Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew
Abstract
:1. Introduction
2. Emergence of Henipaviruses
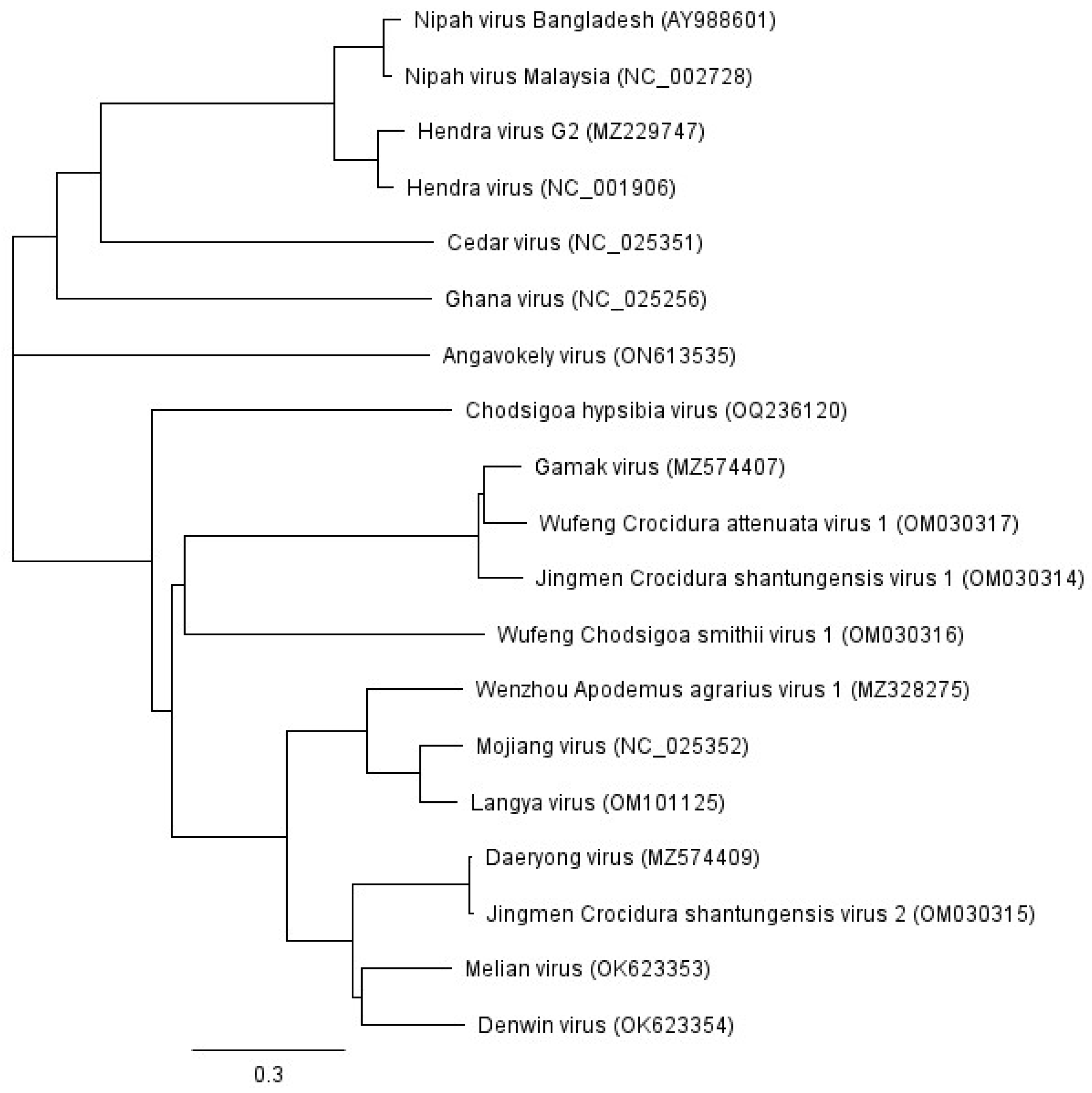
3. The Unclassified Henipa-like Viruses
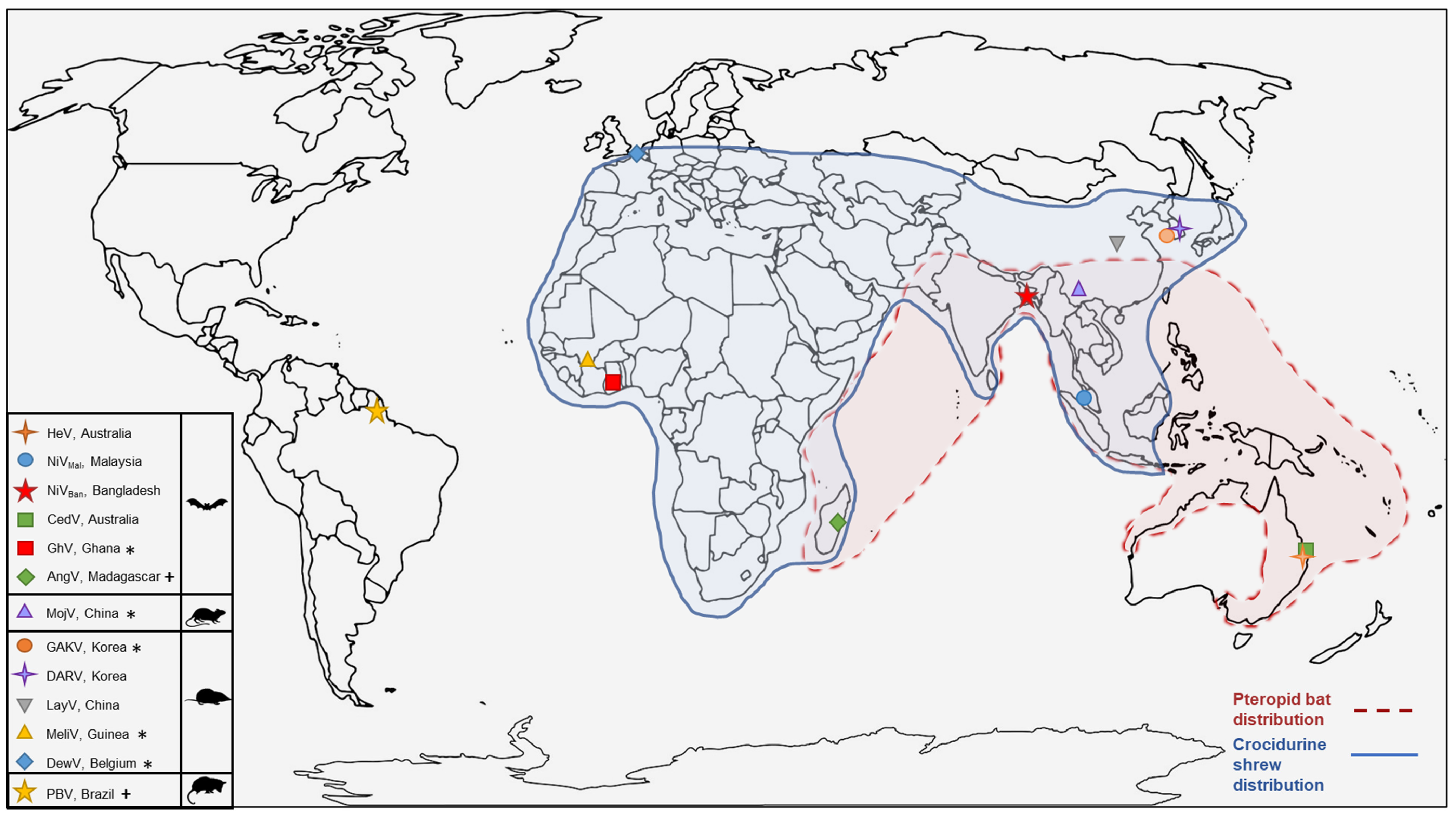
4. Natural Reservoirs for Henipaviruses
5. Shrews: A Henipavirus Reservoir Host
6. Shrews as Reservoir Hosts for Viruses
6.1. Interactions between Shrews and Humans
6.2. Novel Henipaviruses and Shrews
7. Summary
Author Contributions
Funding
Conflicts of Interest
References
- ICTV Genus: Henipavirus. Available online: https://ictv.global/report/chapter/paramyxoviridae/paramyxoviridae/henipavirus (accessed on 18 August 2023).
- Wang, L.F.; Harcourt, B.H.; Yu, M.; Tamin, A.; Rota, P.A.; Bellini, W.J.; Eaton, B.T. Molecular Biology of Hendra and Nipah Viruses. Microbes Infect. 2001, 3, 279–287. [Google Scholar] [CrossRef]
- O’Sullivan, J.D.; Allworth, A.M.; Paterson, D.L.; Snow, T.M.; Boots, R.; Gleeson, L.J.; Gould, A.R.; Hyatt, A.D.; Bradfield, J. Fatal Encephalitis Due to Novel Paramyxovirus Transmitted from Horses. Lancet 1997, 349, 93–95. [Google Scholar] [CrossRef]
- Hsu, V.P.; Hossain, M.J.; Parashar, U.D.; Ali, M.M.; Ksiazek, T.G.; Kuzmin, I.; Niezgoda, M.; Rupprecht, C.; Bresee, J.; Breiman, R.F. Nipah Virus Encephalitis Reemergence, Bangladesh. Emerg. Infect. Dis. 2004, 10, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention Outbreak of Hendra-Like Virus—Malaysia and Singapore, 1998–1999. Available online: https://www.cdc.gov/mmwr/preview/mmwrhtml/00056866.htm (accessed on 24 April 2023).
