IFITM1 and IFITM3 Proteins Inhibit the Infectivity of Progeny HIV-1 without Disrupting Envelope Glycoprotein Clusters
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines, Plasmids, and Reagents
2.2. Pseudovirus Production and Characterization
2.3. Western Blotting
2.4. Infectivity Assay
2.5. Single Virus-Based IFITM and Env Incorporation Analysis
2.6. Immunostaining and Sample Preparation for dSTORM
2.7. Wide-Field Fluorescence and dSTORM Imaging
2.8. DBCAN Analysis
2.9. Data Processing and Statistical Analyses
3. Results
3.1. IFITM1 and IFITM3 Incorporation into HIV-1 Pseudoviruses Inhibits Infectivity but Only IFITM3 Interferes with Processing and Incorporation of Sensitive Envs
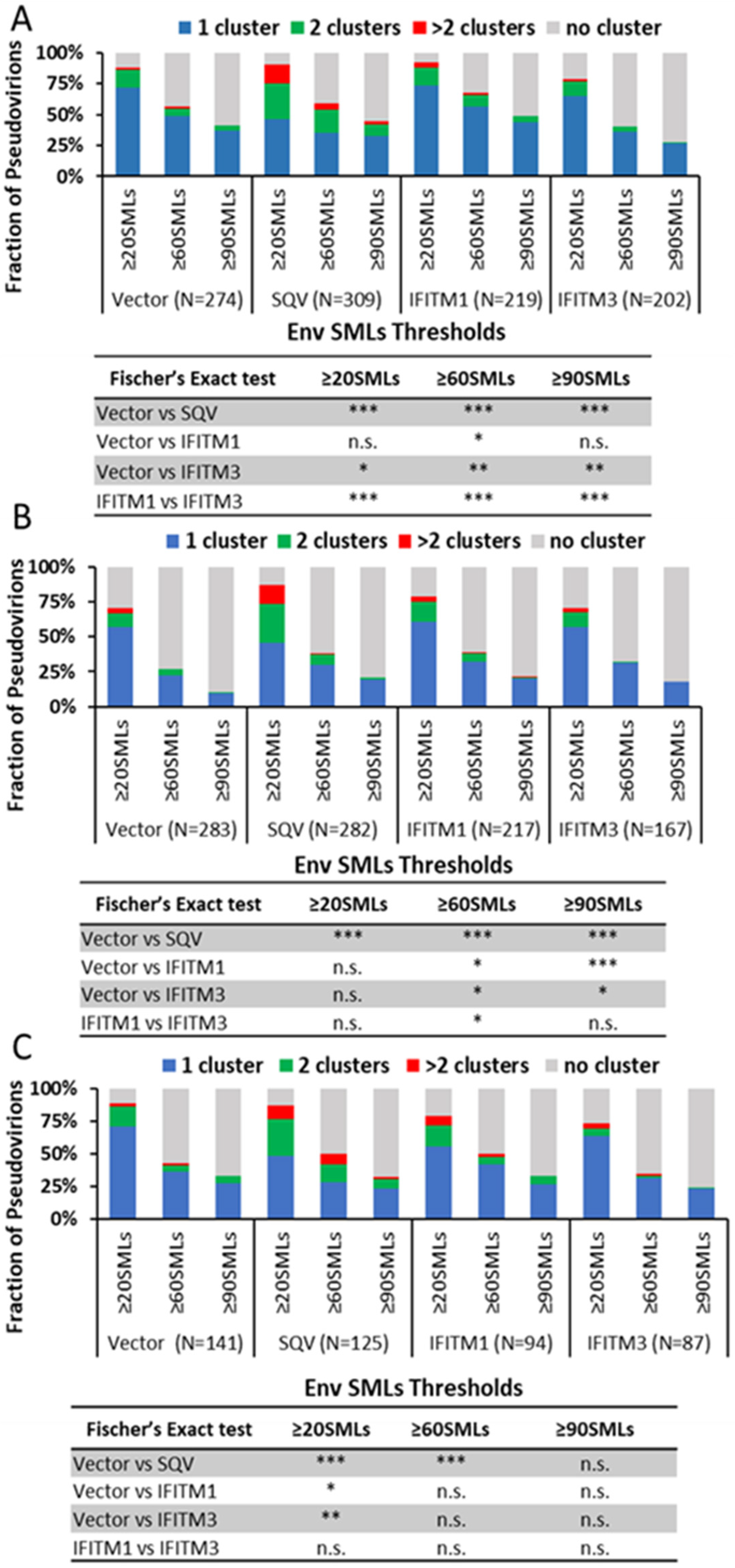
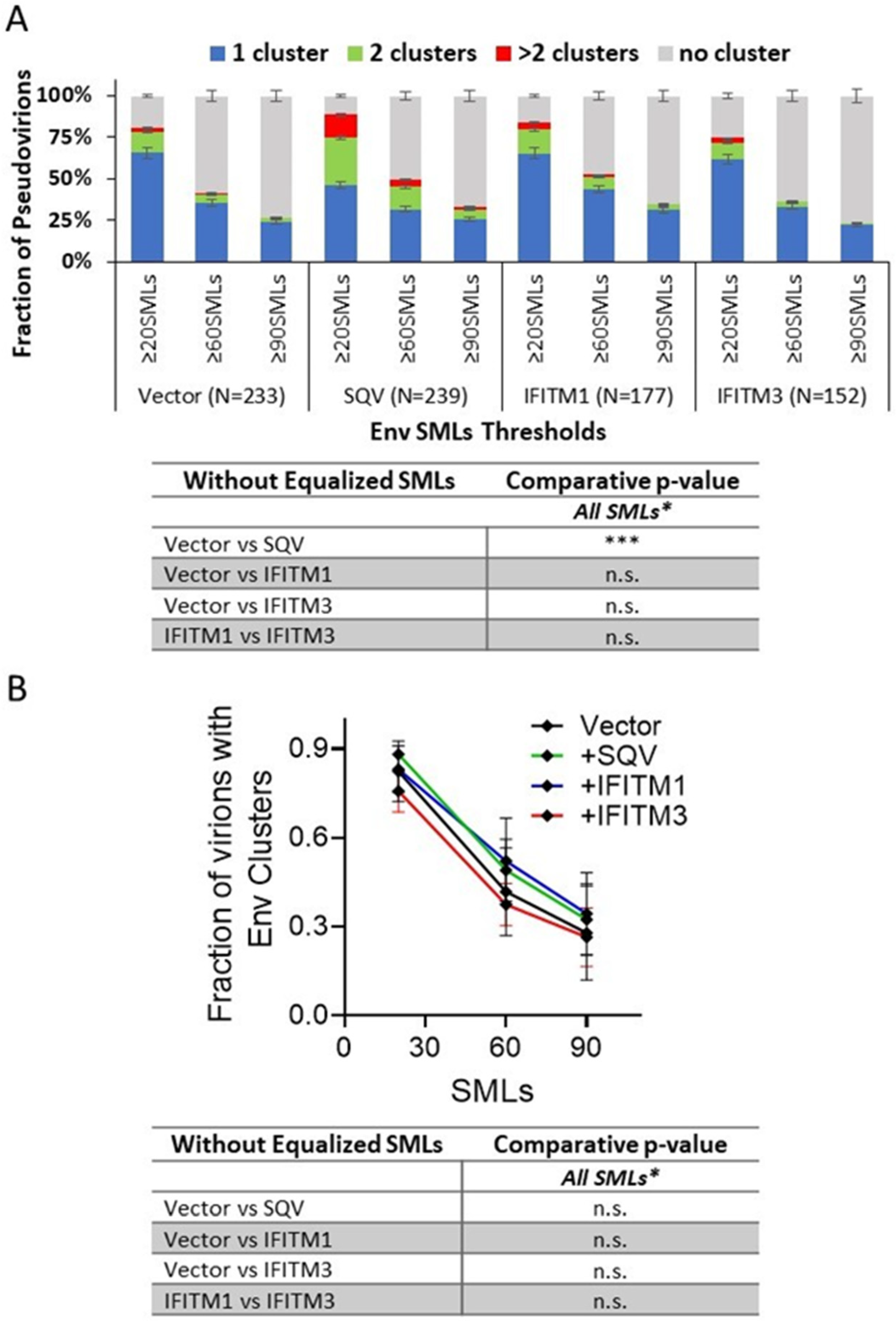
3.2. IFITM Incorporation Does Not Consistently Perturb Env Clustering on HIV-1 Particles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegrist, F.; Ebeling, M.; Certa, U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 2011, 31, 183–197. [Google Scholar] [CrossRef]
- Perreira, J.M.; Chin, C.R.; Feeley, E.M.; Brass, A.L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 2013, 425, 4937–4955. [Google Scholar] [CrossRef]
- Bailey, C.C.; Zhong, G.; Huang, I.C.; Farzan, M. IFITM-Family Proteins: The Cell’s First Line of Antiviral Defense. Annu. Rev. Virol. 2014, 1, 261–283. [Google Scholar] [CrossRef]
- Shi, G.; Schwartz, O.; Compton, A.A. More than meets the I: The diverse antiviral and cellular functions of interferon-induced transmembrane proteins. Retrovirology 2017, 14, 53. [Google Scholar] [CrossRef] [PubMed]
- Marziali, F.; Cimarelli, A. Membrane Interference Against HIV-1 by Intrinsic Antiviral Factors: The Case of IFITMs. Cells 2021, 10, 1171. [Google Scholar] [CrossRef] [PubMed]
- Majdoul, S.; Compton, A.A. Lessons in self-defence: Inhibition of virus entry by intrinsic immunity. Nat. Rev. Immunol. 2022, 22, 339–352. [Google Scholar] [CrossRef] [PubMed]
- Diamond, M.S.; Farzan, M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat. Rev. Immunol. 2013, 13, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Friedlova, N.; Zavadil Kokas, F.; Hupp, T.R.; Vojtesek, B.; Nekulova, M. IFITM protein regulation and functions: Far beyond the fight against viruses. Front. Immunol. 2022, 13, 1042368. [Google Scholar] [CrossRef]
