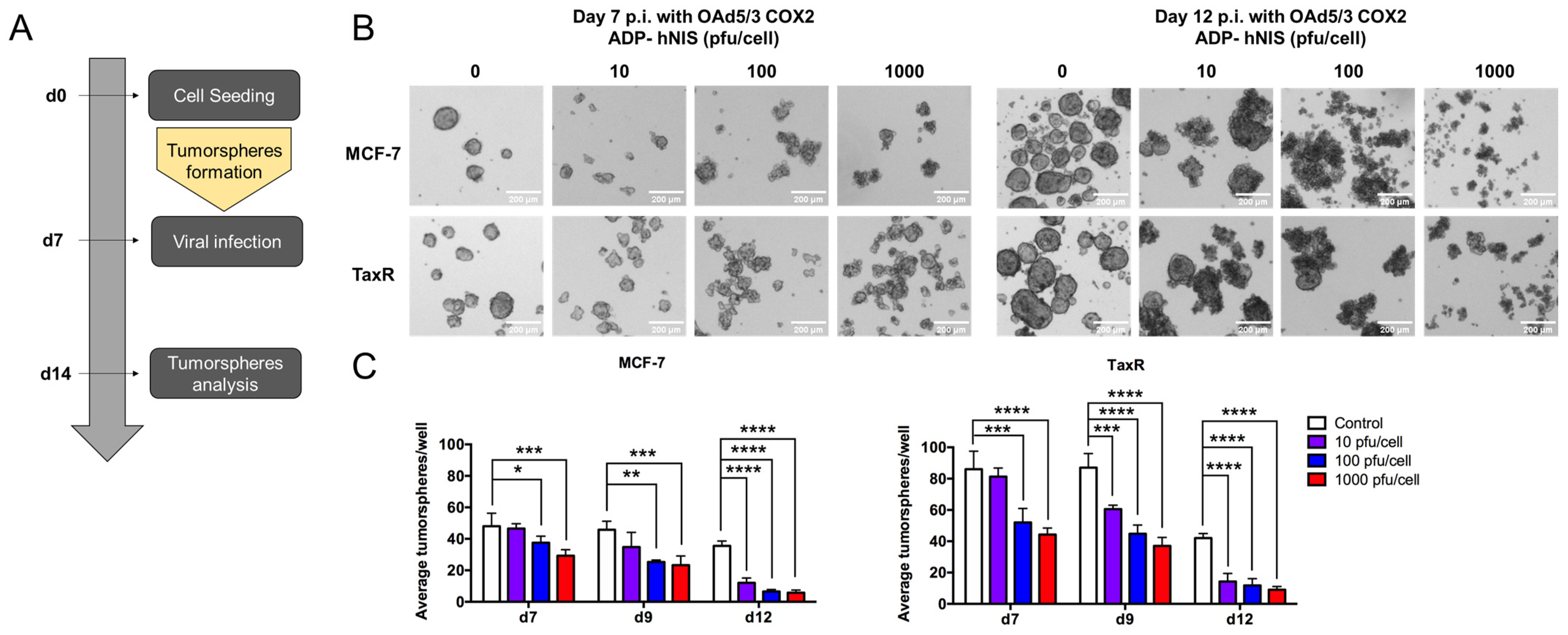
- 6.
- Evaluation of OAd5/3 Cox-2 ADP(−) hNIS efficiency to target paclitaxel-resistant BCSCs in tumorsphere assay.
- 7.
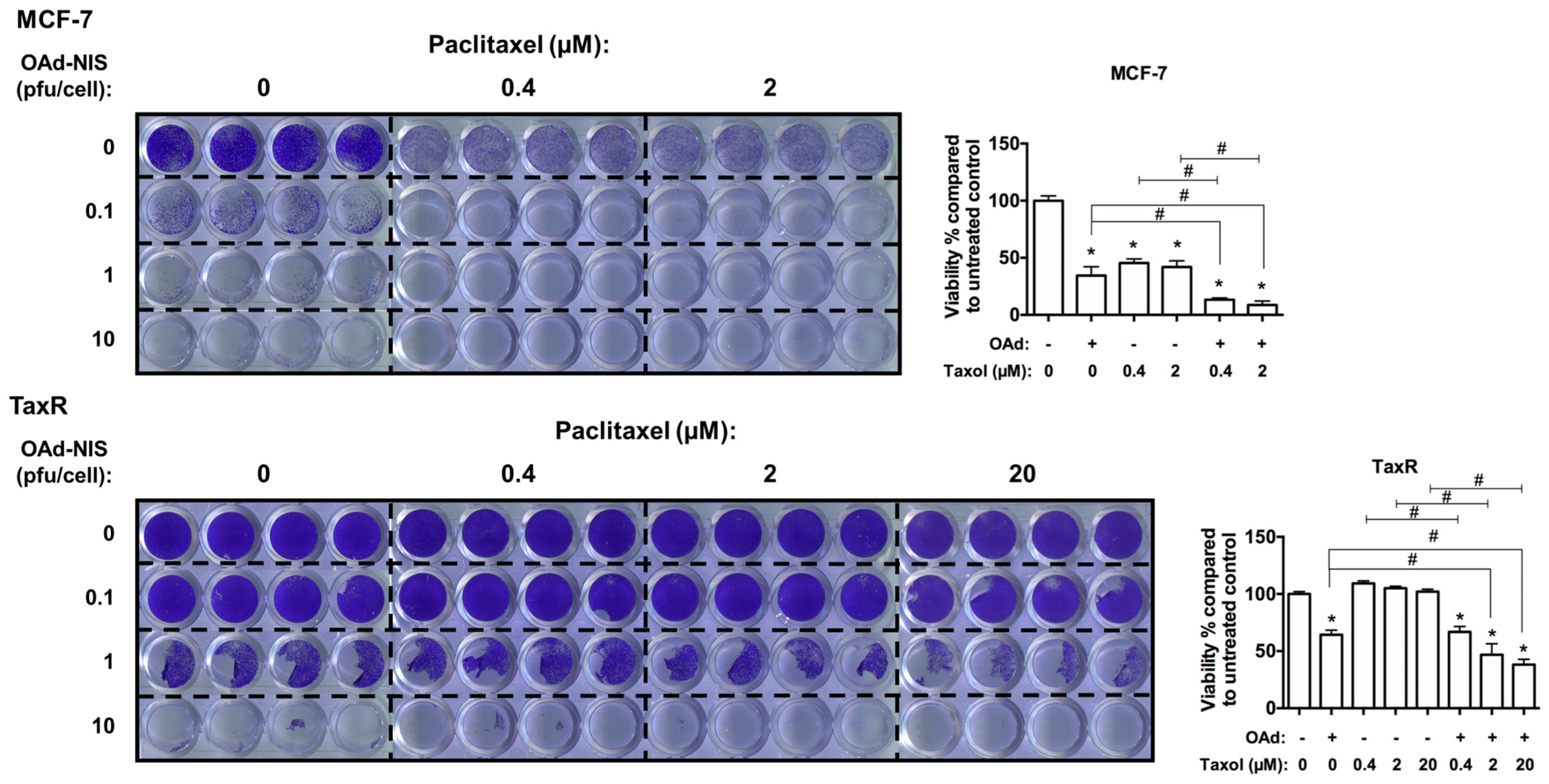
- Evaluation of OAd5/3 Cox-2 ADP(−) hNIS in combination treatment with paclitaxel on chemoresistant and chemosensitive ER+ BCSCs.
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Sleightholm, R.; Neilsen, B.K.; Elkhatib, S.; Flores, L.; Dukkipati, S.; Zhao, R.; Choudhury, S.; Gardner, B.; Carmichael, J.; Smith, L.; et al. Percentage of Hormone Receptor Positivity in Breast Cancer Provides Prognostic Value: A Single-Institute Study. J. Clin. Med. Res. 2021, 13, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Wu, V.S.; Kanaya, N.; Lo, C.; Mortimer, J.; Chen, S. From Bench to Bedside: What Do We Know about Hormone Receptor-Positive and Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer? J. Steroid Biochem. Mol. Biol. 2015, 153, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Gray, R.; Braybrooke, J.; Davies, C.; Taylor, C.; McGale, P.; Peto, R.; Pritchard, K.I.; Bergh, J.; Dowsett, M.; et al. 20-Year Risks of Breast-Cancer Recurrence after Stopping Endocrine Therapy at 5 Years. N. Engl. J. Med. 2017, 377, 1836–1846. [Google Scholar] [CrossRef] [PubMed]
- Davies, C.; Pan, H.; Godwin, J.; Gray, R.; Arriagada, R.; Raina, V.; Abraham, M.; Medeiros Alencar, V.H.; Badran, A.; Bonfill, X.; et al. Long-Term Effects of Continuing Adjuvant Tamoxifen to 10 Years versus Stopping at 5 Years after Diagnosis of Oestrogen Receptor-Positive Breast Cancer: ATLAS, a Randomised Trial. Lancet Lond. Engl. 2013, 381, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Early Breast Cancer Trialists’ Collaborative Group (EBCTCG); Davies, C.; Godwin, J.; Gray, R.; Clarke, M.; Cutter, D.; Darby, S.; McGale, P.; Pan, H.C.; Taylor, C.; et al. Relevance of Breast Cancer Hormone Receptors and Other Factors to the Efficacy of Adjuvant Tamoxifen: Patient-Level Meta-Analysis of Randomised Trials. Lancet Lond. Engl. 2011, 378, 771–784. [Google Scholar] [CrossRef]
- Gomis, R.R.; Gawrzak, S. Tumor Cell Dormancy. Mol. Oncol. 2017, 11, 62–78. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Velázquez, M.A.; Popov, V.M.; Lisanti, M.P.; Pestell, R.G. The Role of Breast Cancer Stem Cells in Metastasis and Therapeutic Implications. Am. J. Pathol. 2011, 179, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Yee, D.; Isaacs, C.; Wolf, D.M.; Yau, C.; Haluska, P.; Giridhar, K.V.; Forero-Torres, A.; Jo Chien, A.; Wallace, A.M.; Pusztai, L.; et al. Ganitumab and Metformin plus Standard Neoadjuvant Therapy in Stage 2/3 Breast Cancer. npj Breast Cancer 2021, 7, 131. [Google Scholar] [CrossRef]