- Uppal, P.K. Emergence of Nipah Virus in Malaysia. Ann. N. Y. Acad. Sci. 2000, 916, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.T. Emerging Epidemic Viral Encephalitides with a Special Focus on Henipaviruses. Acta Neuropathol. 2010, 120, 317–325. [Google Scholar] [CrossRef]
- Wong, K.T.; Tan, C.T. Clinical and Pathological Manifestations of Human Henipavirus Infection. Curr. Top. Microbiol. Immunol. 2012, 359, 95–104. [Google Scholar] [CrossRef]
- Murray, K.; Selleck, P.; Hooper, P.; Hyatt, A.; Gould, A.; Gleeson, L.; Westbury, H.; Hiley, L.; Selvey, L.; Rodwell, B.; et al. A Morbillivirus That Caused Fatal Disease in Horses and Humans. Science 1995, 268, 94–97. [Google Scholar] [CrossRef]
- Selvey, L.A.; Wells, R.M.; McCormack, J.G.; Ansford, A.J.; Murray, K.; Rogers, R.J.; Lavercombe, P.S.; Selleck, P.; Sheridan, J.W. Infection of Humans and Horses by a Newly Described Morbillivirus. Med. J. Aust. 1995, 162, 642–645. [Google Scholar] [CrossRef]
- Wang, L.-F.; Michalski, W.P.; Yu, M.; Pritchard, L.I.; Crameri, G.; Shiell, B.; Eaton, B.T. A Novel P/V/C Gene in a New Member of the Paramyxoviridae Family, Which Causes Lethal Infection in Humans, Horses, and Other Animals. J. Virol. 1998, 72, 1482–1490. [Google Scholar] [CrossRef]
- NSW Health Summary of Human Cases of Hendra Virus Infection. Available online: https://www.health.nsw.gov.au/Infectious/controlguideline/Pages/hendra-case-summary.aspx (accessed on 24 April 2023).
- Queensland, B. Summary of Hendra Virus Incidents in Horses. Available online: https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary (accessed on 24 April 2023).