- Gomez-Herranz, M.; Taylor, J.; Sloan, R.D. IFITM proteins: Understanding their diverse roles in viral infection, cancer, and immunity. J. Biol. Chem. 2023, 299, 102741. [Google Scholar] [CrossRef]
- Lu, J.; Pan, Q.; Rong, L.; He, W.; Liu, S.L.; Liang, C. The IFITM proteins inhibit HIV-1 infection. J. Virol. 2011, 85, 2126–2137. [Google Scholar] [CrossRef]
- Compton, A.A.; Bruel, T.; Porrot, F.; Mallet, A.; Sachse, M.; Euvrard, M.; Liang, C.; Casartelli, N.; Schwartz, O. IFITM proteins incorporated into HIV-1 virions impair viral fusion and spread. Cell Host Microbe 2014, 16, 736–747. [Google Scholar] [CrossRef]
- Tartour, K.; Appourchaux, R.; Gaillard, J.; Nguyen, X.N.; Durand, S.; Turpin, J.; Beaumont, E.; Roch, E.; Berger, G.; Mahieux, R.; et al. IFITM proteins are incorporated onto HIV-1 virion particles and negatively imprint their infectivity. Retrovirology 2014, 11, 103. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.; Weston, S.; Kellam, P.; Marsh, M. IFITM proteins-cellular inhibitors of viral entry. Curr. Opin. Virol. 2014, 4, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, Y.; Helenius, A. Virus entry at a glance. J. Cell Sci. 2013, 126 Pt 6, 1289–1295. [Google Scholar] [CrossRef]
- Amini-Bavil-Olyaee, S.; Choi, Y.J.; Lee, J.H.; Shi, M.; Huang, I.C.; Farzan, M.; Jung, J.U. The antiviral effector IFITM3 disrupts intracellular cholesterol homeostasis to block viral entry. Cell Host Microbe 2013, 13, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Markosyan, R.M.; Zheng, Y.M.; Golfetto, O.; Bungart, B.; Li, M.; Ding, S.; He, Y.; Liang, C.; Lee, J.C.; et al. IFITM proteins restrict viral membrane hemifusion. PLoS Pathog. 2013, 9, e1003124. [Google Scholar] [CrossRef]