- Potter, D.A.; Herrera-Ponzanelli, C.A.; Hinojosa, D.; Castillo, R.; Hernandez-Cruz, I.; Arrieta, V.A.; Franklin, M.J.; Yee, D. Recent Advances in Neoadjuvant Therapy for Breast Cancer. Fac. Rev. 2021, 10, 2. [Google Scholar] [CrossRef]
- Rahman, M.M.; McFadden, G. Oncolytic Viruses: Newest Frontier for Cancer Immunotherapy. Cancers 2021, 13, 5452. [Google Scholar] [CrossRef] [PubMed]
- Larson, C.; Oronsky, B.; Scicinski, J.; Fanger, G.R.; Stirn, M.; Oronsky, A.; Reid, T.R. Going Viral: A Review of Replication-Selective Oncolytic Adenoviruses. Oncotarget 2015, 6, 19976–19989. [Google Scholar] [CrossRef] [PubMed]
- Hemminki, O.; Dos Santos, J.M.; Hemminki, A. Oncolytic Viruses for Cancer Immunotherapy. J. Hematol. Oncol. 2020, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Peter, M.; Kühnel, F. Oncolytic Adenovirus in Cancer Immunotherapy. Cancers 2020, 12, 3354. [Google Scholar] [CrossRef] [PubMed]
- Carter, M.E.; Koch, A.; Lauer, U.M.; Hartkopf, A.D. Clinical Trials of Oncolytic Viruses in Breast Cancer. Front. Oncol. 2021, 11, 803050. [Google Scholar] [CrossRef] [PubMed]
- Ravera, S.; Reyna-Neyra, A.; Ferrandino, G.; Amzel, L.M.; Carrasco, N. The Sodium/Iodide Symporter (NIS): Molecular Physiology and Preclinical and Clinical Applications. Annu. Rev. Physiol. 2017, 79, 261–289. [Google Scholar] [CrossRef] [PubMed]
- Oneal, M.J.; Trujillo, M.A.; Davydova, J.; McDonough, S.; Yamamoto, M.; Morris, J.C. Characterization of Infectivity-Enhanced Conditionally Replicating Adenovectors for Prostate Cancer Radiovirotherapy. Hum. Gene Ther. 2012, 23, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Robertson, M.G.; Eidenschink, B.B.; Iguchi, E.; Zakharkin, S.O.; LaRocca, C.J.; Tolosa, E.J.; Truty, M.J.; Jacobsen, K.; Fernandez-Zapico, M.E.; Davydova, J. Cancer Imaging and Therapy Utilizing a Novel NIS-Expressing Adenovirus: The Role of Adenovirus Death Protein Deletion. Mol. Ther. Oncolytics 2021, 20, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Winzer, K.-J.; Müller, B.-M.; Weichert, W.; Pest, S.; Köbel, M.; Kristiansen, G.; Reles, A.; Siegert, A.; Guski, H.; et al. Elevated Expression of Cyclooxygenase-2 Is a Negative Prognostic Factor for Disease Free Survival and Overall Survival in Patients with Breast Carcinoma. Cancer 2003, 97, 2978–2987. [Google Scholar] [CrossRef]
- Fleischli, C.; Sirena, D.; Lesage, G.; Havenga, M.J.E.; Cattaneo, R.; Greber, U.F.; Hemmi, S. Species B Adenovirus Serotypes 3, 7, 11 and 35 Share Similar Binding Sites on the Membrane Cofactor Protein CD46 Receptor. J. Gen. Virol. 2007, 88, 2925–2934. [Google Scholar] [CrossRef]
- Wang, H.; Li, Z.-Y.; Liu, Y.; Persson, J.; Beyer, I.; Möller, T.; Koyuncu, D.; Drescher, M.R.; Strauss, R.; Zhang, X.-B.; et al. Desmoglein 2 Is a Receptor for Adenovirus Serotypes 3, 7, 11 and 14. Nat. Med. 2011, 17, 96–104. [Google Scholar] [CrossRef]