- NSW Government Hendra Virus Confirmed in Unvaccinated Horse near Newcastle. Available online: https://www.dpi.nsw.gov.au/about-us/media-centre/releases/2023/general2/hendra-virus-confirmed-in-unvaccinated-horse-near-newcastle (accessed on 24 April 2023).
- Wang, J.; Anderson, D.E.; Halpin, K.; Hong, X.; Chen, H.; Walker, S.; Valdeter, S.; van der Heide, B.; Neave, M.J.; Bingham, J.; et al. A New Hendra Virus Genotype Found in Australian Flying Foxes. Virol. J. 2021, 18, 197. [Google Scholar] [CrossRef]
- Marsh, G.A.; Todd, S.; Foord, A.; Hansson, E.; Davies, K.; Wright, L.; Morrissy, C.; Halpin, K.; Middleton, D.; Field, H.E.; et al. Genome Sequence Conservation of Hendra Virus Isolates during Spillover to Horses, Australia. Emerg. Infect. Dis. 2010, 16, 1767–1769. [Google Scholar] [CrossRef]
- Chua, K.B. Nipah Virus: A Recently Emergent Deadly Paramyxovirus. Science 2000, 288, 1432–1435. [Google Scholar] [CrossRef]
- Paton, N.I.; Leo, Y.S.; Zaki, S.R.; Auchus, A.P.; Lee, K.E.; Ling, A.E.; Chew, S.K.; Ang, B.; Rollin, P.E.; Umapathi, T.; et al. Outbreak of Nipah-Virus Infection among Abattoir Workers in Singapore. Lancet 1999, 354, 1253–1256. [Google Scholar] [CrossRef]
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Seow, P.; Tan, K.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; et al. Fatal Encephalitis Due to Nipah Virus among Pig-Farmers in Malaysia. Lancet 1999, 354, 1257–1259. [Google Scholar] [CrossRef]
- Harcourt, B.H.; Lowe, L.; Tamin, A.; Liu, X.; Bankamp, B.; Bowden, N.; Rollin, P.E.; Comer, J.A.; Ksiazek, T.G.; Hossain, M.J.; et al. Genetic Characterization of Nipah Virus, Bangladesh, 2004. Emerg. Infect. Dis. 2005, 11, 1594–1597. [Google Scholar] [CrossRef]
- Gurley, E.S.; Montgomery, J.M.; Hossain, M.J.; Bell, M.; Azad, A.K.; Islam, M.R.; Molla, M.A.R.; Carroll, D.S.; Ksiazek, T.G.; Rota, P.A.; et al. Person-to-Person Transmission of Nipah Virus in a Bangladeshi Community. Emerg. Infect. Dis. 2007, 13, 1031–1037. [Google Scholar] [CrossRef]
- Ching, P.K.G.; de Los Reyes, V.C.; Sucaldito, M.N.; Tayag, E.; Columna-Vingno, A.B.; Malbas, F.F.; Bolo, G.C.; Sejvar, J.J.; Eagles, D.; Playford, G.; et al. Outbreak of Henipavirus Infection, Philippines, 2014. Emerg. Infect. Dis. 2015, 21, 328–331. [Google Scholar] [CrossRef]
- Chadha, M.S.; Comer, J.A.; Lowe, L.; Rota, P.A.; Rollin, P.E.; Bellini, W.J.; Ksiazek, T.G.; Mishra, A.C. Nipah Virus-Associated Encephalitis Outbreak, Siliguri, India. Emerg. Infect. Dis. 2006, 12, 235–240. [Google Scholar] [CrossRef]
- IEDCR Yearly Distribution of Nipah Cases in Bangladesh 2001–2022. Available online: https://www.iedcr.gov.bd/sites/default/files/files/iedcr.portal.gov.bd/page/ac4bc0d9_027e_48b2_99ab_16cca255bb14/2023-02-01-08-12-5df8e17975c8e134b6f35fc08f79dd1d.pdf (accessed on 10 August 2023).
- Arankalle, V.A.; Bandyopadhyay, B.T.; Ramdasi, A.Y.; Jadi, R.; Patil, D.R.; Rahman, M.; Majumdar, M.; Banerjee, P.S.; Hati, A.K.; Goswami, R.P.; et al. Genomic Characterization of Nipah Virus, West Bengal, India. Emerg. Infect. Dis. J. 2011, 17, 907. [Google Scholar] [CrossRef]
- Thomas, B.; Chandran, P.; Lilabi, M.; George, B.; Sivakumar, C.; Jayadev, V.; Bindu, V.; Rajasi, R.; Vijayan, B.; Mohandas, A.; et al. Nipah Virus Infection in Kozhikode, Kerala, South India, in 2018: Epidemiology of an Outbreak of an Emerging Disease. Indian J. Community Med. 2019, 44, 383. [Google Scholar] [CrossRef] [PubMed]
- Sudeep, A.B.; Yadav, P.D.; Gokhale, M.D.; Balasubramanian, R.; Gupta, N.; Shete, A.; Jain, R.; Patil, S.; Sahay, R.R.; Nyayanit, D.A.; et al. Detection of Nipah Virus in Pteropus Medius in 2019 Outbreak from Ernakulam District, Kerala, India. BMC Infect. Dis. 2021, 21, 162. [Google Scholar] [CrossRef]
- World Health Organisation Nipah Virus Disease—India. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/nipah-virus-disease---india (accessed on 10 August 2023).
- World Health Organization Disease Outbreak News; Nipah Virus Disease—Bangladesh. Available online: https://www.who.int/emergencies/disease-outbreak-news/item/2023-DON442 (accessed on 9 August 2023).