- Desai, T.M.; Marin, M.; Chin, C.R.; Savidis, G.; Brass, A.L.; Melikyan, G.B. IFITM3 restricts influenza A virus entry by blocking the formation of fusion pores following virus-endosome hemifusion. PLoS Pathog. 2014, 10, e1004048. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Li, M.; Wilkins, J.; Ding, S.; Swartz, T.H.; Esposito, A.M.; Zheng, Y.M.; Freed, E.O.; Liang, C.; Chen, B.K.; et al. IFITM Proteins Restrict HIV-1 Infection by Antagonizing the Envelope Glycoprotein. Cell Rep. 2015, 13, 145–156. [Google Scholar] [CrossRef]
- Kuhnl, A.; Musiol, A.; Heitzig, N.; Johnson, D.E.; Ehrhardt, C.; Grewal, T.; Gerke, V.; Ludwig, S.; Rescher, U. Late Endosomal/Lysosomal Cholesterol Accumulation Is a Host Cell-Protective Mechanism Inhibiting Endosomal Escape of Influenza A Virus. mBio 2018, 9, e01345-18. [Google Scholar] [CrossRef]
- Appourchaux, R.; Delpeuch, M.; Zhong, L.; Burlaud-Gaillard, J.; Tartour, K.; Savidis, G.; Brass, A.; Etienne, L.; Roingeard, P.; Cimarelli, A. Functional Mapping of Regions Involved in the Negative Imprinting of Virion Particle Infectivity and in Target Cell Protection by Interferon-Induced Transmembrane Protein 3 against HIV-1. J. Virol. 2019, 93, e01716-18. [Google Scholar] [CrossRef]
- Suddala, K.C.; Lee, C.C.; Meraner, P.; Marin, M.; Markosyan, R.M.; Desai, T.M.; Cohen, F.S.; Brass, A.L.; Melikyan, G.B. Interferon-induced transmembrane protein 3 blocks fusion of sensitive but not resistant viruses by partitioning into virus-carrying endosomes. PLoS Pathog. 2019, 15, e1007532. [Google Scholar] [CrossRef]
- Guo, X.; Steinkuhler, J.; Marin, M.; Li, X.; Lu, W.; Dimova, R.; Melikyan, G.B. Interferon-Induced Transmembrane Protein 3 Blocks Fusion of Diverse Enveloped Viruses by Altering Mechanical Properties of Cell Membranes. ACS Nano 2021, 15, 8155–8170. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Du, S.; Tian, M.; Wang, Y.; Bai, J.; Tan, P.; Liu, W.; Yin, R.; Wang, M.; Jiang, Y.; et al. The Host Restriction Factor Interferon-Inducible Transmembrane Protein 3 Inhibits Vaccinia Virus Infection. Front. Immunol. 2018, 9, 228. [Google Scholar] [CrossRef] [PubMed]