- Maciejczyk, A.; Szelachowska, J.; Szynglarewicz, B.; Szulc, R.; Szulc, A.; Wysocka, T.; Jagoda, E.; Lage, H.; Surowiak, P. CD46 Expression Is an Unfavorable Prognostic Factor in Breast Cancer Cases. Appl. Immunohistochem. Mol. Morphol. 2011, 19, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-H.; Chen, M.-C.; Tsai, Y.-P.; Tan, G.Y.T.; Hsu, P.-H.; Jeng, Y.-M.; Tsai, Y.-F.; Yang, M.-H.; Hwang-Verslues, W.W. Interplay between Desmoglein2 and Hypoxia Controls Metastasis in Breast Cancer. Proc. Natl. Acad. Sci. USA 2021, 118, e2014408118. [Google Scholar] [CrossRef] [PubMed]
- Aponte, P.M.; Caicedo, A. Stemness in Cancer: Stem Cells, Cancer Stem Cells, and Their Microenvironment. Stem Cells Int. 2017, 2017, 5619472. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.-J.; Qiu, W.; Xu, S.-L.; Wang, B.; Ye, X.-Z.; Ping, Y.-F.; Zhang, X.; Bian, X.-W.; Yu, S.-C. Strategies for Isolating and Enriching Cancer Stem Cells: Well Begun Is Half Done. Stem Cells Dev. 2013, 22, 2221–2239. [Google Scholar] [CrossRef]
- Regan Anderson, T.M.; Ma, S.; Perez Kerkvliet, C.; Peng, Y.; Helle, T.M.; Krutilina, R.I.; Raj, G.V.; Cidlowski, J.A.; Ostrander, J.H.; Schwertfeger, K.L.; et al. Taxol Induces Brk-Dependent Prosurvival Phenotypes in TNBC Cells through an AhR/GR/HIF-Driven Signaling Axis. Mol. Cancer Res. 2018, 16, 1761–1772. [Google Scholar] [CrossRef]
- Truong, T.H.; Benner, E.A.; Hagen, K.M.; Temiz, N.A.; Kerkvliet, C.P.; Wang, Y.; Cortes-Sanchez, E.; Yang, C.-H.; Trousdell, M.C.; Pengo, T.; et al. PELP1/SRC-3-Dependent Regulation of Metabolic PFKFB Kinases Drives Therapy Resistant ER+ Breast Cancer. Oncogene 2021, 40, 4384–4397. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective Identification of Tumorigenic Breast Cancer Cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Liu, S.; Chen, C. Targeting Breast Cancer Stem Cells. Int. J. Biol. Sci. 2023, 19, 552–570. [Google Scholar] [CrossRef]
- Post, D.E.; Khuri, F.R.; Simons, J.W.; Van Meir, E.G. Replicative Oncolytic Adenoviruses in Multimodal Cancer Regimens. Hum. Gene Ther. 2003, 14, 933–946. [Google Scholar] [CrossRef]
- Liu, T.-C.; Galanis, E.; Kirn, D. Clinical Trial Results with Oncolytic Virotherapy: A Century of Promise, a Decade of Progress. Nat. Clin. Pract. Oncol. 2007, 4, 101–117. [Google Scholar] [CrossRef]
- LaRocca, C.J.; Han, J.; Gavrikova, T.; Armstrong, L.; Oliveira, A.R.; Shanley, R.; Vickers, S.M.; Yamamoto, M.; Davydova, J. Oncolytic Adenovirus Expressing Interferon Alpha in a Syngeneic Syrian Hamster Model for the Treatment of Pancreatic Cancer. Surgery 2015, 157, 888–898. [Google Scholar] [CrossRef]