- World Health Organization. Nipah Virus Infection in India. Disease Outbreak News, 17 February 2023. [Google Scholar]
- Marsh, G.A.; de Jong, C.; Barr, J.A.; Tachedjian, M.; Smith, C.; Middleton, D.; Yu, M.; Todd, S.; Foord, A.J.; Haring, V.; et al. Cedar Virus: A Novel Henipavirus Isolated from Australian Bats. PLoS Pathog. 2012, 8, e1002836. [Google Scholar] [CrossRef]
- Burroughs, A.L.; Durr, P.A.; Boyd, V.; Graham, K.; White, J.R.; Todd, S.; Barr, J.; Smith, I.; Baverstock, G.; Meers, J.; et al. Hendra Virus Infection Dynamics in the Grey-Headed Flying Fox (Pteropus poliocephalus) at the Southern-Most Extent of Its Range: Further Evidence This Species Does Not Readily Transmit the Virus to Horses. PLoS ONE 2016, 11, e0155252. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Gloza-Rausch, F.; Seebens, A.; Annan, A.; Ipsen, A.; Kruppa, T.; Müller, M.A.; Kalko, E.K.V.; Adu-Sarkodie, Y.; et al. Henipavirus RNA in African Bats. PLoS ONE 2009, 4, e6367. [Google Scholar] [CrossRef]
- Drexler, J.F.; Corman, V.M.; Müller, M.A.; Maganga, G.D.; Vallo, P.; Binger, T.; Gloza-Rausch, F.; Rasche, A.; Yordanov, S.; Seebens, A.; et al. Bats Host Major Mammalian Paramyxoviruses. Nat. Commun. 2012, 3, 796. [Google Scholar] [CrossRef]
- Wu, Z.; Yang, L.; Yang, F.; Ren, X.; Jiang, J.; Dong, J.; Sun, L.; Zhu, Y.; Zhou, H.; Jin, Q. Novel Henipa-like Virus, Mojiang Paramyxovirus, in Rats, China, 2012. Emerg. Infect. Dis. 2014, 20, 1064–1066. [Google Scholar] [CrossRef]
- Lee, S.H.; Kim, K.; Kim, J.; No, J.S.; Park, K.; Budhathoki, S.; Lee, S.H.; Lee, J.; Cho, S.H.; Cho, S.; et al. Discovery and Genetic Characterization of Novel Paramyxoviruses Related to the Genus Henipavirus in Crocidura Species in the Republic of Korea. Viruses 2021, 13, 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.-A.; Li, H.; Jiang, F.-C.; Zhu, F.; Zhang, Y.-F.; Chen, J.-J.; Tan, C.-W.; Anderson, D.E.; Fan, H.; Dong, L.-Y.; et al. A Zoonotic Henipavirus in Febrile Patients in China. N. Engl. J. Med. 2022, 387, 470–472. [Google Scholar] [CrossRef] [PubMed]
- Madera, S.; Kistler, A.; Ranaivoson, H.C.; Ahyong, V.; Andrianiaina, A.; Andry, S.; Raharinosy, V.; Randriambolamanantsoa, T.H.; Ravelomanantsoa, N.A.F.; Tato, C.M.; et al. Discovery and Genomic Characterization of a Novel Henipavirus, Angavokely Virus, from Fruit Bats in Madagascar. J. Virol. 2022, 96, e00921-22. [Google Scholar] [CrossRef] [PubMed]
- Vanmechelen, B.; Meurs, S.; Horemans, M.; Loosen, A.; Maes, T.J.; Laenen, L.; Vergote, V.; Koundouno, F.R.; Magassouba, N.; Konde, M.K.; et al. The Characterization of Multiple Novel Paramyxoviruses Highlights the Diverse Nature of the Subfamily Orthoparamyxovirinae. Virus Evol. 2022, 8, veac061. [Google Scholar] [CrossRef]
- Hernández, L.H.A.; da Paz, T.Y.B.; Silva, S.P.d.; Silva, F.S.d.; Barros, B.C.V.d.; Nunes, B.T.D.; Casseb, L.M.N.; Medeiros, D.B.A.; Vasconcelos, P.F.d.C.; Cruz, A.C.R. First Genomic Evidence of a Henipa-like Virus in Brazil. Viruses 2022, 14, 2167. [Google Scholar] [CrossRef]
- Timmiss, L.A.; Martin, J.M.; Murray, N.J.; Welbergen, J.A.; Westcott, D.; McKeown, A.; Kingsford, R.T. Threatened but Not Conserved: Flying-Fox Roosting and Foraging Habitat in Australia. Aust. J. Zool. 2021, 68, 226–233. [Google Scholar] [CrossRef]
- Churchfield, S. The Natural History of Shrews, 1st ed.; Cornell University Press: New York, NY, USA, 1990; ISBN 0-8014-2595-6. [Google Scholar]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra Virus from Pteropid Bats: A Natural Reservoir of Hendra Virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef] [PubMed]
- Young, P.L.; Halpin, K.; Selleck, P.W.; Field, H.; Gravel, J.L.; Kelly, M.A.; Mackenzie, J.S. Serologic Evidence for the Presence in Pteropus Bats of a Paramyxovirus Related to Equine Morbillivirus. Emerg. Infect. Dis. 1996, 2, 239–240. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Lek Koh, C.; Hooi, P.S.; Wee, K.F.; Khong, J.H.; Chua, B.H.; Chan, Y.P.; Lim, M.E.; Lam, S.K. Isolation of Nipah Virus from Malaysian Island Flying-Foxes. Microbes Infect. 2002, 4, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Hahn, M.B.; Epstein, J.H.; Gurley, E.S.; Islam, M.S.; Luby, S.P.; Daszak, P.; Patz, J.A. Roosting Behaviour and Habitat Selection of Pteropus Giganteus Reveals Potential Links to Nipah Virus Epidemiology. J. Appl. Ecol. 2014, 51, 376–387. [Google Scholar] [CrossRef]
- Eby, P.; Peel, A.J.; Hoegh, A.; Madden, W.; Giles, J.R.; Hudson, P.J.; Plowright, R.K. Pathogen Spillover Driven by Rapid Changes in Bat Ecology. Nature 2023, 63, 340–344. [Google Scholar] [CrossRef]