- Brass, A.L.; Huang, I.C.; Benita, Y.; John, S.P.; Krishnan, M.N.; Feeley, E.M.; Ryan, B.J.; Weyer, J.L.; van der Weyden, L.; Fikrig, E.; et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139, 1243–1254. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Griffin, L.M.; Little, A.S.; Huang, I.C.; Farzan, M.; Pyeon, D. The antiviral restriction factors IFITM1, 2 and 3 do not inhibit infection of human papillomavirus, cytomegalovirus and adenovirus. PLoS ONE 2014, 9, e96579. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa-Sasaki, K.; Murata, T.; Sasaki, J. IFITM1 enhances nonenveloped viral RNA replication by facilitating cholesterol transport to the Golgi. PLoS Pathog. 2023, 19, e1011383. [Google Scholar] [CrossRef]
- Zhao, X.; Guo, F.; Liu, F.; Cuconati, A.; Chang, J.; Block, T.M.; Guo, J.T. Interferon induction of IFITM proteins promotes infection by human coronavirus OC43. Proc. Natl. Acad. Sci. USA 2014, 111, 6756–6761. [Google Scholar] [CrossRef]
- Prelli Bozzo, C.; Nchioua, R.; Volcic, M.; Koepke, L.; Kruger, J.; Schutz, D.; Heller, S.; Sturzel, C.M.; Kmiec, D.; Conzelmann, C.; et al. IFITM proteins promote SARS-CoV-2 infection and are targets for virus inhibition in vitro. Nat. Commun. 2021, 12, 4584. [Google Scholar] [CrossRef]
- Tartour, K.; Nguyen, X.N.; Appourchaux, R.; Assil, S.; Barateau, V.; Bloyet, L.M.; Burlaud Gaillard, J.; Confort, M.P.; Escudero-Perez, B.; Gruffat, H.; et al. Interference with the production of infectious viral particles and bimodal inhibition of replication are broadly conserved antiviral properties of IFITMs. PLoS Pathog. 2017, 13, e1006610. [Google Scholar] [CrossRef]
- Wang, Y.; Pan, Q.; Ding, S.; Wang, Z.; Yu, J.; Finzi, A.; Liu, S.L.; Liang, C. The V3 Loop of HIV-1 Env Determines Viral Susceptibility to IFITM3 Impairment of Viral Infectivity. J. Virol. 2017, 91, e02441-16. [Google Scholar] [CrossRef]
- Ahi, Y.S.; Yimer, D.; Shi, G.; Majdoul, S.; Rahman, K.; Rein, A.; Compton, A.A. IFITM3 Reduces Retroviral Envelope Abundance and Function and Is Counteracted by glycoGag. mBio 2020, 11, e03088-19. [Google Scholar] [CrossRef]
- Drouin, A.; Migraine, J.; Durand, M.A.; Moreau, A.; Burlaud-Gaillard, J.; Beretta, M.; Roingeard, P.; Bouvin-Pley, M.; Braibant, M. Escape of HIV-1 envelope glycoprotein from the restriction of infection by IFITM3. J. Virol. 2021, 95, e01994-20. [Google Scholar] [CrossRef] [PubMed]