- Farrera-Sal, M.; Fillat, C.; Alemany, R. Effect of Transgene Location, Transcriptional Control Elements and Transgene Features in Armed Oncolytic Adenoviruses. Cancers 2020, 12, 1034. [Google Scholar] [CrossRef] [PubMed]
- Cody, J.J.; Hurst, D.R. Promising Oncolytic Agents for Metastatic Breast Cancer Treatment. Oncolytic Virotherapy 2015, 4, 63–73. [Google Scholar] [CrossRef]
- Hemminki, O.; Diaconu, I.; Cerullo, V.; Pesonen, S.K.; Kanerva, A.; Joensuu, T.; Kairemo, K.; Laasonen, L.; Partanen, K.; Kangasniemi, L.; et al. Ad3-hTERT-E1A, a Fully Serotype 3 Oncolytic Adenovirus, in Patients with Chemotherapy Refractory Cancer. Mol. Ther. J. Am. Soc. Gene Ther. 2012, 20, 1821–1830. [Google Scholar] [CrossRef] [PubMed]
- Nemunaitis, J.; Senzer, N.; Sarmiento, S.; Zhang, Y.-A.; Arzaga, R.; Sands, B.; Maples, P.; Tong, A.W. A Phase I Trial of Intravenous Infusion of ONYX-015 and Enbrel in Solid Tumor Patients. Cancer Gene Ther. 2007, 14, 885–893. [Google Scholar] [CrossRef]
- Nemunaitis, J.; Tong, A.W.; Nemunaitis, M.; Senzer, N.; Phadke, A.P.; Bedell, C.; Adams, N.; Zhang, Y.-A.; Maples, P.B.; Chen, S.; et al. A Phase I Study of Telomerase-Specific Replication Competent Oncolytic Adenovirus (Telomelysin) for Various Solid Tumors. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 429–434. [Google Scholar] [CrossRef] [PubMed]
- Machiels, J.-P.; Salazar, R.; Rottey, S.; Duran, I.; Dirix, L.; Geboes, K.; Wilkinson-Blanc, C.; Pover, G.; Alvis, S.; Champion, B.; et al. A Phase 1 Dose Escalation Study of the Oncolytic Adenovirus Enadenotucirev, Administered Intravenously to Patients with Epithelial Solid Tumors (EVOLVE). J. Immunother. Cancer 2019, 7, 20. [Google Scholar] [CrossRef]
- Garcia-Carbonero, R.; Bazan-Peregrino, M.; Gil-Martín, M.; Álvarez, R.; Macarulla, T.; Riesco-Martinez, M.C.; Verdaguer, H.; Guillén-Ponce, C.; Farrera-Sal, M.; Moreno, R.; et al. Phase I, Multicenter, Open-Label Study of Intravenous VCN-01 Oncolytic Adenovirus with or without Nab-Paclitaxel plus Gemcitabine in Patients with Advanced Solid Tumors. J. Immunother. Cancer 2022, 10, e003255. [Google Scholar] [CrossRef]
- Li, J.-L.; Liu, H.-L.; Zhang, X.-R.; Xu, J.-P.; Hu, W.-K.; Liang, M.; Chen, S.-Y.; Hu, F.; Chu, D.-T. A Phase I Trial of Intratumoral Administration of Recombinant Oncolytic Adenovirus Overexpressing HSP70 in Advanced Solid Tumor Patients. Gene Ther. 2009, 16, 376–382. [Google Scholar] [CrossRef]
- Lim, B. Binary Oncolytic Adenovirus in Combination with HER2-Specific Autologous CAR VST, Advanced HER2 Positive Solid Tumors (VISTA). NIH U.S. National Library of Medicine. ClinicalTrials.gov Identifier number NCT03740256. 2018. Available online: https://classic.clinicaltrials.gov/ct2/show/NCT03740256 (accessed on 1 April 2024).