- Edson, D.; Field, H.; McMichael, L.; Vidgen, M.; Goldspink, L.; Broos, A.; Melville, D.; Kristoffersen, J.; De Jong, C.; McLaughlin, A.; et al. Routes of Hendra Virus Excretion in Naturally-Infected Flying-Foxes: Implications for Viral Transmission and Spillover Risk. PLoS ONE 2015, 10, e0140670. [Google Scholar] [CrossRef]
- Edson, D.; Field, H.; McMichael, L.; Jordan, D.; Kung, N.; Mayer, D.; Smith, C. Flying-Fox Roost Disturbance and Hendra Virus Spillover Risk. PLoS ONE 2015, 10, e0125881. [Google Scholar] [CrossRef]
- Pernet, O.; Schneider, B.S.; Beaty, S.M.; Lebreton, M.; Yun, T.E.; Park, A.; Zachariah, T.T.; Bowden, T.A.; Hitchens, P.; Ramirez, C.M.; et al. Evidence for Henipavirus Spillover into Human Populations in Africa. Nat. Commun. 2014, 5, 5342. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Hossain, M.J.; Sultana, S.; Homaira, N.; Khan, S.U.; Rahman, M.; Gurley, E.S.; Rollin, P.E.; Lo, M.K.; Comer, J.A.; et al. Date Palm Sap Linked to Nipah Virus Outbreak in Bangladesh, 2008. Vector-Borne Zoonotic Dis. 2012, 12, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Luby, S.P.; Rahman, M.; Hossain, M.J.; Blum, L.S.; Husain, M.M.; Gurley, E.; Khan, R.; Ahmed, B.N.; Rahman, S.; Nahar, N.; et al. Foodborne Transmission of Nipah Virus, Bangladesh. Emerg. Infect. Dis. 2006, 12, 1888–1894. [Google Scholar] [CrossRef] [PubMed]
- Salah Uddin Khan, M.; Hossain, J.; Gurley, E.S.; Nahar, N.; Sultana, R.; Luby, S.P. Use of Infrared Camera to Understand Bats’ Access to Date Palm Sap: Implications for Preventing Nipah Virus Transmission. EcoHealth 2010, 7, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Clayton, B.A.; Marsh, G.A. Nipah Viruses from Malaysia and Bangladesh: Two of a Kind? Future Virol. 2014, 9, 935–946. [Google Scholar] [CrossRef]
- Middleton, D.J.; Morrissy, C.J.; van der Heide, B.M.; Russell, G.M.; Braun, M.A.; Westbury, H.A.; Halpin, K.; Daniels, P.W. Experimental Nipah Virus Infection in Pteropid Bats (Pteropus poliocephalus). J. Comp. Pathol. 2007, 136, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Barr, J.; Smith, C.; Smith, I.; De Jong, C.; Todd, S.; Melville, D.; Broos, A.; Crameri, S.; Haining, J.; Marsh, G.; et al. Isolation of Multiple Novel Paramyxoviruses from Pteropid Bat Urine. J. Gen. Virol. 2015, 96, 24–29. [Google Scholar] [CrossRef] [PubMed]
- Peel, A.J.; Yinda, C.K.; Annand, E.J.; Dale, A.S.; Eby, P.; Eden, J.S.; Jones, D.N.; Kessler, M.K.; Lunn, T.J.; Pearson, T.; et al. Novel Hendra Virus Variant Circulating in Black Flying Foxes and Grey-Headed Flying Foxes, Australia. Emerg. Infect. Dis. 2022, 28, 1043–1047. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention Nipah Virus Distribution Map. Available online: https://www.cdc.gov/vhf/nipah/outbreaks/distribution-map.html (accessed on 18 January 2023).
- Schulz, J.E.; Seifert, S.N.; Thompson, J.T.; Avanzato, V.; Sterling, S.L.; Yan, L.; Letko, M.C.; Matson, M.J.; Fischer, R.J.; Tremeau-Bravard, A.; et al. Serological Evidence for Henipa-like and Filo-like Viruses in Trinidad Bats. J. Infect. Dis. 2020, 221, S375–S382. [Google Scholar] [CrossRef]
- Li, Y.; Wang, J.; Hickey, A.C.; Zhang, Y.; Li, Y.; Wu, Y.; Zhang, H.; Yuan, J.; Han, Z.; McEachern, J.; et al. Antibodies to Nipah or Nipah-Like Viruses in Bats, China. Emerg. Infect. Dis. 2008, 14, 1974–1976. [Google Scholar] [CrossRef]
- Hayman, D.T.S.; Suu-Ire, R.; Breed, A.C.; McEachern, J.A.; Wang, L.; Wood, J.L.N.; Cunningham, A.A. Evidence of Henipavirus Infection in West African Fruit Bats. PLoS ONE 2008, 3, e2739. [Google Scholar] [CrossRef]
- Brook, C.E.; Ranaivoson, H.C.; Broder, C.C.; Cunningham, A.A.; Héraud, J.M.; Peel, A.J.; Gibson, L.; Wood, J.L.N.; Metcalf, C.J.; Dobson, A.P. Disentangling Serology to Elucidate Henipa- and Filovirus Transmission in Madagascar Fruit Bats. J. Anim. Ecol. 2019, 88, 1001–1016. [Google Scholar] [CrossRef]
- Willows-Munro, S.; Matthee, C.A. Exploring the Diversity and Molecular Evolution of Shrews (Family Soricidae) Using mtDNA Cytochrome b Data. Afr. Zool. 2011, 46, 246–262. [Google Scholar] [CrossRef]
- Hutterer, R. Anatomical Adaptations of Shrews. Mammal Rev. 1985, 15, 43–55. [Google Scholar] [CrossRef]
- Ginsberg, J.R. Biodiversity of Mammals. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.A., Ed.; Academic Press: Waltham, MA, USA, 2013; pp. 681–707. ISBN 978-0-12-384720-1. [Google Scholar]
- Novacek, M.J. Mammalian Phytogeny: Shaking the Tree. Nature 1992, 356, 121–125. [Google Scholar] [CrossRef] [PubMed]
- Repenning, Charles A Subfamilies and Genera of the Soricidae; Geological survey professional paper; United States Government printing Office: Washington, DC, USA, 1967.