- Beitari, S.; Pan, Q.; Finzi, A.; Liang, C. Differential Pressures of SERINC5 and IFITM3 on HIV-1 Envelope Glycoprotein over the Course of HIV-1 Infection. J. Virol. 2020, 94, e00514-20. [Google Scholar] [CrossRef] [PubMed]
- Sood, C.; Marin, M.; Chande, A.; Pizzato, M.; Melikyan, G.B. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J. Biol. Chem. 2017, 292, 6014–6026. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.C.; Sood, C.; Marin, M.; Aaron, J.; Gratton, E.; Salaita, K.; Melikyan, G.B. Super-Resolution Fluorescence Imaging Reveals That Serine Incorporator Protein 5 Inhibits Human Immunodeficiency Virus Fusion by Disrupting Envelope Glycoprotein Clusters. ACS Nano 2020, 14, 10929–10943. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Le Duff, Y.; Wang, Y.; Pan, Q.; Ding, S.; Zheng, Y.M.; Liu, S.L.; Liang, C. Primate lentiviruses are differentially inhibited by interferon-induced transmembrane proteins. Virology 2015, 474, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, J.; Zheng, Y.M.; Yu, J.; Liang, C.; Liu, S.L. Nonhuman Primate IFITM Proteins Are Potent Inhibitors of HIV and SIV. PLoS ONE 2016, 11, e0156739. [Google Scholar] [CrossRef] [PubMed]
- Foster, T.L.; Wilson, H.; Iyer, S.S.; Coss, K.; Doores, K.; Smith, S.; Kellam, P.; Finzi, A.; Borrow, P.; Hahn, B.H.; et al. Resistance of Transmitted Founder HIV-1 to IFITM-Mediated Restriction. Cell Host Microbe 2016, 20, 429–442. [Google Scholar] [CrossRef]
- Zhu, P.; Chertova, E.; Bess, J., Jr.; Lifson, J.D.; Arthur, L.O.; Liu, J.; Taylor, K.A.; Roux, K.H. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc. Natl. Acad. Sci. USA 2003, 100, 15812–15817. [Google Scholar] [CrossRef]
- Zhu, P.; Liu, J.; Bess, J., Jr.; Chertova, E.; Lifson, J.D.; Grise, H.; Ofek, G.A.; Taylor, K.A.; Roux, K.H. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 2006, 441, 847–852. [Google Scholar] [CrossRef]