- Chang, J. SBRT and Oncolytic Virus Therapy Before Pembrolizumab for Metastatic TNBC and NSCLC (STOMP). NIH U.S. National Library of Medicine. ClinicalTrials.gov Identifier number NCT03004183. 2016. Available online: https://classic.clinicaltrials.gov/ct2/show/study/NCT03004183 (accessed on 1 April 2024).
- Kanerva, A.; Nokisalmi, P.; Diaconu, I.; Koski, A.; Cerullo, V.; Liikanen, I.; Tähtinen, S.; Oksanen, M.; Heiskanen, R.; Pesonen, S.; et al. Antiviral and Antitumor T-Cell Immunity in Patients Treated with GM-CSF-Coding Oncolytic Adenovirus. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 2734–2744. [Google Scholar] [CrossRef]
- Bramante, S.; Koski, A.; Liikanen, I.; Vassilev, L.; Oksanen, M.; Siurala, M.; Heiskanen, R.; Hakonen, T.; Joensuu, T.; Kanerva, A.; et al. Oncolytic Virotherapy for Treatment of Breast Cancer, Including Triple-Negative Breast Cancer. Oncoimmunology 2016, 5, e1078057. [Google Scholar] [CrossRef]
- Hemminki, O.; Parviainen, S.; Juhila, J.; Turkki, R.; Linder, N.; Lundin, J.; Kankainen, M.; Ristimäki, A.; Koski, A.; Liikanen, I.; et al. Immunological Data from Cancer Patients Treated with Ad5/3-E2F-Δ24-GMCSF Suggests Utility for Tumor Immunotherapy. Oncotarget 2015, 6, 4467–4481. [Google Scholar] [CrossRef]
- Cerullo, V.; Pesonen, S.; Diaconu, I.; Escutenaire, S.; Arstila, P.T.; Ugolini, M.; Nokisalmi, P.; Raki, M.; Laasonen, L.; Särkioja, M.; et al. Oncolytic Adenovirus Coding for Granulocyte Macrophage Colony-Stimulating Factor Induces Antitumoral Immunity in Cancer Patients. Cancer Res. 2010, 70, 4297–4309. [Google Scholar] [CrossRef]
- Cerullo, V.; Diaconu, I.; Kangasniemi, L.; Rajecki, M.; Escutenaire, S.; Koski, A.; Romano, V.; Rouvinen, N.; Tuuminen, T.; Laasonen, L.; et al. Immunological Effects of Low-Dose Cyclophosphamide in Cancer Patients Treated with Oncolytic Adenovirus. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 1737–1746. [Google Scholar] [CrossRef]
- Pesonen, S.; Diaconu, I.; Cerullo, V.; Escutenaire, S.; Raki, M.; Kangasniemi, L.; Nokisalmi, P.; Dotti, G.; Guse, K.; Laasonen, L.; et al. Integrin Targeted Oncolytic Adenoviruses Ad5-D24-RGD and Ad5-RGD-D24-GMCSF for Treatment of Patients with Advanced Chemotherapy Refractory Solid Tumors. Int. J. Cancer 2012, 130, 1937–1947. [Google Scholar] [CrossRef]
- Nokisalmi, P.; Pesonen, S.; Escutenaire, S.; Särkioja, M.; Raki, M.; Cerullo, V.; Laasonen, L.; Alemany, R.; Rojas, J.; Cascallo, M.; et al. Oncolytic Adenovirus ICOVIR-7 in Patients with Advanced and Refractory Solid Tumors. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010, 16, 3035–3043. [Google Scholar] [CrossRef]
- Xu, W.; Yang, Y.; Hu, Z.; Head, M.; Mangold, K.A.; Sullivan, M.; Wang, E.; Saha, P.; Gulukota, K.; Helseth, D.L.; et al. LyP-1-Modified Oncolytic Adenoviruses Targeting Transforming Growth Factor β Inhibit Tumor Growth and Metastases and Augment Immune Checkpoint Inhibitor Therapy in Breast Cancer Mouse Models. Hum. Gene Ther. 2020, 31, 863–880. [Google Scholar] [CrossRef]
- Trujillo, M.A.; Oneal, M.J.; Davydova, J.; Bergert, E.; Yamamoto, M.; Morris, J.C. Construction of an MUC-1 Promoter Driven, Conditionally Replicating Adenovirus That Expresses the Sodium Iodide Symporter for Gene Therapy of Breast Cancer. Breast Cancer Res. 2009, 11, R53. [Google Scholar] [CrossRef]
- Stepanenko, A.A.; Sosnovtseva, A.O.; Valikhov, M.P.; Chernysheva, A.A.; Cherepanov, S.A.; Yusubalieva, G.M.; Ruzsics, Z.; Lipatova, A.V.; Chekhonin, V.P. Superior Infectivity of the Fiber Chimeric Oncolytic Adenoviruses Ad5/35 and Ad5/3 over Ad5-Delta-24-RGD in Primary Glioma Cultures. Mol. Ther. Oncolytics 2022, 24, 230–248. [Google Scholar] [CrossRef]