- Fons, R.; Sender, S.; Peters, T.; Jürgens, K.D. Rates of Rewarming, Heart and Respiratory Rates and Their Significance for Oxygen Transport During Arousal from Torpor in the Smallest Mammal, the Etruscan Shrew Suncus Etruscus. J. Exp. Biol. 1997, 200, 1451–1458. [Google Scholar] [CrossRef] [PubMed]
- Louch, C.D.; Ghosh, A.K.; Pal, B.C. Seasonal Changes in Weight and Reproductive Activity of Suncus Murinus in West Bengal, India. J. Mammal. 1966, 47, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Gliwicz, J.; Taylor, J.R.E. Comparing Life Histories of Shrews and Rodents. Acta Theriol. 2002, 47, 185–208. [Google Scholar] [CrossRef]
- Cantoni, D.; Vogel, P. Social Organization and Mating System of Free-Ranging, Greater White-Toothed Shrews, Crocidura Russula. Anim. Behav. 1989, 38, 205–214. [Google Scholar] [CrossRef]
- Hilbe, M.; Herrsche, R.; Kolodziejek, J.; Nowotny, N.; Zlinszky, K.; Ehrensperger, F. Shrews as Reservoir Hosts of Borna Disease Virus. Emerg. Infect. Dis. 2006, 12, 675–677. [Google Scholar] [CrossRef]
- Goethert, H.K.; Mather, T.N.; Johnson, R.W.; Telford, S.R. Incrimination of Shrews as a Reservoir for Powassan Virus. Commun. Biol. 2021, 4, 1319. [Google Scholar] [CrossRef]
- Song, J.-W.; Kang, H.J.; Gu, S.H.; Moon, S.S.; Bennett, S.N.; Song, K.-J.; Baek, L.J.; Kim, H.-C.; O’Guinn, M.L.; Chong, S.-T.; et al. Characterization of Imjin Virus, a Newly Isolated Hantavirus from the Ussuri White-Toothed Shrew (Crocidura lasiura). J. Virol. 2009, 83, 6184–6191. [Google Scholar] [CrossRef]
- Gu, S.H.; Nicolas, V.; Lalis, A.; Sathirapongsasuti, N.; Yanagihara, R. Complete Genome Sequence and Molecular Phylogeny of a Newfound Hantavirus Harbored by the Doucet’s Musk Shrew (Crocidura douceti) in Guinea. Infect. Genet. Evol. 2013, 20, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kadjo, B.; Dubey, S.; Jacquet, F.; Yanagihara, R. Molecular Evolution of Azagny Virus, a Newfound Hantavirus Harbored by the West African Pygmy Shrew (Crocidura obscurior) in Côte d’Ivoire. Virol. J. 2011, 8, 373. [Google Scholar] [CrossRef]
- Onyuok, S.O.; Hu, B.; Li, B.; Fan, Y.; Kering, K.; Ochola, G.O.; Zheng, X.S.; Obanda, V.; Ommeh, S.; Yang, X.L.; et al. Molecular Detection and Genetic Characterization of Novel RNA Viruses in Wild and Synanthropic Rodents and Shrews in Kenya. Front. Microbiol. 2019, 10, 2696. [Google Scholar] [CrossRef]
- Sasaki, M.; Muleya, W.; Ishii, A.; Orba, Y.; Hang’ombe, B.M.; Mweene, A.S.; Moonga, L.; Thomas, Y.; Kimura, T.; Sawa, H. Molecular Epidemiology of Paramyxoviruses in Zambian Wild Rodents and Shrews. J. Gen. Virol. 2014, 95, 325–330. [Google Scholar] [CrossRef] [PubMed]
- Low, D.H.W.; Ch’ng, L.; Su, Y.C.F.; Linster, M.; Zhang, R.; Zhuang, Y.; Kwak, M.L.; Borthwick, S.A.; Hitch, A.T.; Smith, G.J.D.; et al. Cencurut Virus: A Novel Orthonairovirus from Asian House Shrews (Suncus murinus) in Singapore. One Health 2023, 16, 100529. [Google Scholar] [CrossRef] [PubMed]
- El Khoury, M.Y.; Hull, R.C.; Bryant, P.W.; Escuyer, K.L.; St George, K.; Wong, S.J.; Nagaraja, A.; Kramer, L.; Dupuis, A.P.; Purohit, T.; et al. Diagnosis of Acute Deer Tick Virus Encephalitis. Clin. Infect. Dis. 2013, 56, e40–e47. [Google Scholar] [CrossRef]
- Happold, M.; Happold, D.C.D. Mammals of Africa, Volume IV: Hedgehogs, Shrews and Bats; Birds and Mammals of Africa; Bloomsbury Publishing: London, UK, 2016; Volume IV, ISBN 978-1-4081-2254-9. [Google Scholar]
- Rasche, A.; Lehmann, F.; König, A.; Goldmann, N.; Corman, V.M.; Moreira-Soto, A.; Geipel, A.; van Riel, D.; Vakulenko, Y.A.; Sander, A.-L.; et al. Highly Diversified Shrew Hepatitis B Viruses Corroborate Ancient Origins and Divergent Infection Patterns of Mammalian Hepadnaviruses. Proc. Natl. Acad. Sci. USA 2019, 116, 17007–17012. [Google Scholar] [CrossRef]
- Rasche, A.; Sander, A.-L.; Corman, V.M.; Drexler, J.F. Evolutionary Biology of Human Hepatitis Viruses. J. Hepatol. 2019, 70, 501–520. [Google Scholar] [CrossRef]
- Klempa, B.; Finchet-Calvet, E.; Lecompte, E.; Auste, B.; Aniskin, V.; Meisel, H.; Barriere, P.; Koivogui, L.; ter Meulen, J.; Krüger, D.H. Novel Hantavirus Sequences in Shrew, Guinea. Emerg. Infect. Dis. 2007, 13, 520–522. [Google Scholar] [CrossRef]