- Zhu, P.; Winkler, H.; Chertova, E.; Taylor, K.A.; Roux, K.H. Cryoelectron tomography of HIV-1 envelope spikes: Further evidence for tripod-like legs. PLoS Pathog. 2008, 4, e1000203. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Waithe, D.; Carravilla, P.; Huarte, N.; Galiani, S.; Enderlein, J.; Eggeling, C. Envelope glycoprotein mobility on HIV-1 particles depends on the virus maturation state. Nat. Commun. 2017, 8, 545. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, P.; DeVico, A.L.; Foulke, J.S., Jr.; Lewis, G.K.; Pazgier, M.; Ray, K. Stoichiometric Analyses of Soluble CD4 to Native-like HIV-1 Envelope by Single-Molecule Fluorescence Spectroscopy. Cell Rep. 2019, 29, 176–186.e4. [Google Scholar] [CrossRef] [PubMed]
- Brandenberg, O.F.; Magnus, C.; Rusert, P.; Regoes, R.R.; Trkola, A. Different infectivity of HIV-1 strains is linked to number of envelope trimers required for entry. PLoS Pathog. 2015, 11, e1004595. [Google Scholar] [CrossRef]
- Magnus, C.; Regoes, R.R. Estimating the stoichiometry of HIV neutralization. PLoS Comput. Biol. 2010, 6, e1000713. [Google Scholar] [CrossRef] [PubMed]
- Chojnacki, J.; Staudt, T.; Glass, B.; Bingen, P.; Engelhardt, J.; Anders, M.; Schneider, J.; Muller, B.; Hell, S.W.; Krausslich, H.G. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science 2012, 338, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Roy, N.H.; Chan, J.; Lambele, M.; Thali, M. Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion. J. Virol. 2013, 87, 7516–7525. [Google Scholar] [CrossRef]
- Zhang, Z.; Liu, J.; Li, M.; Yang, H.; Zhang, C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS ONE 2012, 7, e49265. [Google Scholar] [CrossRef]
- Wei, X.; Decker, J.M.; Liu, H.; Zhang, Z.; Arani, R.B.; Kilby, J.M.; Saag, M.S.; Wu, X.; Shaw, G.M.; Kappes, J.C. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 2002, 46, 1896–1905. [Google Scholar] [CrossRef]
- Miyauchi, K.; Kim, Y.; Latinovic, O.; Morozov, V.; Melikyan, G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 2009, 137, 433–444. [Google Scholar] [CrossRef]
- Hammonds, J.; Chen, X.; Zhang, X.; Lee, F.; Spearman, P. Advances in methods for the production, purification, and characterization of HIV-1 Gag-Env pseudovirion vaccines. Vaccine 2007, 25, 8036–8048. [Google Scholar] [CrossRef]
- Nikon N-STORM Protocol-Sample Preparation. Available online: http://www.mvi-inc.com/wp-content/uploads/N-STORM+Protocol.pdf (accessed on 1 September 2023).