- Davydova, J.; Le, L.P.; Gavrikova, T.; Wang, M.; Krasnykh, V.; Yamamoto, M. Infectivity-Enhanced Cyclooxygenase-2-Based Conditionally Replicative Adenoviruses for Esophageal Adenocarcinoma Treatment. Cancer Res. 2004, 64, 4319–4327. [Google Scholar] [CrossRef]
- Armstrong, L.; Arrington, A.; Han, J.; Gavrikova, T.; Brown, E.; Yamamoto, M.; Vickers, S.M.; Davydova, J. Generation of a Novel, Cyclooxygenase-2-Targeted, Interferon-Expressing, Conditionally Replicative Adenovirus for Pancreatic Cancer Therapy. Am. J. Surg. 2012, 204, 741–750. [Google Scholar] [CrossRef]
- Seidlin, S.M.; Marinelli, L.D.; Oshry, E. Radioactive Iodine Therapy; Effect on Functioning Metastases of Adenocarcinoma of the Thyroid. J. Am. Med. Assoc. 1946, 132, 838–847. [Google Scholar] [CrossRef]
- Spitzweg, C.; Nelson, P.J.; Wagner, E.; Bartenstein, P.; Weber, W.A.; Schwaiger, M.; Morris, J.C. The Sodium Iodide Symporter (NIS): Novel Applications for Radionuclide Imaging and Treatment. Endocr. Relat. Cancer 2021, 28, T193–T213. [Google Scholar] [CrossRef]
- Barton, K.N.; Stricker, H.; Brown, S.L.; Elshaikh, M.; Aref, I.; Lu, M.; Pegg, J.; Zhang, Y.; Karvelis, K.C.; Siddiqui, F.; et al. Phase I Study of Noninvasive Imaging of Adenovirus-Mediated Gene Expression in the Human Prostate. Mol. Ther. J. Am. Soc. Gene Ther. 2008, 16, 1761–1769. [Google Scholar] [CrossRef]
- Oneal, M.J.; Trujillo, M.A.; Davydova, J.; McDonough, S.; Yamamoto, M.; Morris, J.C. Effect of Increased Viral Replication and Infectivity Enhancement on Radioiodide Uptake and Oncolytic Activity of Adenovirus Vectors Expressing the Sodium Iodide Symporter. Cancer Gene Ther. 2013, 20, 195–200. [Google Scholar] [CrossRef]
- Zhang, L.; Suksanpaisan, L.; Jiang, H.; DeGrado, T.R.; Russell, S.J.; Zhao, M.; Peng, K.-W. Dual-Isotope SPECT Imaging with NIS Reporter Gene and Duramycin to Visualize Tumor Susceptibility to Oncolytic Virus Infection. Mol. Ther. Oncolytics 2019, 15, 178–185. [Google Scholar] [CrossRef]
- Dwyer, R.M.; Bergert, E.R.; O’connor, M.K.; Gendler, S.J.; Morris, J.C. In Vivo Radioiodide Imaging and Treatment of Breast Cancer Xenografts after MUC1-Driven Expression of the Sodium Iodide Symporter. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2005, 11, 1483–1489. [Google Scholar] [CrossRef]
- Montiel-Equihua, C.A.; Martín-Duque, P.; de la Vieja, A.; Quintanilla, M.; Burnet, J.; Vassaux, G.; Lemoine, N.R. Targeting Sodium/Iodide Symporter Gene Expression for Estrogen-Regulated Imaging and Therapy in Breast Cancer. Cancer Gene Ther. 2008, 15, 465–473. [Google Scholar] [CrossRef]
- Riesco-Eizaguirre, G.; De la Vieja, A.; Rodríguez, I.; Miranda, S.; Martín-Duque, P.; Vassaux, G.; Santisteban, P. Telomerase-Driven Expression of the Sodium Iodide Symporter (NIS) for in Vivo Radioiodide Treatment of Cancer: A New Broad-Spectrum NIS-Mediated Antitumor Approach. J. Clin. Endocrinol. Metab. 2011, 96, E1435–E1443. [Google Scholar] [CrossRef]
- Boland, A.; Ricard, M.; Opolon, P.; Bidart, J.M.; Yeh, P.; Filetti, S.; Schlumberger, M.; Perricaudet, M. Adenovirus-Mediated Transfer of the Thyroid Sodium/Iodide Symporter Gene into Tumors for a Targeted Radiotherapy. Cancer Res. 2000, 60, 3484–3492. [Google Scholar]
- Davydova, J.; Gavrikova, T.; Brown, E.J.; Luo, X.; Curiel, D.T.; Vickers, S.M.; Yamamoto, M. In Vivo Bioimaging Tracks Conditionally Replicative Adenoviral Replication and Provides an Early Indication of Viral Antitumor Efficacy. Cancer Sci. 2010, 101, 474–481. [Google Scholar] [CrossRef]
- Doronin, K.; Toth, K.; Kuppuswamy, M.; Krajcsi, P.; Tollefson, A.E.; Wold, W.S.M. Overexpression of the ADP (E3-11.6K) Protein Increases Cell Lysis and Spread of Adenovirus. Virology 2003, 305, 378–387. [Google Scholar] [CrossRef]
- Fisher, K.D.; Stallwood, Y.; Green, N.K.; Ulbrich, K.; Mautner, V.; Seymour, L.W. Polymer-Coated Adenovirus Permits Efficient Retargeting and Evades Neutralising Antibodies. Gene Ther. 2001, 8, 341–348. [Google Scholar] [CrossRef]