- Song, J.-W.; Baek, L.J.; Schmaljohn, C.S.; Yanagihara, R. Thottapalayam Virus, a Prototype Shrewborne Hantavirus. Emerg. Infect. Dis. 2007, 13, 980–985. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-W.; Gu, S.; Bennett, S.N.; Arai, S.; Puorger, M.; Hilbe, M.; Yanagihara, R. Seewis Virus, a Genetically Distinct Hantavirus in the Eurasian Common Shrew (Sorex araneus). Virol. J. 2007, 4, 114. [Google Scholar] [CrossRef] [PubMed]
- Lwande, O.W.; Mohamed, N.; Bucht, G.; Ahlm, C.; Olsson, G.; Evander, M. Seewis Hantavirus in Common Shrew (Sorex araneus) in Sweden. Virol. J. 2020, 17, 198. [Google Scholar] [CrossRef] [PubMed]
- Schlegel, M.; Radosa, L.; Rosenfeld, U.M.; Schmidt, S.; Triebenbacher, C.; Löhr, P.-W.; Fuchs, D.; Heroldová, M.; Jánová, E.; Stanko, M.; et al. Broad Geographical Distribution and High Genetic Diversity of Shrew-Borne Seewis Hantavirus in Central Europe. Virus Genes 2012, 45, 48–55. [Google Scholar] [CrossRef]
- Luby, S.P.; Hossain, M.J.; Gurley, E.S.; Ahmed, B.-N.; Banu, S.; Khan, S.U.; Homaira, N.; Rota, P.A.; Rollin, P.E.; Comer, J.A.; et al. Recurrent Zoonotic Transmission of Nipah Virus into Humans, Bangladesh, 2001–2007. Emerg. Infect. Dis. J. 2009, 15, 1229. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.-W.; Lee, H.-J.; Kim, Y.-K.; Oh, H.-S.; Han, S.-H. Genetic Identification of Prey Species from Teeth in Faeces from the Endangered Leopard Cat Prionailurus Bengalensis Using Mitochondrial Cytochrome b Gene Sequence. Mitochondrial DNA Part A 2018, 29, 170–174. [Google Scholar] [CrossRef]
- Tosh, D.G.; Lusby, J.; Montgomery, W.I.A.N.; O’Halloran, J. First Record of Greater White-Toothed Shrew Crocidura russula in Ireland. Mammal Rev. 2008, 38, 321–326. [Google Scholar] [CrossRef]
- McDevitt, A.D.; Montgomery, W.I.; Tosh, D.G.; Lusby, J.; Reid, N.; White, T.A.; McDevitt, C.D.; O’Halloran, J.; Searle, J.B.; Yearsley, J.M. Invading and Expanding: Range Dynamics and Ecological Consequences of the Greater White-Toothed Shrew (Crocidura russula) Invasion in Ireland. PLoS ONE 2014, 9, e0100403. [Google Scholar] [CrossRef]
- Nally, J.E.; Arent, Z.; Bayles, D.O.; Hornsby, R.L.; Gilmore, C.; Regan, S.; McDevitt, A.D.; Yearsley, J.; Fanning, S.; McMahon, B.J. Emerging Infectious Disease Implications of Invasive Mammalian Species: The Greater White-Toothed Shrew (Crocidura russula) Is Associated with a Novel Serovar of Pathogenic Leptospira in Ireland. PLoS Negl. Trop. Dis. 2016, 10, e0005174. [Google Scholar] [CrossRef]
- Ehinger, M.; Fontanillas, P.; Petit, E.; Perrin, N. Mitochondrial DNA Variation along an Altitudinal Gradient in the Greater White-Toothed Shrew, Crocidura Russula. Mol. Ecol. 2002, 11, 939–945. [Google Scholar] [CrossRef]
- Igbokwe, J.; Nicolas, V.; Oyeyiola, A.; Obadare, A.; Adesina, A.S.; Awodiran, M.O.; Van Houtte, N.; Fichet-Calvet, E.; Verheyen, E.; Olayemi, A. Molecular Taxonomy of Crocidura Species (Eulipotyphla: Soricidae) in a Key Biogeographical Region for African Shrews, Nigeria. Comptes Rendus Biol. 2019, 342, 108–117. [Google Scholar] [CrossRef]
- von Merten, S.; Oliveira, F.G.; Tapisso, J.T.; Pustelnik, A.; da Luz Mathias, M.; Rychlik, L. Urban Populations of Shrews Show Larger Behavioural Differences among Individuals than Rural Populations. Anim. Behav. 2022, 187, 35–46. [Google Scholar] [CrossRef]
- Weatherman, S.; Feldmann, H.; de Wit, E. Transmission of Henipaviruses. Curr. Opin. Virol. 2018, 28, 7–11. [Google Scholar] [CrossRef]
- Field, H.E. Hendra Virus Ecology and Transmission. Curr. Opin. Virol. 2016, 16, 120–125. [Google Scholar] [CrossRef] [PubMed]
- McDevitt, A.D.; Vega, R.; Rambau, R.V.; Yannic, G.; Herman, J.S.; Hayden, T.J.; Searle, J.B. Colonization of Ireland: Revisiting ‘the Pygmy Shrew Syndrome’ Using Mitochondrial, Y Chromosomal and Microsatellite Markers. Heredity 2011, 107, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Ohdachi, S.D.; Iwasa, M.A.; Nesterenko, V.A.; Abe, H.; Masuda, R.; Haberl, W. Molecular Phylogenetics of Crocidura Shrews (Insectivora) in East and Central Asia. J. Mammal. 2004, 85, 396–403. [Google Scholar] [CrossRef]