- Heris, M.K. DBSCAN Clustering in MATLAB. Available online: https://yarpiz.com/255/ypml110-dbscan-clustering (accessed on 1 September 2023).
- Izenman, A.J. Recent Developments in Nonparametric Density Estimation. J. Am. Stat. Assoc. 1991, 86, 205–224. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clusters in Large Spatial Databases with Noise. Knowl. Discov. Data Min. 1996, 96, 226–231. [Google Scholar]
- Xu, X.; Ester, M.; Kriegel, H.-P.; Sander, J. A distribution-based clustering algorithm for mining in large spatial databases. In Proceedings of the 14th International Conference on Data Engineering, Orlando, FL, USA, 23–27 February 1998; pp. 324–331. [Google Scholar]
- Smithson, M.; Verkuilen, J. A better lemon squeezer? Maximum-likelihood regression with beta-distributed dependent variables. Psychol. Methods 2006, 11, 54–71. [Google Scholar] [CrossRef]
- Montefiori, D.C. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr. Protoc. Immunol. 2005, 64, 12.11.1–12.11.17. [Google Scholar] [CrossRef] [PubMed]
- Platt, E.J.; Wehrly, K.; Kuhmann, S.E.; Chesebro, B.; Kabat, D. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 1998, 72, 2855–2864. [Google Scholar] [CrossRef]
- Rahman, K.; Datta, S.A.K.; Beaven, A.H.; Jolley, A.A.; Sodt, A.J.; Compton, A.A. Cholesterol Binds the Amphipathic Helix of IFITM3 and Regulates Antiviral Activity. J. Mol. Biol. 2022, 434, 167759. [Google Scholar] [CrossRef]
- Das, T.; Yang, X.; Lee, H.; Garst, E.H.; Valencia, E.; Chandran, K.; Im, W.; Hang, H.C. S-Palmitoylation and Sterol Interactions Mediate Antiviral Specificity of IFITMs. ACS Chem. Biol. 2022, 17, 2109–2120. [Google Scholar] [CrossRef]
- Klein, S.; Golani, G.; Lolicato, F.; Lahr, C.; Beyer, D.; Herrmann, A.; Wachsmuth-Melm, M.; Reddmann, N.; Brecht, R.; Hosseinzadeh, M.; et al. IFITM3 blocks influenza virus entry by sorting lipids and stabilizing hemifusion. Cell Host Microbe 2023, 31, 616–633.e20. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Verma, S.; Chen, Y.-C.; Marin, M.; Gillespie, S.E.; Melikyan, G.B. IFITM1 and IFITM3 Proteins Inhibit the Infectivity of Progeny HIV-1 without Disrupting Envelope Glycoprotein Clusters. Viruses 2023, 15, 2390. https://doi.org/10.3390/v15122390
Verma S, Chen Y-C, Marin M, Gillespie SE, Melikyan GB. IFITM1 and IFITM3 Proteins Inhibit the Infectivity of Progeny HIV-1 without Disrupting Envelope Glycoprotein Clusters. Viruses. 2023; 15(12):2390. https://doi.org/10.3390/v15122390
Chicago/Turabian StyleVerma, Smita, Yen-Cheng Chen, Mariana Marin, Scott E. Gillespie, and Gregory B. Melikyan. 2023. "IFITM1 and IFITM3 Proteins Inhibit the Infectivity of Progeny HIV-1 without Disrupting Envelope Glycoprotein Clusters" Viruses 15, no. 12: 2390. https://doi.org/10.3390/v15122390