- Nemunaitis, J.; Ganly, I.; Khuri, F.; Arseneau, J.; Kuhn, J.; McCarty, T.; Landers, S.; Maples, P.; Romel, L.; Randlev, B.; et al. Selective Replication and Oncolysis in P53 Mutant Tumors with ONYX-015, an E1B-55kD Gene-Deleted Adenovirus, in Patients with Advanced Head and Neck Cancer: A Phase II Trial. Cancer Res. 2000, 60, 6359–6366. [Google Scholar]
- Zhu, W.; Zhang, H.; Shi, Y.; Song, M.; Zhu, B.; Wei, L. Oncolytic Adenovirus Encoding Tumor Necrosis Factor-Related Apoptosis Inducing Ligand (TRAIL) Inhibits the Growth and Metastasis of Triple-Negative Breast Cancer. Cancer Biol. Ther. 2013, 14, 1016–1023. [Google Scholar] [CrossRef]
- Li, W.; Li, Y.; Cui, Y.; Li, S.; Zhu, Y.; Shang, C.; Song, G.; Liu, Z.; Xiu, Z.; Cong, J.; et al. Anti-Tumour Effects of a Dual Cancer-Specific Oncolytic Adenovirus on Breast Cancer Stem Cells. J. Cell. Mol. Med. 2021, 25, 666–676. [Google Scholar] [CrossRef]
- Zhu, W.; Wei, L.; Zhang, H.; Chen, J.; Qin, X. Oncolytic Adenovirus Armed with IL-24 Inhibits the Growth of Breast Cancer in Vitro and in Vivo. J. Exp. Clin. Cancer Res. CR 2012, 31, 51. [Google Scholar] [CrossRef]
- Hernandez-Alcoceba, R.; Pihalja, M.; Qian, D.; Clarke, M.F. New Oncolytic Adenoviruses with Hypoxia- and Estrogen Receptor-Regulated Replication. Hum. Gene Ther. 2002, 13, 1737–1750. [Google Scholar] [CrossRef] [PubMed]
- Bauerschmitz, G.J.; Ranki, T.; Kangasniemi, L.; Ribacka, C.; Eriksson, M.; Porten, M.; Herrmann, I.; Ristimäki, A.; Virkkunen, P.; Tarkkanen, M.; et al. Tissue-Specific Promoters Active in CD44+CD24-/Low Breast Cancer Cells. Cancer Res. 2008, 68, 5533–5539. [Google Scholar] [CrossRef] [PubMed]
- Kurihara, T.; Brough, D.E.; Kovesdi, I.; Kufe, D.W. Selectivity of a Replication-Competent Adenovirus for Human Breast Carcinoma Cells Expressing the MUC1 Antigen. J. Clin. Investig. 2000, 106, 763–771. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.-K.; Seong, Y.R.; Lee, Y.H.; Kim, M.J.; Hwang, K.-S.; Yoo, J.; Choi, S.; Jung, C.-R.; Im, D.-S. Oncolytic Effects of Adenovirus Mutant Capable of Replicating in Hypoxic and Normoxic Regions of Solid Tumor. Mol. Ther. J. Am. Soc. Gene Ther. 2004, 10, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Gomes, E.M.; Rodrigues, M.S.; Phadke, A.P.; Butcher, L.D.; Starling, C.; Chen, S.; Chang, D.; Hernandez-Alcoceba, R.; Newman, J.T.; Stone, M.J.; et al. Antitumor Activity of an Oncolytic Adenoviral-CD40 Ligand (CD154) Transgene Construct in Human Breast Cancer Cells. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 1317–1325. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.B.; Makhija, S.K.; Lu, B.; Wang, M.; Rivera, A.A.; Kim-Park, S.; Ulasov, I.V.; Zhou, F.; Alvarez, R.D.; Siegal, G.P.; et al. Incorporating the Survivin Promoter in an Infectivity Enhanced CRAd-Analysis of Oncolysis and Anti-Tumor Effects In Vitro and In Vivo. Int. J. Oncol. 2005, 27, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Akbulut, H.; Tang, Y.; Peng, X.; Pizzorno, G.; Sapi, E.; Manegold, S.; Deisseroth, A. Adenoviral Vectors with E1A Regulated by Tumor-Specific Promoters Are Selectively Cytolytic for Breast Cancer and Melanoma. Mol. Ther. J. Am. Soc. Gene Ther. 2002, 6, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Jana, D.; Sarkar, D.K.; Ganguly, S.; Saha, S.; Sa, G.; Manna, A.K.; Banerjee, A.; Mandal, S. Role of Cyclooxygenase 2 (COX-2) in Prognosis of Breast Cancer. Indian J. Surg. Oncol. 2014, 5, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Gavrikova, T.; Nakamura, N.; Davydova, J.; Antonarakis, E.S.; Yamamoto, M. Infectivity-Enhanced, Conditionally Replicative Adenovirus for COX-2-Expressing Castration-Resistant Prostate Cancer. Viruses 2023, 15, 901. [Google Scholar] [CrossRef] [PubMed]