- Rofes, J.; Cuenca-Bescós, G. Evolutionary History and Biogeography of the Genus Crocidura (Mammalia, Soricidae) in Europe, with Emphasis on Crocidura Kornfeldi. Mamm. Biol. 2011, 76, 64–78. [Google Scholar] [CrossRef]
- Guo, W.-P.; Lin, X.-D.; Wang, W.; Tian, J.-H.; Cong, M.-L.; Zhang, H.-L.; Wang, M.-R.; Zhou, R.-H.; Wang, J.-B.; Li, M.-H.; et al. Phylogeny and Origins of Hantaviruses Harbored by Bats, Insectivores, and Rodents. PLoS Pathog. 2013, 9, e1003159. [Google Scholar] [CrossRef] [PubMed]
- Nobach, D.; Bourg, M.; Herzog, S.; Lange-Herbst, H.; Encarnação, J.A.; Eickmann, M.; Herden, C.; Kuhn, J.H. Shedding of Infectious Borna Disease Virus-1 in Living Bicolored White-Toothed Shrews. PLoS ONE 2015, 10, e0137018. [Google Scholar] [CrossRef]
- Sauder, C.; Staeheli, P. Rat Model of Borna Disease Virus Transmission: Epidemiological Implications. J. Virol. 2003, 77, 12886–12890. [Google Scholar] [CrossRef]
| Year (s) | Country | Confirmed Cases | Deaths | % Case Fatality | Reference |
|---|---|---|---|---|---|
| 1998–1999 | Malaysia | 265 | 105 | 40 | [19] |
| 1999 | Singapore | 11 | 1 | 9 | [18] |
| 2001 | India | 66 | 45 | 68 | [23] |
| 2001 | Bangladesh | 13 | 9 | 69 | [24] |
| 2003 | 12 | 8 | 67 | ||
| 2004 | 67 | 50 | 75 | ||
| 2005 | 13 | 11 | 85 | ||
| 2007 | 18 | 9 | 50 | ||
| 2007 | India | 5 | 5 | 100 | [25] |
| 2008 | Bangladesh | 11 | 9 | 82 | [24] |
| 2009 | 4 | 0 | 0 | ||
| 2010 | 18 | 16 | 89 | ||
| 2011 | 42 | 36 | 86 | ||
| 2012 | 18 | 13 | 72 | ||
| 2013 | 26 | 22 | 85 | ||
| 2014 | 38 | 15 | 39 | ||
| 2014 | Philippines | 17 | 9 | 53 | [22] |
| 2015 | Bangladesh | 18 | 11 | 61 | [24] |
| 2017 | 3 | 2 | 67 | ||
| 2018 | India | 18 | 16 | 89 | [26] |
| 2018 | Bangladesh | 4 | 3 | 75 | [24] |
| 2019 | 8 | 7 | 88 | ||
| 2019 | India | 1 | 0 | 0 | [27] |
| 2020 | Bangladesh | 6 | 4 | 67 | [24] |
| 2021 | India | 1 | 1 | 100 | [28] |
| 2021 | Bangladesh | 2 | 0 | 0 | [24] |
| 2022 | 3 | 2 | 67 | [29] | |
| 2023 | 13 | 8 | 73 | ||
| 2023 | India | 6 | 2 | 34 | [30] |
| Malaysia, Singapore, and Philippines | 293 | 115 | 34 | ||
| India | 95 | 69 | 73 | ||
| Bangladesh | 335 | 235 | 65 | ||
| Total | 723 | 412 | 58 | ||
| Henipa/Henipa-like Virus | Primary Site of Detection | Isolate (I) or Sequence Only (S) | Proposed Reservoir | Reference (or GenBank Accession Number) |
|---|---|---|---|---|
| Hendra virus | Australia | I | Bat | [9] |
| Nipah virus Malaysia | Malaysia | I | Bat | [19] |
| Nipah virus Bangladesh | Bangladesh | I | Bat | [4] |
| Cedar virus | Australia | I | Bat | [31] |
| Mòjiāng virus | China | S | Cave rat | [35] |
| Ghanian bat virus | Ghana | S | Bat | [33] |
| Angavokely henipavirus | Madagascar | S | Bat | [38] |
| Denwin virus | Belgium | S | Shrew | [39] |
| Gamak virus | Korea | I | Shrew | [36] |
| Daeryong virus | Korea | S | Shrew | [36] |
| Langya virus | China | I | Shrew | [37] |
| Melian virus | Guinea | S | Shrew | [39] |
| Jingmen Crocidura shantungensis henipavirus 1 | China | S | Shrew | OM030314.1 |
| Jingmen Crocidura shantungensis henipavirus 2 | China | S | Shrew | OM030315.1 |
| Wufeng Chodsigoa smithii henipavirus 1 | China | S | Shrew | OM030316.1 |
| Wufeng Crocidura attenuata henipavirus 1 | China | S | Shrew | OM030317.1 |
| Wenzhou Apodemus agrarius henipavirus 1 | China | S | Striped field mouse | MZ328275.1 |
| Peixe-Boi virus | Brazil | S | Opossum | [40] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caruso, S.; Edwards, S.J. Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew. Viruses 2023, 15, 2407. https://doi.org/10.3390/v15122407
Caruso S, Edwards SJ. Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew. Viruses. 2023; 15(12):2407. https://doi.org/10.3390/v15122407
Chicago/Turabian StyleCaruso, Sarah, and Sarah J. Edwards. 2023. "Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew" Viruses 15, no. 12: 2407. https://doi.org/10.3390/v15122407
APA StyleCaruso, S., & Edwards, S. J. (2023). Recently Emerged Novel Henipa-like Viruses: Shining a Spotlight on the Shrew. Viruses, 15(12), 2407. https://doi.org/10.3390/v15122407