- LaRocca, C.J.; Salzwedel, A.O.; Sato-Dahlman, M.; Romanenko, M.V.; Andrade, R.; Davydova, J.; Yamamoto, M. Interferon Alpha-Expressing Oncolytic Adenovirus for Treatment of Esophageal Adenocarcinoma. Ann. Surg. Oncol. 2021, 28, 8556–8564. [Google Scholar] [CrossRef]
- Tong, Y.; You, L.; Liu, H.; Li, L.; Meng, H.; Qian, Q.; Qian, W. Potent Antitumor Activity of Oncolytic Adenovirus Expressing Beclin-1 via Induction of Autophagic Cell Death in Leukemia. Oncotarget 2013, 4, 860–874. [Google Scholar] [CrossRef]
- Jiang, H.; Gomez-Manzano, C.; Aoki, H.; Alonso, M.M.; Kondo, S.; McCormick, F.; Xu, J.; Kondo, Y.; Bekele, B.N.; Colman, H.; et al. Examination of the Therapeutic Potential of Delta-24-RGD in Brain Tumor Stem Cells: Role of Autophagic Cell Death. J. Natl. Cancer Inst. 2007, 99, 1410–1414. [Google Scholar] [CrossRef]
- Zhang, X.; Meng, S.; Zhang, R.; Ma, B.; Liu, T.; Yang, Y.; Xie, W.; Liu, X.; Huang, F.; Liu, T.; et al. GP73-Regulated Oncolytic Adenoviruses Possess Potent Killing Effect on Human Liver Cancer Stem-like Cells. Oncotarget 2016, 7, 29346–29358. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, H.; Huang, W.; Ding, M.; Xiao, J.; Yang, D.; Li, H.; Liu, X.-Y.; Chu, L. Targeting Lung Cancer Stem-like Cells with TRAIL Gene Armed Oncolytic Adenovirus. J. Cell. Mol. Med. 2015, 19, 915–923. [Google Scholar] [CrossRef] [PubMed]
- Yano, S.; Tazawa, H.; Hashimoto, Y.; Shirakawa, Y.; Kuroda, S.; Nishizaki, M.; Kishimoto, H.; Uno, F.; Nagasaka, T.; Urata, Y.; et al. A Genetically Engineered Oncolytic Adenovirus Decoys and Lethally Traps Quiescent Cancer Stem-like Cells in S/G2/M Phases. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6495–6505. [Google Scholar] [CrossRef] [PubMed]
- Walker, O.L.; Dahn, M.L.; Power Coombs, M.R.; Marcato, P. The Prostaglandin E2 Pathway and Breast Cancer Stem Cells: Evidence of Increased Signaling and Potential Targeting. Front. Oncol. 2022, 11, 791696. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, P.C.; Guttridge, D.C.; Funkhouser, W.K.; Baldwin, A.S. Selective Activation of NF-Kappa B Subunits in Human Breast Cancer: Potential Roles for NF-Kappa B2/P52 and for Bcl-3. Oncogene 2000, 19, 1123–1131. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Kudo, I. Recent Advances in Molecular Biology and Physiology of the Prostaglandin E2-Biosynthetic Pathway. Prog. Lipid Res. 2004, 43, 3–35. [Google Scholar] [CrossRef]
- Eriksson, M.; Guse, K.; Bauerschmitz, G.; Virkkunen, P.; Tarkkanen, M.; Tanner, M.; Hakkarainen, T.; Kanerva, A.; Desmond, R.A.; Pesonen, S.; et al. Oncolytic Adenoviruses Kill Breast Cancer Initiating CD44+CD24-/Low Cells. Mol. Ther. J. Am. Soc. Gene Ther. 2007, 15, 2088–2093. [Google Scholar] [CrossRef]
- Hyland, C.J.; Varghese, F.; Yau, C.; Beckwith, H.; Khoury, K.; Varnado, W.; Hirst, G.L.; Flavell, R.R.; Chien, A.J.; Yee, D.; et al. Use of 18F-FDG PET/CT as an Initial Staging Procedure for Stage II–III Breast Cancer: A Multicenter Value Analysis. J. Natl. Compr. Cancer Netw. 2020, 18, 1510–1517. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robert, S.; Roman Ortiz, N.I.; LaRocca, C.J.; Ostrander, J.H.; Davydova, J. Oncolytic Adenovirus for the Targeting of Paclitaxel-Resistant Breast Cancer Stem Cells. Viruses 2024, 16, 567. https://doi.org/10.3390/v16040567
Robert S, Roman Ortiz NI, LaRocca CJ, Ostrander JH, Davydova J. Oncolytic Adenovirus for the Targeting of Paclitaxel-Resistant Breast Cancer Stem Cells. Viruses. 2024; 16(4):567. https://doi.org/10.3390/v16040567
Chicago/Turabian StyleRobert, Sacha, Natasha Ivelisse Roman Ortiz, Christopher J. LaRocca, Julie Hanson Ostrander, and Julia Davydova. 2024. "Oncolytic Adenovirus for the Targeting of Paclitaxel-Resistant Breast Cancer Stem Cells" Viruses 16, no. 4: 567. https://doi.org/10.3390/v16040567





